Abstract
Personalized medicine is nowadays a paradigm in lung cancer management, offering important benefits to patients. This study aimed to test the feasibility and utility of embedding two multiplexed genomic platforms as the routine workup of advanced non-squamous non-small cell lung cancer (NSCLC) patients. Two parallel multiplexed approaches were performed based on DNA sequencing and direct digital detection of RNA with nCounter® technology to evaluate gene mutations and fusions. The results were used to guide genotype-directed therapies and patient outcomes were collected. A total of 224 advanced non-squamous NSCLC patients were prospectively included in the study. Overall, 85% of samples were successfully characterized at DNA and RNA levels and oncogenic drivers were found in 68% of patients, with KRAS, EGFR, METΔex14, BRAF, and ALK being the most frequent (31%, 19%, 5%, 4%, and 4%, respectively). Among all patients with complete genotyping results and follow-up data (n = 156), the median overall survival (OS) was 1.90 years (confidence interval (CI) 95% 1.69–2.10) for individuals harbouring an actionable driver treated with a matched therapy, compared with 0.59 years (CI 95% 0.39–0.79) in those not eligible for any targeted therapy and 0.61 years (CI 95% 0.12–1.10) in patients with no drivers identified (p < 0.001). Integrating DNA and RNA multiplexing technologies into the routine molecular testing of advanced NSCLC patients is feasible and useful and highlights the necessity of widespread integrating comprehensive molecular diagnosis into lung cancer care.
1. Introduction
The emergence of targeted therapy has revolutionized the management of patients with lung cancer by incorporating tumour molecular analyses into the routine diagnostic and therapeutic process [1]. At this time, non-small cell lung cancer (NSCLC) has undergone a paradigm shift from a purely histological to a molecular-based treatment approach, allowing for the selection of patients that would most benefit from targeted therapies, and thus impacting lung cancer care overall.
Adenocarcinoma is probably the lung cancer subtype that has most benefited from this molecular-based strategy, with an estimated frequency of oncogenic drivers higher than 50% [2]. About 25%–30% of these patients can benefit from a targeted therapy that has already been approved, and another 25%–30% are able to enroll in clinical trials. In order to personalize treatment decisions, several guidelines currently endorse routine genetic testing of four leading oncogenic drivers—EGFR, ALK, ROS1, and BRAF—in newly metastatic non-squamous NSCLC patients [3,4,5,6,7], while a number of other emerging molecular targets, such as RET and NTRK gene rearrangements, MET exon 14 skipping mutations (METΔex14), and activating HER2 mutations, are likely approaching clinical practice [8]. However, real-world data indicate that comprehensive biomarker testing is still not universal and remains suboptimal in daily clinical practice [8,9]. This is the case of Europeans, who face extreme inequality in access to appropriate molecular diagnosis, even within the same country, with huge differences in the national policies for reimbursement [10].
Our understanding of the molecular events that drive lung cancer pathogenesis has come from the progress made in highly efficient next-generation sequencing (NGS) technologies, which allow for a blanket testing of multiple potentially actionable driver genes from a single sample [11]. Several groups have already demonstrated the practicality of routine genetic testing in large cohorts of patients nationwide and the potential use of this information to guide treatment decisions [4,12,13,14,15,16,17,18,19,20,21,22]. However, the vast majority of those groups used DNA workflows to identify somatic variants—deletions, insertions, inversions, and substitutions—present in selected regions, whereas RNA-based multiplex analysis, which enables testing of several rearrangements and fusion partners, is still not in such common use [8,14] (Table S1).
In a previous study, we retrospectively validated a multiplexed RNA-based nCounter codeset for the detection of ALK, ROS1, and RET fusion transcripts in formalin-fixed and paraffin embedded (FFPE) samples from patients with advanced NSCLC and proved its advantage compared with standard diagnostic assays—immunohistochemistry (IHC) and fluorescent in situ hybridization (FISH) [23].
Here, we aimed to prospectively demonstrate the feasibility and usefulness of embedding two DNA and RNA tissue-based multiplex technologies in the real-life context of advanced lung cancer patients. The co-primary objectives set for this work were to determine the frequency of oncogenic alterations in advanced non-squamous NSCLC patients from our community and to measure overall survival (OS) in major molecular subgroups (driver/no-driver). The secondary objectives were to describe the characteristics of genotyped patients (age, gender, Eastern Cooperative Oncology Group (ECOG) performance status, and smoking history) and compare the outcomes according to treatment strategy (targeted/no-targeted therapy).
2. Results
2.1. Patient Cohort and Clinical Data
Between June 2017 and August 2019, a total of 224 advanced NSCLC patients were prospectively diagnosed at our institution. The last follow-up and update of the database was done in November 2019. Molecular DNA and/or RNA testing yielded an informative result in 207 patients (92%). Among them, 191 (85%) had both DNA and RNA informative results. All successfully genotyped patients had available clinical data and follow-up (Figure 1).
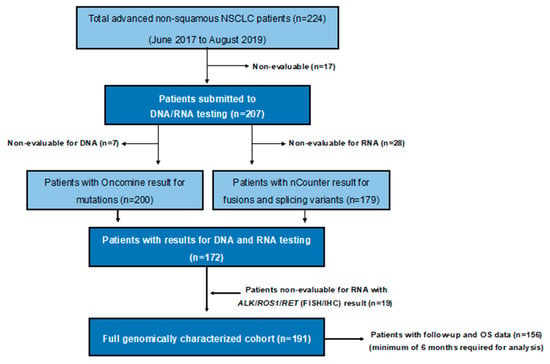
Figure 1.
