53BP1 Accumulation in Circulating Tumor Cells Identifies Chemotherapy-Responsive Metastatic Breast Cancer Patients
Abstract
1. Introduction
2. Results
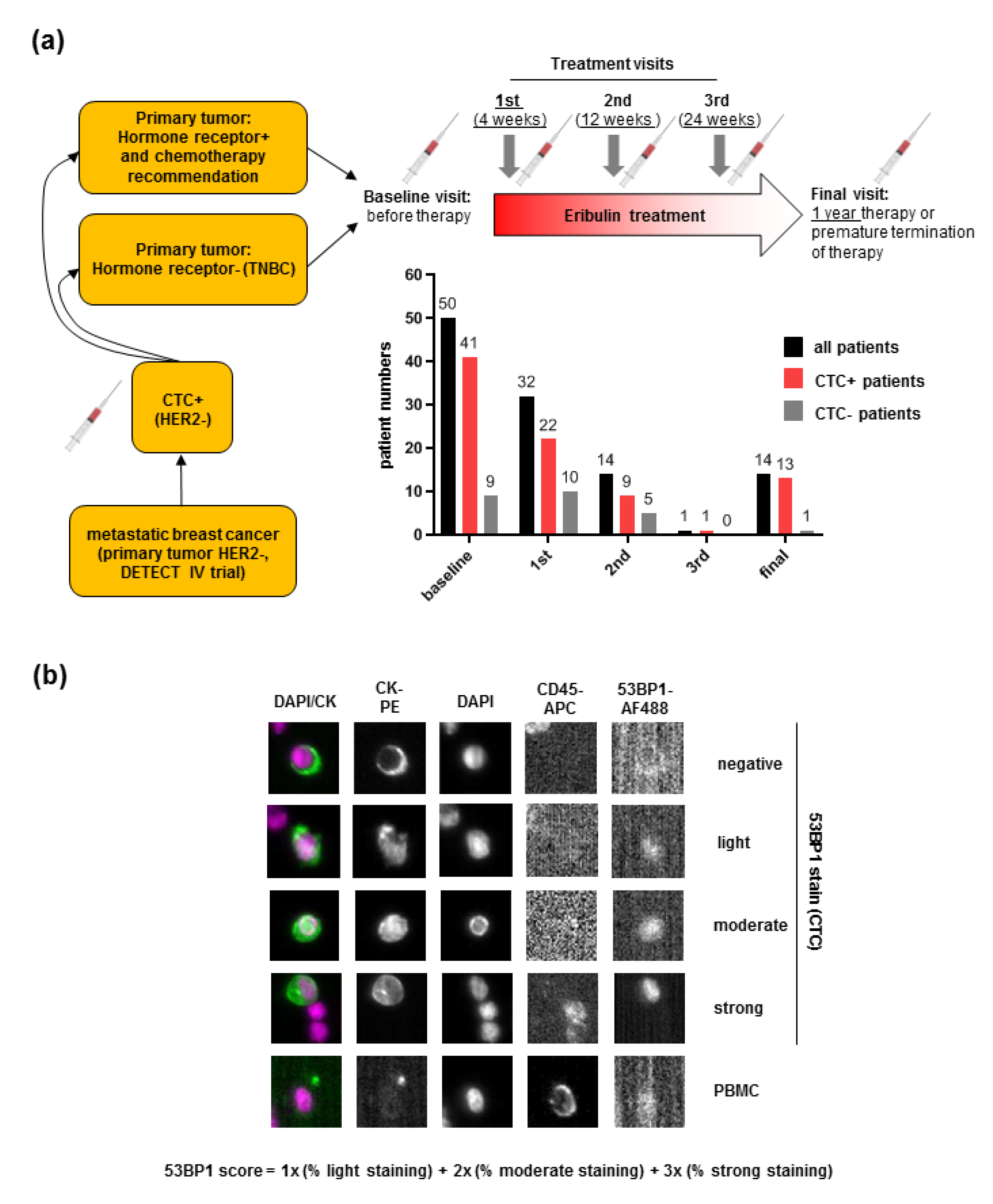
2.1. Detection of 53BP1 Signals in CTCs from MBC Patients
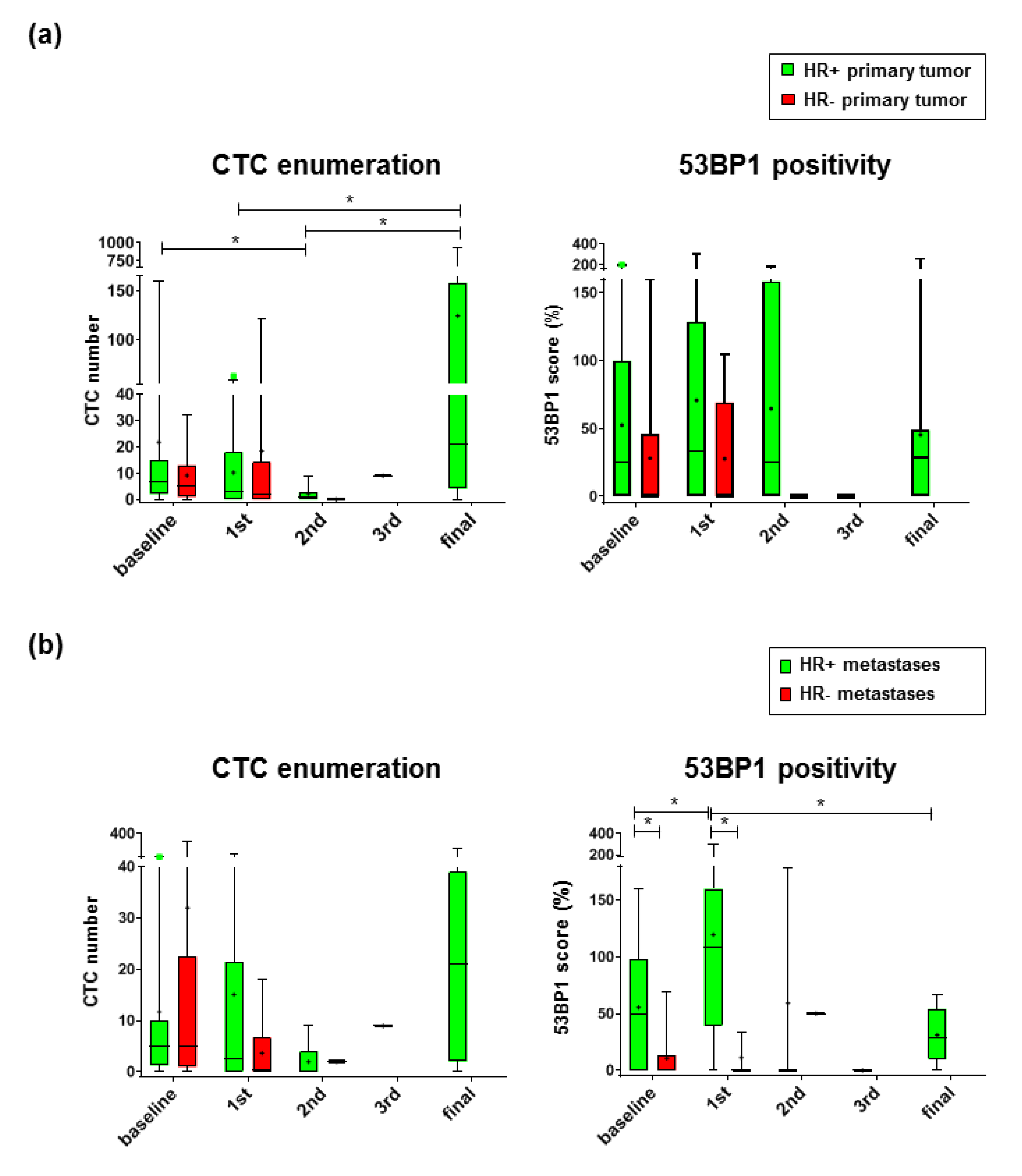
2.2. 53 BP1 Accumulates in CTCs from HR+ MBC Patients During Eribulin Treatment
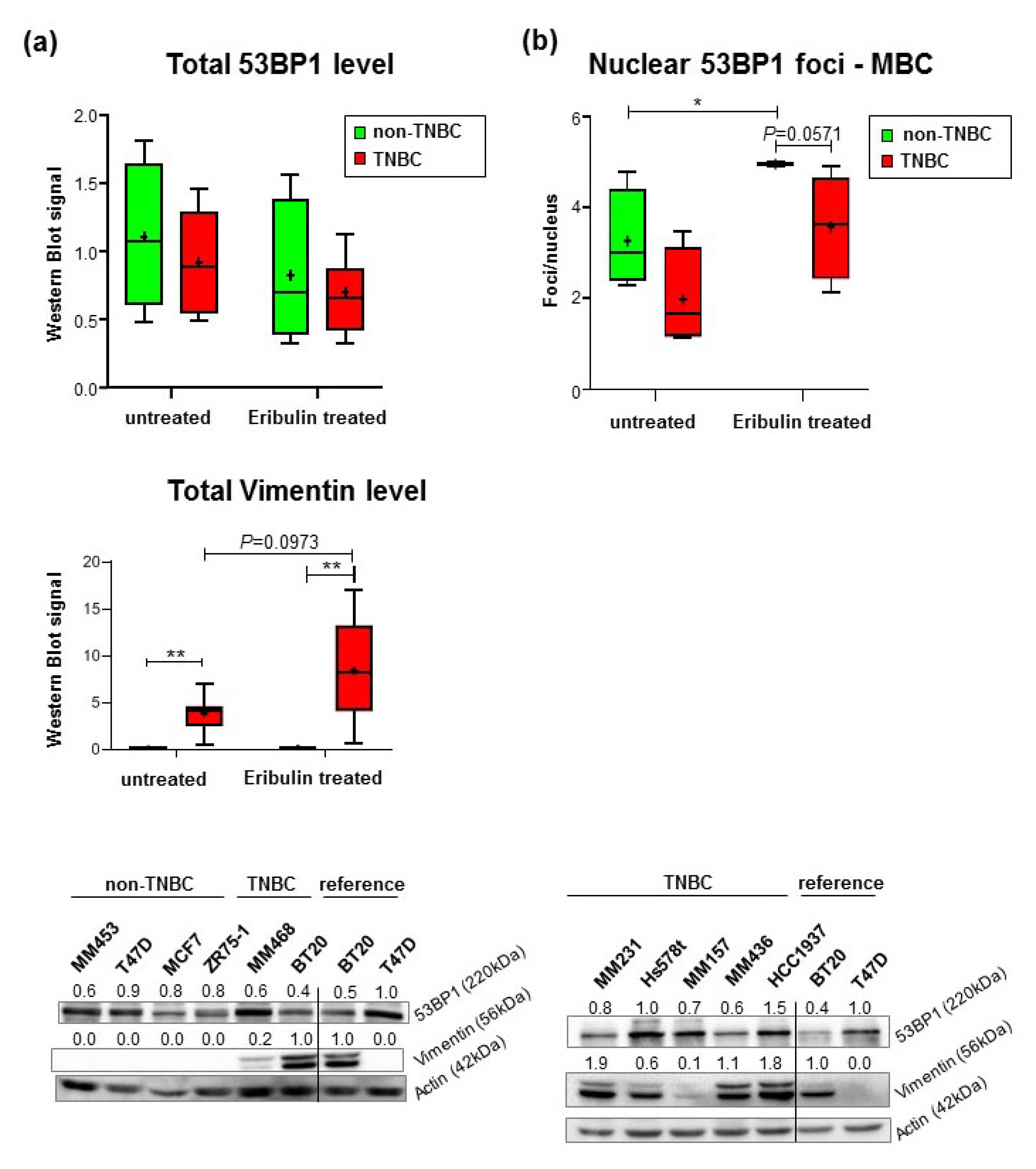
