Circulating miR-21 as a Potential Biomarker for the Diagnosis of Oral Cancer: A Systematic Review with Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
- Studies investigating the expression in whole blood, plasma, or serum of miR-21 in OSCC patients with oral cancer and healthy controls.
- Articles reporting data related to the diagnostic prediction and predictive performance, including parameters such as specificity, sensitivity, and area under the receiver operating characteristic (ROC) curve.
- Case reports, reviews, and in vitro studies, as well as studies on animal models and human cell lines were excluded from this study.
- We excluded all articles that did not focus on the expression of miR-21 or that did not report adequate data related to this.
2.2. Research Methodology
2.3. Screening Methodology
2.4. Statistical Analysis Protocol
3. Results
3.1. Study Characteristics and Data Extraction
3.2. Risk of Bias
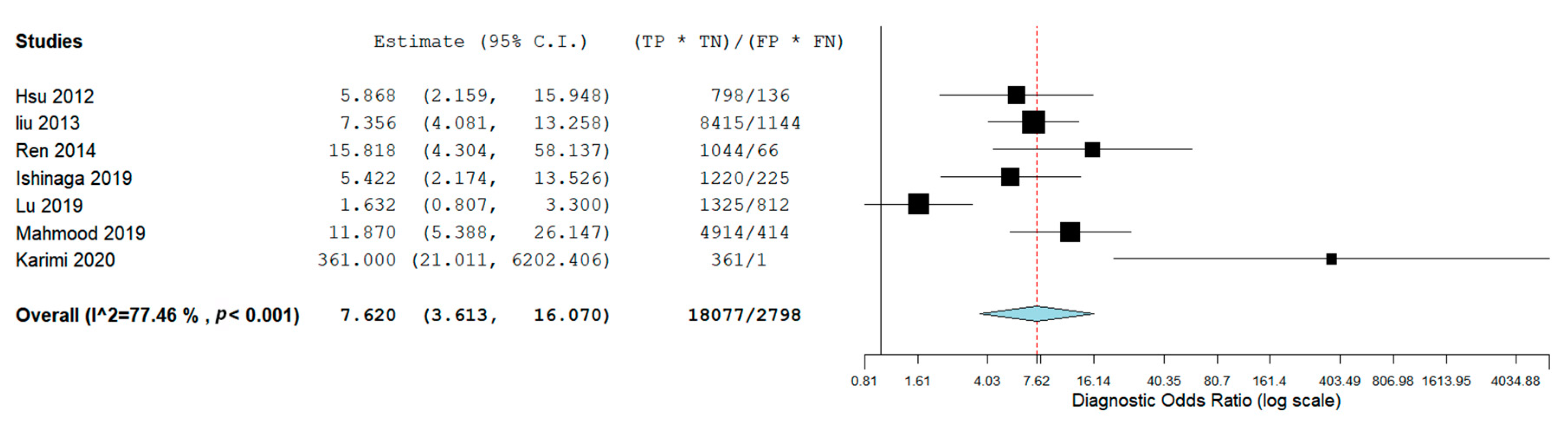
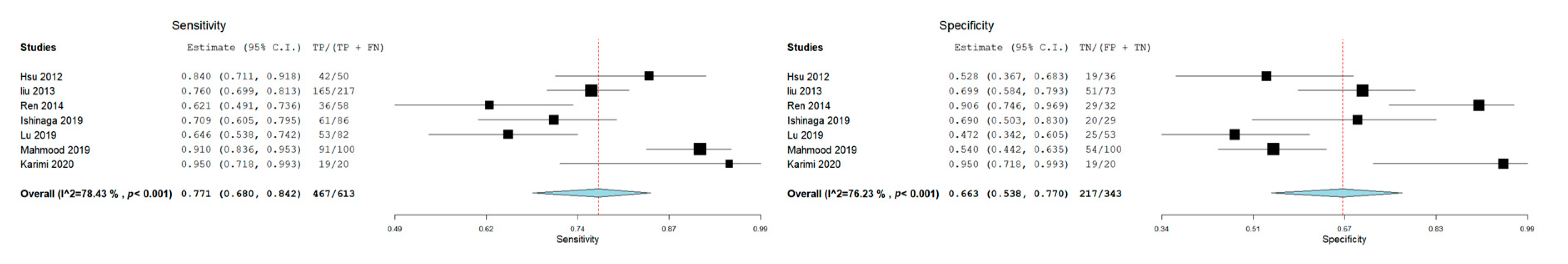
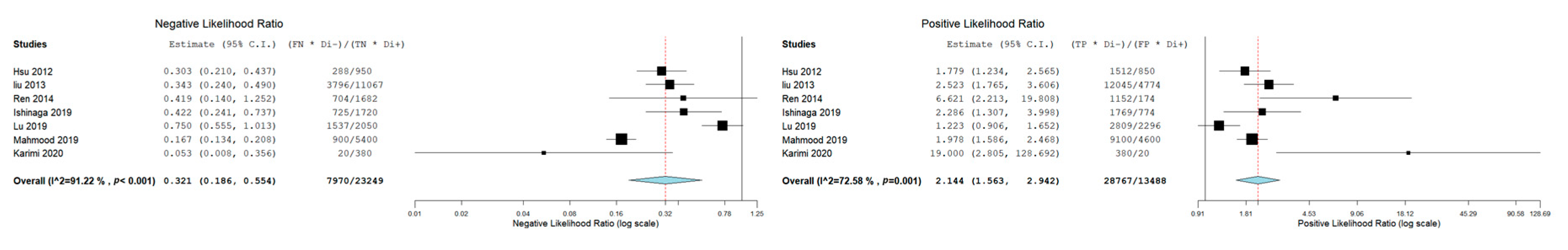
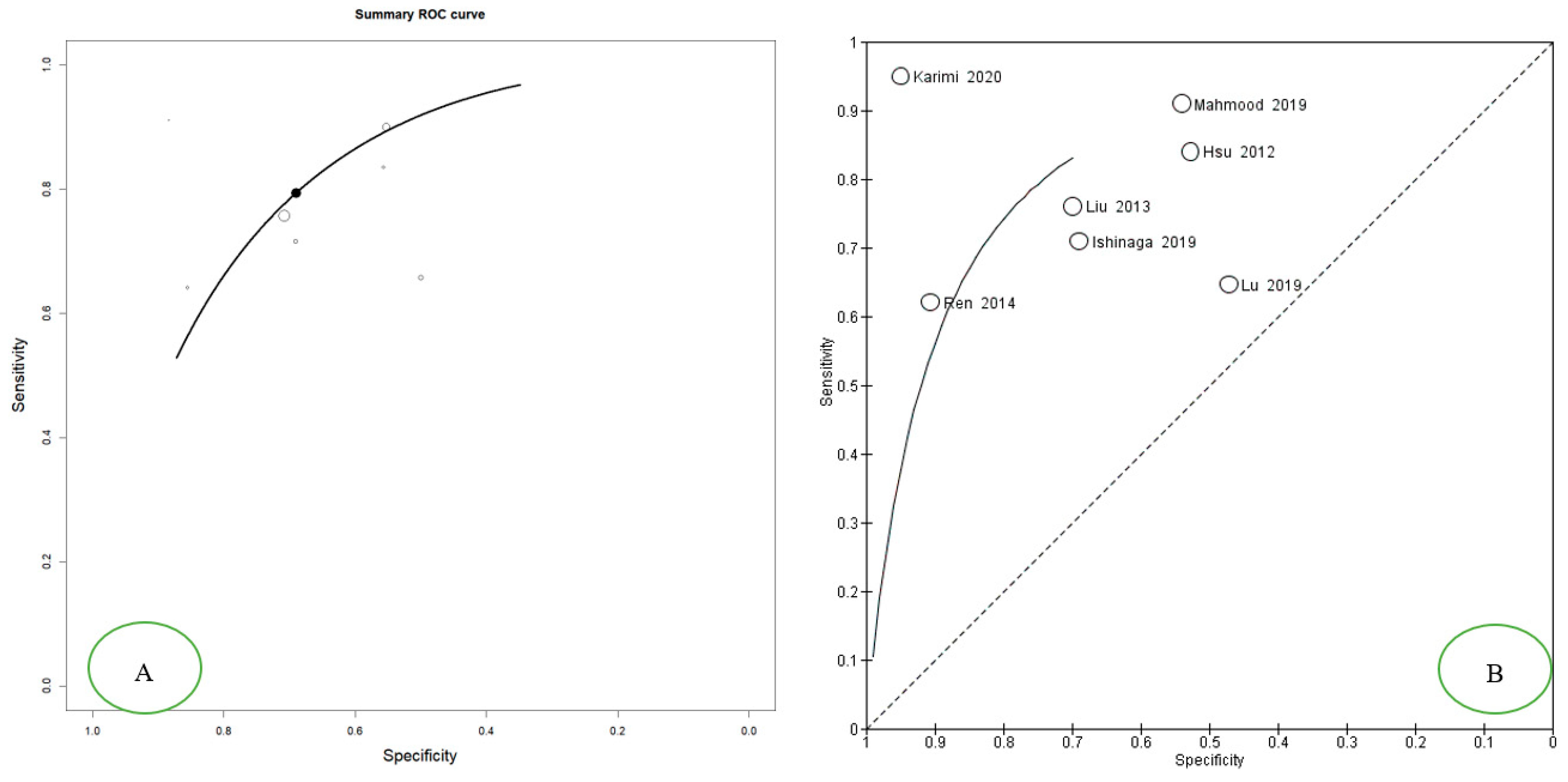
3.3. Meta-Analysis: Diagnostic Accuracy of miR-21 for OSCC and HNSCC
4. Discussion
Limits of the Study
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jou, A.; Hess, J. Epidemiology and Molecular Biology of Head and Neck Cancer. Oncol. Res. Treat. 2017, 40, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, L.P.; Carvalho, A.L. Natural history of untreated head and neck cancer. Eur. J. Cancer 2000, 36, 1032–1037. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J Clin 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.; Wiesenfeld, D. Oral Cancer. Aust. Dent. J. 2018, 63 (Suppl. 1), S91–S99. [Google Scholar] [CrossRef]