Flow diagram. NSCLC: non-small-cell lung cancer; OS: overall survival; FISH: fluorescence in situ hybridization; IHC: immunohistochemistry.
The primary characteristics of the patients are shown in Table 1. Median age was 70 years (interquartile range of 60–77), 63% of patients were men, and 57% had an ECOG performance status of 0 or 1 at metastatic disease diagnosis. Most patients (81%) were former or current smokers and 72% were diagnosed at advanced stage IV disease. The histology was adenocarcinoma for 92% of patients, while the remaining cases included not otherwise specified (NOS) or mixed NSCLC histologies. The majority of patients (84%) were treatment-naïve at the time of molecular assessment.

Table 1.
Patient characteristics.
2.2. Molecular Characterization
Among the 191 genotyped patients with a complete DNA and RNA molecular characterization, a genetic alteration was recorded in 90% of patients (Table 2). Driver alterations were found in 129 patients (68%), with KRAS (n = 59, 31%), EGFR (n = 36, 19%), METΔex14 (n = 9, 5%), BRAF (n = 7, 4%), and ALK (n = 7, 4%) being the most commonly detected. Other less-common oncogenic driver genes identified were ERBB4, ERBB2, PIK3CA, NRAS, NTRK1, and ROS1. Non-driver alterations were identified in 43 patients (22%), whereas 19 full-wild-type (WT) patients (10%) did not harbour any alteration (Figure 2A).

Table 2.
Genes with mutational or structural changes identified and targeted treatments received for patients with full genotyping (n = 191).

Figure 2.
Genetic alterations identified. Pie charts depicting frequencies of driver alterations (EGFR, BRAF, KRAS, MET∆ex14, ALK, ERBB2, ERBB4, PIK3CA, NTRK1, ROS1, and NRAS) from 191 patients included in the prospective cohort (expressed as the percentage of positive samples for each molecular alteration relative to the total number of cases with an informative molecular result). Non-driver alterations include mutations at AKT1, TP53, STK11, CTNNB1, SMAD4, DDR2, MAPK, METnon∆ex14, ALK mutations, and cases with concomitant mutations (see Table 2 for further details). Wild-type (full-WT) tumours refer to those in which no genetic alteration was identified. (A) Overall population, (B) never smokers, (C) former smokers, (D) smokers, (E) male, and (F) female.
Sensitizing mutations (E19del and L858R) represented the most common EGFR subtype (86%), corresponding to 13% of all genotyped patients, whereas G12C was the most frequent KRAS variant identified (39%), accounting for 12% of all genotyped patients. Co-mutations were found in 51% of cases, with TP53 (38%), PIK3CA (9%), and STK11 (5%) being the most common. Co-mutations with TP53 (32%) and STK11 (10%) were the most common in KRAS-mutant, whereas TP53 (39%) and CTNNB1 (8%) were the most frequently described in EGFR-mutant patients. The most common oncogenic driver mutations were mutually exclusive except for PIK3CA. A co-mutation plot from the full sequencing of genotyped patients is shown in Figure 3.
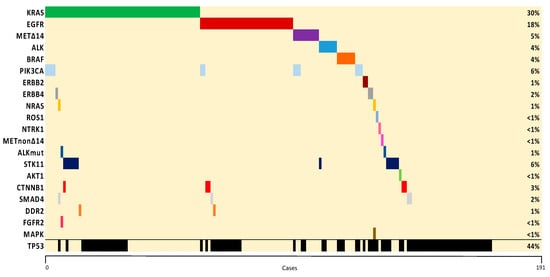
Figure 3.
Co-mutation plot of genetic alterations identified. The percentage of samples with an alteration detected is noted on the right. Samples are displayed as columns and arranged to emphasize mutual exclusivity among mutations.
The frequencies of the molecular alterations according to smoking habits and gender are shown in Figure 2B–F. Oncogenic drivers were more commonly found in women (77%), non-smokers (90%), and former smokers (69%), compared with men and smokers (61% and 51%, respectively). The frequency of KRAS mutation was consistently related to smoking exposure (8% vs. 39% vs. 35% in never-, former-, and current-smokers, respectively), while EGFR mutations were inversely related to smoking exposure (53% vs. 15% vs. 5%, respectively). In our series, two out of five women (35%) and two out of four non-smoking patients (53%) harboured an EGFR mutation. METΔex14 mutations were more frequent in never-smokers (15% vs. 2% in patients with any smoking history). The subset of full-WT tumours (10%, n = 19) did not display differences between the different phenotypes studied, except for the non-smoking group, in which the percentage was slightly lower (3%).
The complete molecular results (DNA and RNA) were used to decide first-line treatments for most patients (172 out of 191, 90%), and 45 out of 191 patients (24%) were selected for targeted therapies (Table 2). Women, non-smokers, and ECOG 0–2 patients were significantly associated with targeted therapies, as well as the presence of EGFR, METΔex14, NTRK1, BRAF, and ALK gene alterations.
2.3. Treatment Strategies and Overall Survival (OS)
Of the 36 patients carrying EGFR mutations, 27 (75%) were treated with a targeted agent, including first-generation (n = 14), second-generation (n = 8), and third-generation (n = 5) EGFR tyrosine-kinase inhibitors (TKIs). All patients with ALK rearrangements (n = 7) were treated with ALK inhibitors (four patients with crizotinib and three patients with new generation TKI). Three out of seven patients (43%) with BRAF mutations were treated with dabrafenib-trametinib (one BRAF-V600E and two non-V600E). Seven out of nine patients (78%) with the METΔex14 alteration were treated with MET inhibitors (four in a clinical trial and three with crizotinib as compassionate use). The only patient diagnosed with an NTRK1 gene rearrangement was treated with entrectinib in a compassionate use program.