2.3. Reduced Formation of Nuclear 53BP1 Foci in TNBC Cell Lines Does Not Reflect Changes in γH2AX-Labeled DNA Damage
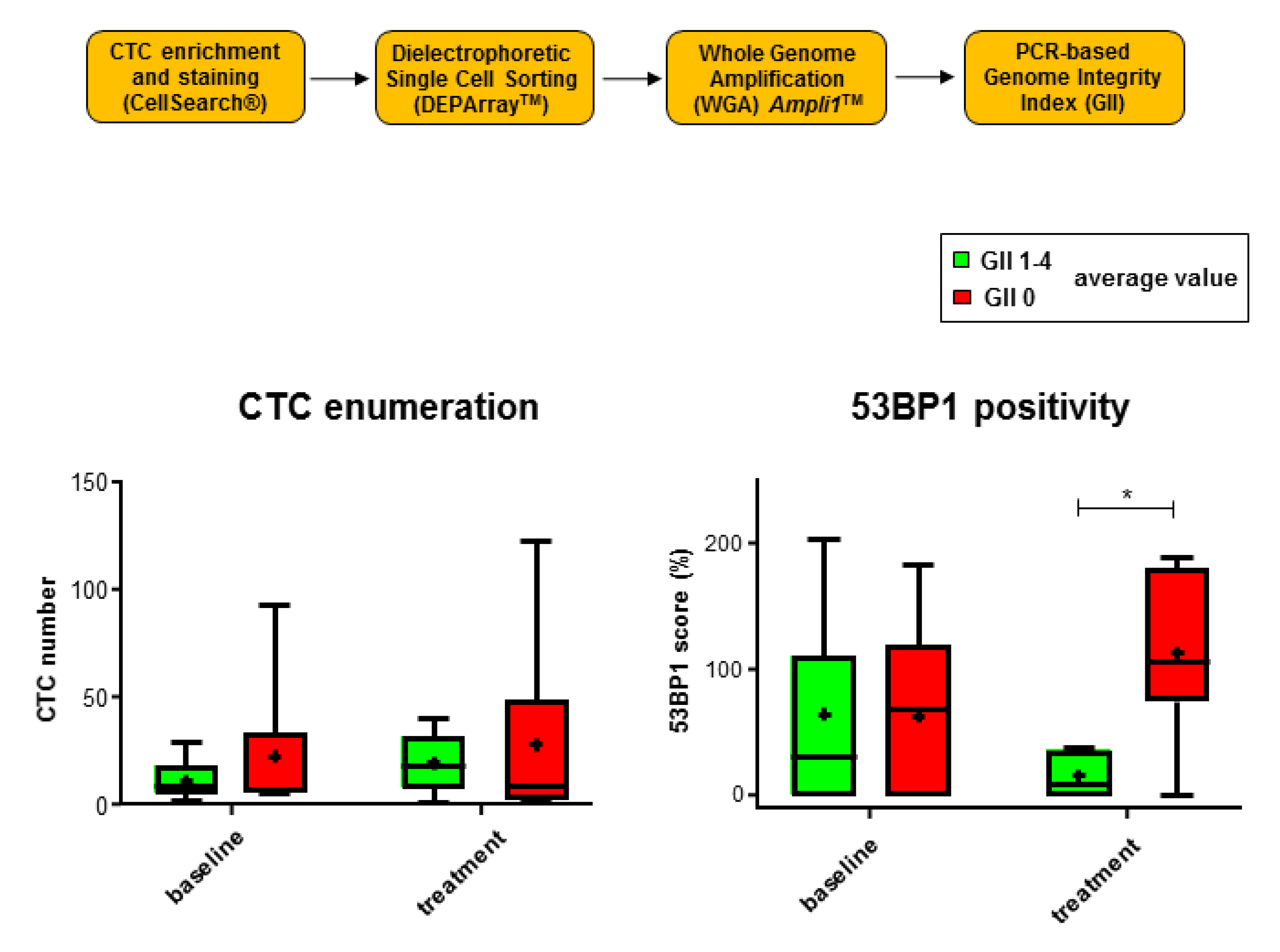
2.4. 53 BP1 Signals Rise in CTCs from MBC Patients with Low Genomic Integrity During Eribulin Treatment
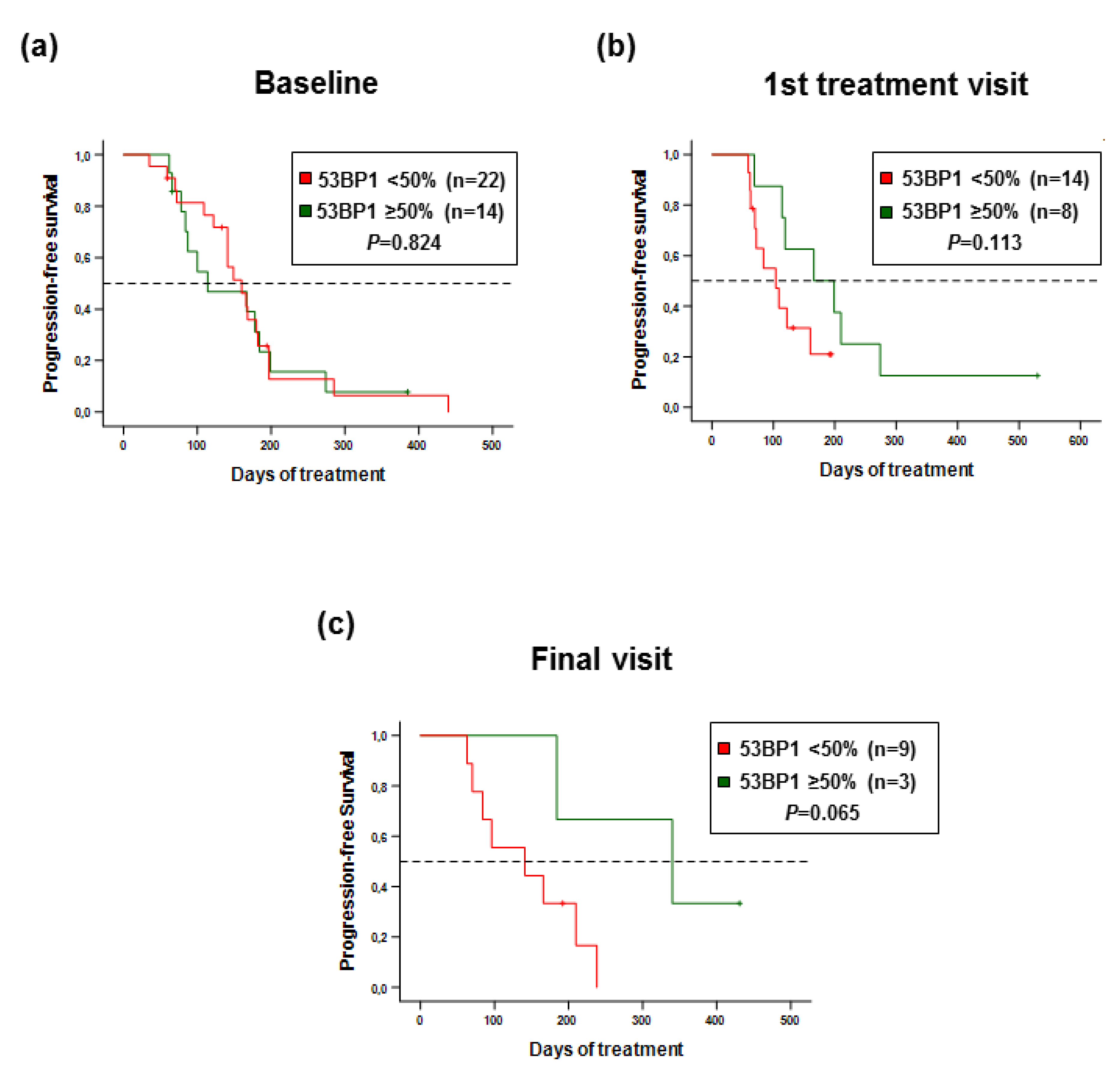
2.5. 53 BP1 Score as a Biomarker for Progression-Free Survival of MBC Patients