- Ali, J.; Sabiha, B.; Jan, H.U.; Haider, S.A.; Khan, A.A.; Ali, S.S. Genetic etiology of oral cancer. Oral Oncol. 2017, 70, 23–28. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Bala, S. Emerging role of non-coding RNA in oral cancer. Cell Signal 2018, 42, 134–143. [Google Scholar] [CrossRef]
- Tomaru, Y.; Hayashizaki, Y. Cancer research with non-coding RNA. Cancer Sci. 2006, 97, 1285–1290. [Google Scholar] [CrossRef]
- Shivdasani, R.A. MicroRNAs: Regulators of gene expression and cell differentiation. Blood 2006, 108, 3646–3653. [Google Scholar] [CrossRef]
- Shukla, S.; Sumaria, C.S.; Pradeepkumar, P.I. Exploring chemical modifications for siRNA therapeutics: A structural and functional outlook. ChemMedChem 2010, 5, 328–349. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Croce, C. MicroRNAs in leukemia. Clin. Adv. Hematol. Oncol. 2006, 4, 577–578. [Google Scholar] [PubMed]
- Mei, Q.; Li, X.; Guo, M.; Fu, X.; Han, W. The miRNA network: Micro-regulator of cell signaling in cancer. Expert Rev. Anticancer Ther. 2014, 14, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Kawa, M.P.; Sobus, A.; Litwinska, Z.; Osowicz-Korolonek, L.; Cymbaluk-Ploska, A.; Stecewicz, I.; Zagrodnik, E.; Romanowska, H.; Walczak, M.; Syrenicz, A.; et al. Expression of selected angiogenesis-related small microRNAs in patients with abnormally increased secretion of glucocorticoids. Endokrynol. Pol. 2019, 70, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Troiano, G.; Mastrangelo, F.; Caponio, V.C.A.; Laino, L.; Cirillo, N.; Lo Muzio, L. Predictive Prognostic Value of Tissue-Based MicroRNA Expression in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-analysis. J. Dent. Res. 2018, 97, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Hirata, Y.; Murai, N.; Yanaihara, N.; Saito, M.; Saito, M.; Urashima, M.; Murakami, Y.; Matsufuji, S.; Okamoto, A. MicroRNA-21 is a candidate driver gene for 17q23-25 amplification in ovarian clear cell carcinoma. BMC Cancer 2014, 14, 799. [Google Scholar] [CrossRef]