At the time of analysis, 87 patients were alive. The median OS of the whole cohort of 156 patients successfully evaluated through April 2019 was 0.93 years (95% confidence interval (CI) 0.63–1.24) (Figure 4A). Median OS was 1.40 years (95% CI 0.75–2.02) in the cohort of patients with an oncogenic driver identified (n = 107) and 0.61 years (95% CI 0.12–1.10) in the cohort of patients with no drivers identified (n = 49) (log-rank p = 0.008) (Figure 5A). Regarding the most common oncogenic drivers, median OS was significantly increased in EGFR-mutant patients (n = 35) compared with KRAS-mutant patients (n = 41) (median OS of 2.02 vs. 0.55 years, respectively, p = 0.001) (Figure 4B). Among METΔex14, the third most common oncogenic driver alteration, the median OS was 1.77 years (95% CI 0.94–2.60).
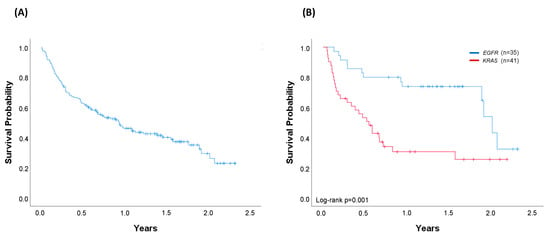
Figure 4.
Overall survival (OS). (A) Entire cohort of successfully genotyped patients diagnosed with metastatic disease through April 2019 (n = 156). Median OS (95% CI): 0.93 years (0.63–1.24); (B) Patients harbouring EGFR (n = 35) or KRAS (n = 41) oncogenic driver mutations. For KRAS-mutated patients, 54% received chemotherapy, 12% immunotherapy, and 34% best supportive care. For EGFR-mutated patients, 77% received a matched therapy, 6% chemotherapy, and 17% best supportive care. Median OS (95%CI): EGFR 2.02 years (1.86–2.19); KRAS 0.55 years (0.36–0.74), log-rank p = 0.001. Vertical tick marks are censoring events.
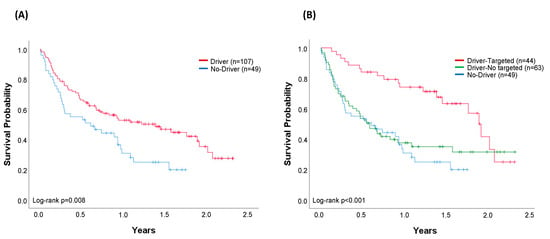
Figure 5.
Comparison of overall survival (OS). (A) Cohort of patients (n = 156) with any oncogenic driver identified (n = 107) and patients with no driver (n = 49). Median OS (95% CI): driver, 1.40 years (0.75–2.02); no-driver, 0.61 years (0.12–1.10), log-rank p = 0.008. (B) Patients with no driver (n = 49) compared with patients with any oncogenic driver treated with targeted (n = 44) and patients with any oncogenic driver not treated with targeted therapy (n = 63). All patients in the driver-targeted group were treated with a genotype-directed therapy. No-driver and driver-no targeted patients were treated with chemotherapy (51% and 48%), immunotherapy (12% and 11%), and best supportive care (37% and 41%), respectively. Median OS (95% CI): driver-targeted 1.90 years (1.69–2.10); driver-no targeted 0.59 years (0.39–0.79); no driver 0.61 years (0.12–1.10), log-rank p < 0.001. Vertical tick marks are censoring events.
When outcomes were compared in accordance with the treatment received (targeted/no-targeted), the median OS was 1.90 years (95% CI 1.69–2.10) in patients with a driver who received a matched genotype-directed therapy, compared with 0.59 years (95% CI 0.39–0.79) in patients with oncogenic drivers but not amenable to any targeted therapy (log-rank p = 0.002). Outcomes in this latter group are very similar to those from patients in whom no driver was identified (log-rank p = 0.559) (Figure 5B).
In the multivariable analysis, two variables were found to be associated with OS differences. Patients receiving targeted therapy had increased OS compared with those who were not amenable (hazard ratio (HR) 0.40, 95% CI 0.21–0.71, p = 0.002) and individuals with higher ECOG grades showed decreased OS values (HR 2.47, 95% CI 1.98–3.09, p < 0.001) (Table 3).

Table 3.
Multivariable Cox proportional hazard model for survival, accounting for confounding variables in patients with full genotyping and diagnosed with metastatic disease through April 2019 (n = 156).
3. Discussion
Our study demonstrates the feasibility of routinely obtaining multiplex genomic and transcriptomic information useful to select therapies from patients with advanced non-squamous NSCLC. The successful implementation of molecular profiling of patients with lung cancer has already been reported (Table S1). However, outside the United States, genetic testing is variable and very little data on prospective genomic surveys in Europe and Spain have been reported. Our study is, to our knowledge, the first presentation of a full prospective real-world data genomic testing with related outcomes in a public healthcare hospital in Spain, and the first worldwide to integrate two parallel, multiplexed approaches based on DNA sequencing and direct digital detection of RNA with nCounter® technology.
Using these two independent multiplexed DNA- and RNA-based platforms, we were able to screen for a total of 27 genes including tumour suppressor genes (TSG). This allowed us to provide a more precise characterization of the genetic alterations underlying non-squamous NSCLC, as compared with other studies in which data w only provided for ten [4] or six [13] of the most common or actionable driver genes (Table S1). In our study, we were able to identify oncogenic drivers in 68% of tumours, a percentage similar to the 64% recorded by The Lung Cancer Mutation Consortium (LCMC) in the USA [12], and higher than the 50% reported by the French Cooperative Thoracic Intergroup (IFCT) [13]. However, in our series, total gene alterations, comprising drivers and non-drivers, were identified in the vast majority of patients (90%). The broader molecular profiling gave us more precise information on the incidence of ‘real’ full-WT tumours, which comprised only 10% of the samples analysed, compared with the 15% previously reported [13].
Our screening failure rates were lower than previously reported, 7.6% in our series, compared with 26% and 35% in biopsies or cytologies for the LCMC [12], reinforcing their practicability in order to support individualised decisions in routine patient care. Although both techniques (NGS and nCounter) require minimum amounts of genetic material, our dropout percentage increased to 15% when considering those samples for which DNA or RNA testing could not be performed. Some groups, which also included combined DNA and RNA in their sequencing workflows, have reported lower invalid test rates of approximately 2%–3% [18,24]. However, these results do not correspond to the total screening failure rates, which should include samples with insufficient material, the major reason for failure in our series.
In our study, the prevalence of oncogenic and TSG (TP53, STK11, or CTNNB1) was aligned with the frequencies reported in other Caucasian cohorts (Table S2). Similarly, the highest incidence of drivers was found in never (90%) and former smokers (69%), endorsing the distinctive molecular signature of NSCLC according to smoking history.
Differences were observed for EGFR mutations, with an incidence (19%) significantly higher than the frequencies reported for other European and Spanish cohorts (11%–13%), but close to the series collected in the USA (23%) (Table S2). However, variations in mutation prevalence may exist depending on the variant filtering and annotation pipeline performed, and they are thus not entirely valid for comparisons. Whereas in some series, adenocarcinoma roughly accounted for 70% of the cases [13,17], in our cohort, we only included non-squamous NSCLC histologies, with a vast majority (90%) of adenocarcinomas. Moreover, some groups considered all tested patients for rating [13], including those partially genotyped, whereas we only considered those patients with a complete informative result. Finally, some studies only consider the most common EGFR sensitizing mutations (E19del and L858R), while in our cohort, we included all EGFR mutations in exons 18–21. Indeed, when considering only E19del and L858R EGFR-sensitizing mutations, our rates of EGFR positivity decrease to 16% (Figure 2).
Regarding the MET gene, we were unexpectedly able to identify 5% of patients carrying METΔex14 skipping mutations, which represented the third most common driver alteration in our cohort. This percentage is over the 2%–3% usually reported [14,16,18,25,26] (Table S2), and endorses the advantage of using an RNA-based detection technique for METΔex14 transcript identification [26]. Actually, commercially available DNA NGS panels have been reported to detect only 63% of literature-described METΔex14 mutations [27]. Interestingly, contrary to other reports [3], our cohort of METΔex14 patients was more frequently related to never-smokers (15%) and women (7%), highlighting the need for more data to properly characterize the phenotype related to this recently documented oncogenic driver.
In our cohort of genotyped patients, the median OS of 0.93 years was inferior to OS reported in previous studies [4]. However, our results reflect more accurately the real-world setting, and include a significant number of patients with ECOG 3–4 (24%) and not eligible for any treatment (16%).
The final goal of molecular testing is to personalise therapies and improve outcomes. Similar to previous reports [4,14], our molecular findings affected the treatment decision for 24% of the genotyped patients and 35% of patients with oncogenic drivers. Significant differences were observed when comparing median OS according to the presence or absence of a driver alteration (median OS 1.40 years vs. 0.61 years, respectively, log-rank p = 0.008). However, this survival advantage was not conditioned by the presence of a driver, but by the ability to receive a matched targeted therapy. Indeed, a significant increase in survival was obtained in patients with a driver who received a targeted therapy (1.90 years), while similar outcomes were observed in the cohort of oncogenic-driven patients not receiving any targeted therapy and those without drivers (median OS 0.59 years vs. 0.61 years, respectively) (Figure 5B). In the multivariable analysis, both treatment with targeted therapy and ECOG were independent variables of survival.
Remarkably, the majority (78%) of patients with the METΔex14 received a targeted therapy with an MET inhibitor, achieving an outstanding median OS of 1.77 years and 12-month OS rate of 80%. These data compare favourably with the poor outcomes of this group of NSCLC patients [28,29], and emphasize the potential for targetability of this not-yet approved biomarker. Recently, the Food and Drug Administration (FDA) has granted a breakthrough therapy designation to capmatinib (INC280) as a first-line treatment for patients with METΔex14 mutated NSCLC [30], based on the primary findings of the phase II GEOMETRY mono-1 study (NCT02414139) [31]. The final outcomes of this study are eagerly awaited, and we anticipate METΔex14 to be soon added to the long list of actionable must-be-tested oncogenes in NSCLC.