- Sayed, D.; Rane, S.; Lypowy, J.; He, M.; Chen, I.Y.; Vashistha, H.; Yan, L.; Malhotra, A.; Vatner, D.; Abdellatif, M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell 2008, 19, 3272–3282. [Google Scholar] [CrossRef]
- Mahmood, N.; Hanif, M.; Ahmed, A.; Jamal, Q.; Mushtaq, S.; Khan, A.; Saqib, M. Circulating miR-21 as a prognostic and predictive biomarker in oral squamous cell carcinoma. Pak. J. Med. Sci. 2019, 35, 1408–1412. [Google Scholar] [CrossRef]
- Karimi, A.; Bahrami, N.; Sayedyahossein, A.; Derakhshan, S. Evaluation of circulating serum 3 types of microRNA as biomarkers of oral squamous cell carcinoma; A pilot study. J. Oral Pathol. Med. 2020, 49, 43–48. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Green, S. Cochrane Collaboration. Cochrane Handbook for Systematic Reviews of Interventions; Wiley-Blackwell: Chichester, UK; Hoboken, NJ, USA, 2008; 649p. [Google Scholar]
- Lo, C.K.; Mertz, D.; Loeb, M. Newcastle-Ottawa Scale: Comparing reviewers’ to authors’ assessments. BMC Med. Res. Methodol. 2014, 14, 45. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Luo, H.N.; Tian, W.D.; Lu, J.; Li, G.; Wang, L.; Zhang, B.; Liang, B.J.; Peng, X.H.; Lin, S.X.; et al. Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol. Ther. 2013, 14, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-M.; Lin, P.-M.; Wang, Y.-M.; Chen, Z.-J.; Lin, S.-F.; Yang, M.-Y. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumor Biol. 2012, 33. [Google Scholar] [CrossRef]