The median OS in our KRAS-mutated cohort was dismal (0.55 years) and might be attributed to the lack of effective targeted therapies. Although this outcome clearly differs from the 2.41 years previously reported in the LCMC cohort [4], it resembles the OS recently reported in a retrospective analysis of chemotherapy-treated patients with advanced NSCLC and KRAS activating mutations (8.8 months vs. 13.5 months, p = 0.038, in KRAS-mutated and –WT, respectively) [32]. Although oncogenic KRAS could serve as an excellent drug target opportunity owing to its high incidence in lung cancer, its direct inhibition has proven to be challenging owing to a lack of traditional small molecule binding pockets on the protein [33]. However, emerging direct inhibitors of the most prevalent G12C variant—overall, the second most frequent driver-subtype alteration in our cohort accounting for 12% of all genotyped patients—have recently shown encouraging results [34], and further highlight the need for comprehensive panels to characterize lung cancer beyond the current biomarker recommendations.
Our single-institution prospective report may be subject to several inherent limitations. Because it was conducted with a rather small sample size, the findings provided on outcomes should be considered cautiously. Moreover, treatment was not randomly assigned, but decided by the physician, and a selection bias cannot be excluded. Finally, we did not adjust for several known confounding variables that might impact the outcomes, such as smoking status, stage at diagnosis, or prior therapies.
4. Materials and Methods
4.1. Patients and Samples
Between June 2017 and August 2019, all of the patients at our institution with newly diagnosed advanced non-squamous NSCLC, as well as those for whom molecular profiling had not been previously completed, were prospectively included in our study with prior full informed patient consent and approval from the Local Ethical Committee (Comité Ético de Investigación Clínica del Hospital Clínic de Barcelona, V2-05/11/2016). The study was conducted in accordance with the principles of the Declaration of Helsinki. The decision to recommend a targeted therapy to a patient harbouring an actionable driver gene was left to the treating physician, who used the genomic data to select an ad-hoc therapy following clinical guidelines or via inclusion in a clinical trial.
Information on therapy and outcomes was collected regardless of whether the patient received targeted therapy. The data obtained were age; gender; histology; ECOG performance status; smoking history (never, former, or current smoker); TNM stage at diagnose, as defined by the seventh edition of the American Joint Committee on Cancer [35]; treatment setting at molecular assessment; the effect of the molecular results on the treatment decision with targeted therapy; and outcomes.
4.2. Molecular Analyses
All molecular analyses were performed on a routine basis at our institution using OncomineTM Solid Tumour DNA Kit and an in-house nCounter-based assay [23]. FFPE sample slides (4 µm) were stained with haematoxylin and eosin and the percentage of tumour cell content was estimated by a pathologist. Samples with tumour cell content higher than 20% were considered assessable for molecular analysis. Macrodissection was performed in those samples with a tumour content superior to 50%. In the case where only one technique could be performed, NGS was prioritized to nCounter.
Purified DNA (10 ng) was used as a template to generate genomic libraries using the Ion Torrent platform. The OncomineTM Solid Tumour DNA Kit (ThermoFisher Scientific, Life Technologies, CA, USA) interrogates somatic mutations on 22 genes, including EGFR, ALK, ERBB2, ERBB4, FGFR1, FGFR2, FGFR3, MET, DDR2, KRAS, PIK3CA, BRAF, AKT1, PTEN, NRAS, MAP2K1, STK11, NOTCH1, CTNNB1, SMAD4, FBXW7, and TP53. Libraries were sequenced using the Ion Personal Genome Machine platform (ThermoFisher Scientific) and analysed by the Ion Reporter Software (ThermoFisher Scientific) according to the manufacturer’s instructions.
For the nCounter assay (Nanostring Technologies, WA, USA), total RNA (25–200 ng) was directly hybridized with a customized codeset designed to detect specific fusion transcripts (7 ALK, 10 ROS1, 6 RET, and 2 NTRK1) and METΔex14 mutations. All processes of hybridization, capture, clean-up, and digital data acquisition were performed as described in previous literature [23]. Regarding METΔex14 testing, log ratios were obtained dividing the normalized counts of the METΔex14 probe by the normalized counts for the MET WT probe. The cut-off for METΔex14 positivity was established as the average log ratio of the sample cohort plus two standard deviations.
Those samples without enough tumour content for a complete nCounter analysis were tested for ALK, ROS1, and RET fusions using alternative recommended tests. ALK status was checked by IHC (anti-ALK rabbit monoclonal antibody D5F3, Ventana, Tuzson, AZ) and FISH (ZytoLight® SPEC ALK Dual Color Break Apart Probe). ROS1 and RET were characterized by FISH (ZytoLight® SPEC ROS1 Dual Color Break Apart Probe and ZytoLight® SPEC RET Dual Color Break Apart Probe, respectively).
4.3. Molecular Subgroups
Molecular alterations—EGFR, KRAS, NRAS, BRAF, MET∆ex14, ERBB2, ERBB4, PIK3CA—and gene fusions—ALK, ROS1, RET, NTRK1—were referred to as oncogenic drivers [36,37,38]. Among the aforementioned, mutations at EGFR, BRAF, and MET∆ex14, and gene fusions at ALK, ROS1, RET, and NTRK1 were considered actionable driver genes. Non-driver mutations included those reported in TP53, STK11, CTNNB1, AKT1, SMAD4, DDR2, FGFR2, MAPK, ALK, and METnon∆ex14. Tumours with an unchanged form were referred to as full WT tumours. Patients carrying co-mutations were considered once and categorized by the driver alteration.
4.4. Statistical Methods
Descriptive statistics, including the median and range for continuous variables and the percentages and frequencies for categorical variables, were tabulated and presented. OS was defined as the time from date of the metastatic disease diagnosis to the date of last follow-up or death. OS probability curves were calculated for all patients diagnosed with metastatic disease until April 2019 and for groups of interest by the Kaplan–Meier method, and compared with a two-sided log-rank test. For OS analysis, the cohort of patients with no oncogenic driver included those with non-driver mutations and full-WT tumours.
A p-value lower than 0.05 was considered for statistical significance. A multivariable Cox proportional hazard model was applied to adjust for potential confounders (clinical or molecular characteristics associated with OS). Adjusted HRs with 95% CI were calculated and the results were interpreted according to the following criteria: lack of association if HR = 1, increased risk of death if HR > 1, and lower risk of death if HR < 1. The SPSS v20.0 program was used for statistical analysis.
5. Conclusions
Our study demonstrates that embedding two DNA and RNA molecular genomic platforms early in the workup of patients with advanced NSCLC provides a complete understanding of the molecular NSCLC subsets that can be used to select patients who are candidates for targeted therapeutic intervention, resulting in significantly improved outcomes. The present paper echoes the need for expanding genomic surveys worldwide to integrate precision medicine into health care in order to guarantee the best outcomes for patients with advanced lung cancer.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/5/1124/s1, Table S1: Summary of comprehensive genotyping studies in NSCLC; Table S2: Summary of the Incidence of Driver Mutations in NSCLC.
Author Contributions
Conceptualization, E.M., C.T., E.C.-R., and N.R.; methodology, E.M., C.T., P.J., and M.Á.M.-V.; software, P.J., C.T., and M.-Á.M.-V.; formal analysis, E.M. and A.P. (acknowledgment); investigation, E.M., C.T., and N.R.; resources, M.S., I.V., M.G., S.A., A.M., P.J., and N.R.; data curation, E.M., C.T., E.C.-R., R.R., A.A., and A.R.; writing—original draft preparation, E.M., C.T., E.R-C., and N.R.; writing—review and editing, E.M., C.T., R.R., N.V., C.C., S.C., S.M., I.G.S., P.J., A.P., M.Á.M.-V., N.R., and K.J. (acknowledgment); visualization, E.M., C.T., E.C.-R., and N.R; supervision, M.Á.M.-V. and N.R.; project administration, E.M., A.A., M.R.-M., and N.R.; funding acquisition, N.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Pfizer, grant number WI230526; Novartis, grant number CINC280AES01T; and Acción Estratégica en Salud, Instituto de Salud Carlos III, Spain, grant number FIS16/00890, PI N.Reguart.
Acknowledgments
We would like to acknowledge Alejandro Pedromingo for his assistance with the statistics used in this report and Kyla M. Juett for improving the use of English in the manuscript.
Conflicts of Interest
E.M. reports no conflicts of interest. C.T. reports grants and personal fees from Pfizer and Novartis during the conduct of the study; personal fees from Roche, Pfizer, Takeda, Novartis, Astrazeneca, MSD, and BMS outside the submitted work. A.P. reports grants and personal fees from Novartis, Pfizer, Lilly, Amgen; grants from Roche; personal fees from Nanostring Technologies, Oncolytics Biotech, Daiichi Sankyo, PUMA, BMS, and Daiichi Sankyo, outside the submitted work. N.R. reports grants from Ministry of Health, Instituto de Salud Carlos III, Spain (FIS16/00890) during the conduct of the study, grants and personal fees from Pfizer and Novartis; personal fees from MSD, BMS, Roche, Boehringer Ingelheim, Guardant Health, Abbvie, Ipsen, Astrazeneca, Lilly, and Takeda outside the submitted work. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Hunter, D.J. Uncertainty in the era of precision medicine. N. Engl. J. Med. 2016, 375, 711–713. [Google Scholar] [CrossRef]
- Pao, W.; Girard, N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011, 12, 175–180. [Google Scholar] [CrossRef]
- Planchard, D.; Hellmann, M.D.; Peters, S.; Popat, S.; Kerr, K.; Novello, S.; Smit, E.F.; Faivre-Finn, C.; Mok, T.S.; Reck, M.; et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, 192–237. [Google Scholar] [CrossRef] [PubMed]
- Kris, M.G.; Johnson, B.E.; Berry, L.D.; Kwiatkowski, D.J.; Iafrate, A.J.; Wistuba, I.I.; Varella-Garcia, M.; Franklin, W.A.; Aronson, S.L.; Su, P.-F.; et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014, 311, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Majem, M.; Juan, O.; Insa, A.; Reguart, N.; Trigo, J.M.; Carcereny, E.; García-Campelo, R.; García, Y.; Guirado, M.; Provencio, M. SEOM clinical guidelines for the treatment of non-small cell lung cancer (2018). Clin. Transl. Oncol. 2019, 21, 3–17. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Aisner, D.L.; Wood, D.E.; Akerley, W.; Bauman, J.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; Dilling, T.J.; Dobelbower, M.; et al. NCCN guidelines insights: Non-Small cell lung cancer, version 5.2018. J. Natl. Compr. Cancer Netw. 2018, 16, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Kalemkerian, G.P.; Narula, N.; Kennedy, E.B.; Biermann, W.A.; Donington, J.; Leighl, N.B.; Lew, M.; Pantelas, J.; Ramalingam, S.S.; Reck, M.; et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the study of lung cancer/Association for Molecular Pathology Clinical Practice guideline update. J. Clin. Oncol. 2018, 36, 911–919. [Google Scholar] [CrossRef]
- Pennell, N.A.; Arcila, M.E.; Gandara, D.R.; West, H. Biomarker testing for patients with advanced non–small cell lung cancer: Real-world issues and tough choices. Am. Soc. Clin. Oncol. Educ. Book 2019, 531–542. [Google Scholar] [CrossRef]
- Remon, J.; Lacroix, L.; Jovelet, C.; Caramella, C.; Howarth, K.; Plagnol, V.; Rosenfeld, N.; Morris, C.; Mezquita, L.; Pannet, C.; et al. Real-World utility of an amplicon-based next-generation sequencing liquid biopsy for broad molecular profiling in patients with advanced non-small-cell lung cancer. JCO Precis. Oncol. 2019, 1–14. [Google Scholar] [CrossRef]
- Europe LuCE. LuCE Report on Lung Cancer. Disparities in Diagnosis, Care and Treatment Access. 2017. Available online: https://www.lungcancereurope.eu/2017/11/07/disparities-in-diagnosis-care-and-500treatment-access/ (accessed on 23 December 2019).