- Ren, W.; Qiang, C.; Gao, L.; Li, S.M.; Zhang, L.M.; Wang, X.L.; Dong, J.W.; Chen, C.; Liu, C.Y.; Zhi, K.Q. Circulating microRNA-21 (MIR-21) and phosphatase and tensin homolog (PTEN) are promising novel biomarkers for detection of oral squamous cell carcinoma. Biomarkers 2014, 19, 590–596. [Google Scholar] [CrossRef]
- Lu, Z.; He, Q.; Liang, J.; Li, W.; Su, Q.; Chen, Z.; Wan, Q.; Zhou, X.; Cao, L.; Sun, J.; et al. miR-31-5p is a Potential Circulating Biomarker and Therapeutic Target for Oral Cancer. Mol. Ther. Nucleic Acids 2019, 16, 471–480. [Google Scholar] [CrossRef]
- Ishinaga, H.; He, F.; Hou, B.; Shah, S.; Murata, M.; Takeuchi, K. A longitudinal study on circulating miR-21 as a therapeutic effect marker in head and neck squamous cell carcinoma. Carcinogenesis 2019. [Google Scholar] [CrossRef]
- Wu, K.; Li, L.; Li, S. Circulating microRNA-21 as a biomarker for the detection of various carcinomas: An updated meta-analysis based on 36 studies. Tumor Biol. 2015, 36, 1973–1981. [Google Scholar] [CrossRef]
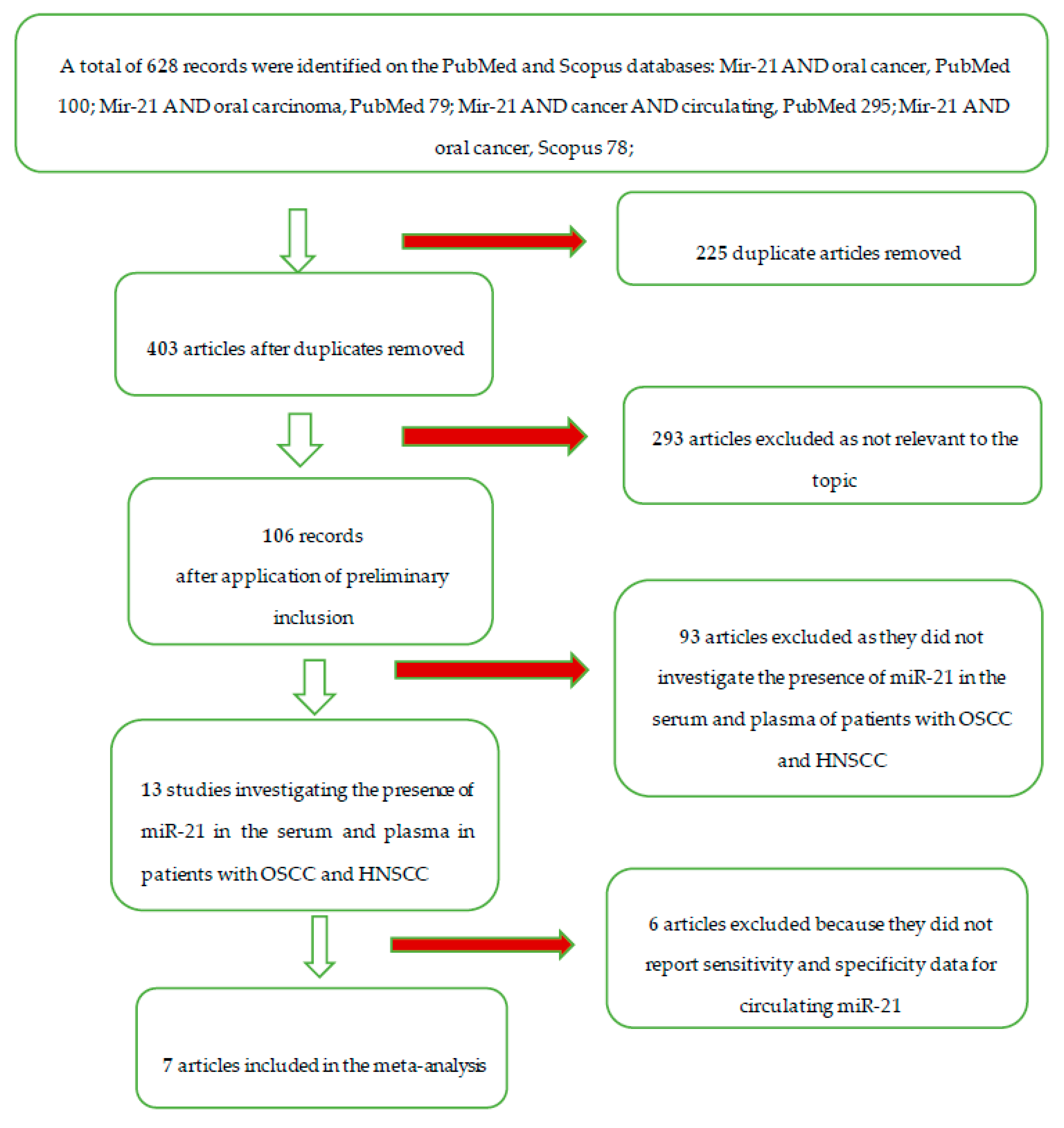
| Database Provider | Keywords | Search Details | Number of Records | Articles after Removal of Duplicates | Number of Records) after Restriction by Year of Publication (Last 20 Years) | Number of Articles that have Investigated the Expression of mir-21 in Relation to Oral Cancer | Number of Articles Investigating the Presence in Serum and Plasma of mir-21, in Patients with Oral Squamous Cancer Cell Carcinoma (OSCC) and Head and Neck Squamous Cell Carcinoma (HNSCC) | Number of Articles included in the Meta-Analysis |
|---|---|---|---|---|---|---|---|---|
| PubMed | Mir-21 AND oral cancer | Mir-21[All Fields] AND ("mouth neoplasms"[MeSH Terms] OR ("mouth"[All Fields] AND "neoplasms"[All Fields]) OR "mouth neoplasms"[All Fields] OR ("oral"[All Fields] AND "cancer"[All Fields]) OR "oral cancer"[All Fields]) | 100 | |||||
| PubMed | Mir-21 AND oral carcinoma | Mir-21[All Fields] AND (("mouth"[MeSH Terms] OR "mouth"[All Fields] OR "oral"[All Fields]) AND ("carcinoma"[MeSH Terms] OR "carcinoma"[All Fields])) | 79 | |||||
| PubMed | Mir-21 AND cancer AND circulating | Mir-21[All Fields] AND ("neoplasms"[MeSH Terms] OR "neoplasms"[All Fields] OR "cancer"[All Fields]) AND circulating [All Fields] | 299 | |||||
| Scopus | Mir-21 AND oral cancer | TITLE-ABS-KEY (mir-21 AND oral AND cancer) | 78 | |||||
| Scopus | Mir-21 AND oral carcinoma | TITLE-ABS-KEY (mir-21 AND oral AND carcinoma) | 72 | |||||
| Total | 628 | 403 | 403 | 106 | 13 | 7 |
| / | / | Reviewer 2 | Reviewer 2 | Reviewer 2 | / |
|---|---|---|---|---|---|
| Include | Exclude | Unsure | Total | ||
| Reviewer 1 | Include | 7 | 0 | 0 | 7 |
| Reviewer 1 | Exclude | 0 | 92 | 2 | 94 |
| Reviewer 1 | Unsure | 1 | 4 | 0 | 5 |
| Total | 8 | 96 | 2 | 106 |
| Autor, Date, Journal | MicroRNA Investigated | Type of Sample | Gender of Patients | Age | Risk Factors | Country | Carcinoma | Control Group | Sensitivity miR-21 | Specificity miR-21 | AUC | Cut-off Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Karimi et al. 2020, J Oral Pathol Med [29] | miR-21, miR-24, miR-29a | Serum | Male (14 OSCC-14 Control) Female (6 OSCC-6 Control) | OSCC (46.60 ± 10.69); Control 47.10 ± 17.66 | Smoking condition (OSCC 10- Control 10) | Iran | OSCC (n = 20) | (n = 20) | 95% (CI: 76.39–99.11) | 95 % (CI: 76.39–99.11) | \ | \ |
| Ishinaga et al. 2019, Carcinogenesis [31] | miR-21 | Plasma | Male (77 HNSCC-23 Control) Female (9 HNSCC-6 Control) | HNSCC (65.5 ± 11.2) Control (61.3 ± 10.6) | / | Japan | HNSCC (n = 86) | (n = 29) | 70.9% | 69.0% | 0.756 (95% CI: 0.661–0.851 | 1.15 |