- Hiley, C.T.; Le Quesne, J.; Santis, G.; Sharpe, R.; de Castro, D.G.; Middleton, G.; Swanton, C. Challenges in molecular testing in non-small-cell lung cancer patients with advanced disease. Lancet 2016, 388, 1002–1011. [Google Scholar] [CrossRef]
- Sholl, L.M.; Aisner, D.L.; Varella-Garcia, M.; Berry, L.D.; Dias-Santagata, D.; Wistuba, I.I.; Chen, H.; Fujimoto, J.; Kugler, K.; Franklin, W.A.; et al. Multi-Institutional oncogenic driver mutation analysis in lung adenocarcinoma: The lung cancer mutation consortium experience. J. Thorac. Oncol. 2015, 10, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Barlesi, F.; Mazieres, J.; Merlio, J.-P.; Debieuvre, D.; Mosser, J.; Lena, H.; Ouafik, L.H.; Besse, B.; Rouquette, I.; Westeel, V.; et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: Results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016, 387, 1415–1426. [Google Scholar] [CrossRef]
- Volckmar, A.L.; Leichsenring, J.; Kirchner, M.; Christopoulos, P.; Neumann, O.; Budczies, J.; Morais de Oliveira, C.M.; Rempel, E.; Buchhalter, I.; Brandt, R.; et al. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: Analysis of the first 3000 Heidelberg cases. Int. J. Cancer 2019, 145, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, L.; Zhu, Y.; Huang, C.; Qin, Y.; Liu, H.; Ren-Heidenreich, L.; Shi, B.; Ren, H.; Chu, X.; et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: A comprehensive mutation profiling from 5125 Chinese cohorts. Br. J. Cancer 2014, 110, 2812–2820. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.H.; Johnson, A.; Albacker, L.; Wang, K.; Chmielecki, J.; Frampton, G.; Gay, L.; Elvin, J.A.; Vergilio, J.-A.; Ali, S.; et al. Comprehensive genomic profiling facilitates implementation of the national comprehensive cancer network guidelines for lung cancer biomarker testing and identifies patients who may benefit from enrollment in mechanism-driven clinical trials. Oncologist 2016, 21, 684–691. [Google Scholar] [CrossRef]
- Chatziandreou, I.; Tsioli, P.; Sakellariou, S.; Mourkioti, I.; Giannopoulou, I.; Levidou, G.; Korkolopoulou, P.; Patsouris, E.; Saetta, A.A. Comprehensive molecular analysis of NSCLC: Clinicopathological Associations. PLoS ONE 2015, 10, e0133859. [Google Scholar] [CrossRef]
- Tsoulos, N.; Papadopoulou, E.; Metaxa-Mariatou, V.; Tsaousis, G.; Efstathiadou, C.; Tounta, G.; Scapeti, A.; Bourkoula, E.; Zarogoulidis, P.; Pentheroudakis, G.; et al. Tumor molecular profiling of NSCLC patients using next generation sequencing. Oncol. Rep. 2017, 38, 3419–3429. [Google Scholar] [CrossRef][Green Version]
- Martín Martorell, P.; Huerta, M.; Compañ Quilis, A.; Abellán, R.; Seda, E.; Blesa, S.; Chaves, F.J.; Dualde Beltrán, D.; Roselló Keränen, S.; Franco, J.; et al. Coexistence of EGFR, KRAS, BRAF, and PIK3CA mutations and ALK rearrangement in a comprehensive cohort of 326 consecutive Spanish nonsquamous NSCLC patients. Clin. Lung Cancer 2017, 18, e395–e402. [Google Scholar] [CrossRef]
- Serizawa, M.; Koh, Y.; Kenmotsu, H.; Isaka, M.; Murakami, H.; Akamatsu, H.; Mori, K.; Abe, M.; Hayashi, I.; Taira, T.; et al. Assessment of mutational profile of Japanese lung adenocarcinoma patients by multitarget assays: A prospective, single-institute study. Cancer 2014, 120, 1471–1481. [Google Scholar] [CrossRef]
- Bast, E.; Morrissey, L.; Tammireddy, S.; Shaw, A.T.; Borger, D.R.; Lennes, I.T.; Baselga, J.; Engelman, J.A.; Temel, J.S.; Sequist, L.V.; et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann. Oncol. 2011, 22, 2616–2624. [Google Scholar] [CrossRef]
- Scheffler, M.; Ihle, M.A.; Hein, R.; Merkelbach-Bruse, S.; Scheel, A.H.; Siemanowski, J.; Brägelmann, J.; Kron, A.; Abedpour, N.; Ueckeroth, F.; et al. K-ras mutation subtypes in NSCLC and associated co-occuring mutations in other oncogenic pathways. J. Thorac. Oncol. 