| Mahmood et al. 2019, Pak J Med Sci. [18] | miR-21 | Plasma | 136 Males and 64 Females | 32.29 ± 4.98 for males, 31.77± 5.4 for female. | Smoking status (66 Yes, 34 No) | Pakistan | Oral cancer (n = 100) | (n = 100) | 91% | 54% | 0.829 | 35 Ct |
| Lu et al. 2019, Molecular Therapy—Nucleic Acids [30] | miR-99a-5p, miR31-5p, miR-138-5p, miR-21-5p, miR-375-3p | Serum | Male (OCSS 61, Control 27) Female (OSCC 21, Control 26) | Age ≥ 60 41 (50%) 12 (22.6%) Age < 60 41 (50%) 41 (77.4%) | / | China | Oral cancer (n = 82) | (n = 53) | 64.2% | 46.3% | 0.579 (95% CI: 0.483-0.675) | \ |
| Ren et al. 2014, Biomarkers [28] | miR-21 | Blood | Male 39 (67.2%); Female 19 (32.8%) | Age Mean (range) 61 (25–92) | Nonsmokers 31 (53.4%). Current smokers 27 (46.6%) | China | OSCC (n = 58) | (n = 32) | 62.1% | 90.6% | 0.788 (95% CI: 0.692–0.883) | 9.646 |
| Liu et al. 2013 Cancer Biol Ther [26] | miR-16, miR -21, miR -24, miR -155 | Plasma | NPC 149 males and 68 females; Control group 50 males and 23 females | NPC, age 45.93 ± 11.78; Control, Age 40.08 ± 10.79. | / | China | Nasopharyngeal carcinoma (NPC) (n= 217) | (N = 73) | 76% | 69,9 | 79.2% DS 0.030(95% CI: 0.733 0.852) | \ |
| Hsu et al. 2012, Tumor Biol [27] | let-7a, miR-21, miR26b, miR-34c, miR-99a, miR133a, miR-137, miR-184, miR-194a, miR-375 | Plasma | HNSCC (48 Male, 2 Female); Control (27 Male, 9 Female) | HNSCC: aged 34–82 years (mean ± SD, 54.61 ± 10.38); Control: aged 23–62 years (mean ± SD, 36.23 ± 2.34) | / | Taiwan | HNSCC (n = 50) | (n = 36) | 83.3% | 51.9% | 0.741 | \ |
| Study | Risk of Bias | Applicability Concerns | |||||
|---|---|---|---|---|---|---|---|
| Patient Selection | Index Test | Reference Standard | Flow and Timing | Patient Selection | Index Test | Reference Standard | |
| Karimi et al. 2020, J Oral Pathol Med [29] | L | L | U | L | L | U | L |
| Ishinaga et al. 2019, Carcinogenesis [31] | L | U | L | L | L | L | L |
| Mahmood et al. 2019, Pak J Med Sci. [18] | L | L | L | L | L | L | L |
| Lu et al. 2019, Molecular Therapy—Nucleic Acids [30] | L | U | L | L | L | L | L |
| Ren et al. 2014, Biomarkers [28] | U | L | L | L | L | L | U |
| Liu et al. 2013 Cancer Biol Ther [26] | L | L | L | L | L | L | L |
| Hsu et al. 2012, Tumor Biol [27] | H | L | L | L | L | L | L |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dioguardi, M.; Caloro, G.A.; Laino, L.; Alovisi, M.; Sovereto, D.; Crincoli, V.; Aiuto, R.; Coccia, E.; Troiano, G.; Lo Muzio, L. Circulating miR-21 as a Potential Biomarker for the Diagnosis of Oral Cancer: A Systematic Review with Meta-Analysis. Cancers 2020, 12, 936. https://doi.org/10.3390/cancers12040936
Dioguardi M, Caloro GA, Laino L, Alovisi M, Sovereto D, Crincoli V, Aiuto R, Coccia E, Troiano G, Lo Muzio L. Circulating miR-21 as a Potential Biomarker for the Diagnosis of Oral Cancer: A Systematic Review with Meta-Analysis. Cancers. 2020; 12(4):936. https://doi.org/10.3390/cancers12040936
Chicago/Turabian StyleDioguardi, Mario, Giorgia Apollonia Caloro, Luigi Laino, Mario Alovisi, Diego Sovereto, Vito Crincoli, Riccardo Aiuto, Erminia Coccia, Giuseppe Troiano, and Lorenzo Lo Muzio. 2020. "Circulating miR-21 as a Potential Biomarker for the Diagnosis of Oral Cancer: A Systematic Review with Meta-Analysis" Cancers 12, no. 4: 936. https://doi.org/10.3390/cancers12040936
APA StyleDioguardi, M., Caloro, G. A., Laino, L., Alovisi, M., Sovereto, D., Crincoli, V., Aiuto, R., Coccia, E., Troiano, G., & Lo Muzio, L. (2020). Circulating miR-21 as a Potential Biomarker for the Diagnosis of Oral Cancer: A Systematic Review with Meta-Analysis. Cancers, 12(4), 936. https://doi.org/10.3390/cancers12040936