2019, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Reguart, N.; Teixidó, C.; Giménez-Capitán, A.; Paré, L.; Galván, P.; Viteri, S.; Rodríguez, S.; Peg, V.; Aldeguer, E.; Viñolas, N.; et al. Identification of ALK, ROS1, and RET fusions by a multiplexed mRNA-based assay in formalin-fixed, paraffin-embedded samples from advanced non-small-cell lung cancer patients. Clin. Chem. 2017, 63, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Simarro, J.; Murria, R.; Pérez-Simó, G.; Llop, M.; Mancheño, N.; Ramos, D.; de Juan, I.; Barragán, E.; Laiz, B.; Cases, E.; et al. Development, implementation and assessment of molecular diagnostics by next generation sequencing in personalized treatment of cancer: Experience of a public reference healthcare hospital. Cancers 2019, 1196. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Peled, N.; Greer, J.; Wu, W.; Choi, P.; Berger, A.H.; Wong, S.; Jen, K.Y.; Seo, Y.; Hann, B.; et al. Exon 14 mutation encodes an actionable therapeutic target in lung adenocarcinoma. Cancer Res. 2017, 77, 4498–4505. [Google Scholar] [CrossRef] [PubMed]
- Reungwetwattana, T.; Liang, Y.; Zhu, V.; Ou, S.-H.I. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: The why, the how, the who, the unknown, and the inevitable. Lung Cancer 2017, 103, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Poirot, B.; Doucet, L.; Benhenda, S.; Champ, J.; Meignin, V.; Lehmann-Che, J. MET exon 14 alterations and new resistance mutations to tyrosine kinase inhibitors: Risk of inadequate detection with current amplicon-based NGS panels. J. Thorac. Oncol. 2017, 12, 1582–1587. [Google Scholar] [CrossRef]
- Tong, J.H.; Yeung, S.F.; Chan, A.W.H.; Chung, L.Y.; Chau, S.L.; Lung, R.W.M.; Tong, C.Y.; Chow, C.; Tin, E.K.Y.; Yu, Y.H.; et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non-small cell lung carcinoma with poor prognosis. Clin. Cancer Res. 2016, 22, 3048–3056. [Google Scholar] [CrossRef]
- Mazza, V. New molecular drivers in NSCLC: The role of MET. Oncol. Res. Rev. 2019, 2, 1–7. [Google Scholar] [CrossRef]
- Novartis. Novartis Investigational Lung Cancer Therapy Capmatinib (INC280) Granted FDA Breakthrough Therapy Designation for Patients with MET-Mutated Advanced Non-Small Cell Lung Cancer (news release). Available online: https://www.novartis.com/news/media-releases/novartis-investigational-lung-cancer-therapy-capmatinib-inc280granted-fda-breakthrough-therapy-designation-patients-met-mutated-advanced-non-small-cell-lung (accessed on 21 December 2019).
- Wolf, J.; Seto, T.; Han, J.-Y.; Reguart, N.; Garon, E.B.; Groen, H.J.M.; Tan, D.S.-W.; Hida, T.; De Jonge, M.J.; Orlov, S.V.; et al. Capmatinib (INC280) in METΔex14-mutated advanced non-small cell lung cancer (NSCLC): Efficacy data from the phase II GEOMETRY mono-1 study. J. Clin. Oncol. 2019, 37, 9004. [Google Scholar] [CrossRef]
- Hames, M.L.; Chen, H.; Iams, W.; Aston, J.; Lovly, C.M.; Horn, L. Correlation between KRAS mutation status and response to chemotherapy in patients with advanced non-small cell lung cancer☆. Lung Cancer 2016, 92, 29–34. [Google Scholar] [CrossRef]
- Yang, H.; Liang, S.-Q.; Schmid, R.A.; Peng, R.-W. New horizons in KRAS-mutant lung cancer: Dawn after darkness. Front. Oncol. 2019, 9, 953. [Google Scholar] [CrossRef] [PubMed]
- Govindan, R.; Fakih, M.G.; Price, T.J.; Falchook, G.S.; Desai, J.; Kuo, J.C.; Strickler, J.H.; Krauss, J.C.; Li, B.T.; Denlinger, C.S.; et al. 446PD phase I study of AMG 510, a novel molecule targeting KRAS G12C mutant solid tumours. Ann. Oncol. 2019, 30, v159–v193. [Google Scholar] [CrossRef]
- Goldstraw, P.; Crowley, J.; Chansky, K.; Giroux, D.J.; Groome, P.A.; Rami-Porta, R.; Postmus, P.E.; Rusch, V.; Sobin, L. The IASLC lung cancer staging project: Proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J. Thorac. Oncol. 2007, 2, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.G.; Zhang, S.M.; Ding, X.X.; He, B.; Zhang, H.Q. Driver genes in non-small cell lung cancer: Characteristics, detection methods, and targeted therapies. Oncotarget 2017, 8, 57680–57692. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Hanker, A.B.; Garrett, J.T. Genomic alterations of ERBB receptors in cancer: Clinical implications. Oncotarget 2017, 8, 114371–114392. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, K.; Sequist, L.V.; Arcila, M.E.; Lovly, C.M.; Chen, X.; Rudin, C.M.; Moran, T.; Camidge, D.R.; Vnencak-Jones, C.L.; Berry, L.; et al. Characteristics of lung cancers harboring NRAS mutations. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 2584–2591. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).