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment and Sample Collection
4.2. CTC Enrichment, Immunostaining, and Image Analysis
4.3. Evaluation of Genomic Integrity in Individual CTCs
4.4. Breast Cancer Cell Lines, Cultivation, and Treatment
4.5. Western Blot Analysis
4.6. Quantitative Immunofluorescence Microscopy
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kassam, F.; Enright, K.; Dent, R.; Dranitsaris, G.; Myers, J.; Flynn, C.; Fralick, M.; Kumar, R.; Clemons, M. Survival Outcomes for Patients with Metastatic Triple-Negative Breast Cancer: Implications for Clinical Practice and Trial Design. Clin. Breast Cancer 2009, 9, 29–33. [Google Scholar] [CrossRef]
- Gonzalez-Angulo, A.M.; Timms, K.M.; Liu, S.; Chen, H.; Litton, J.K.; Potter, J.; Lanchbury, J.S.; Stemke-Hale, K.; Hennessy, B.T.; Arun, B.K.; et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin. Cancer Res. 2011, 17, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, F.C.; van Overeem Hansen, T.; Sørensen, C.S. Hereditary breast and ovarian cancer: New genes in confined pathways. Nat. Rev. Cancer 2016, 16, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, E.; Ganzinelli, M.; Andreis, D.; Bertoni, R.; Giardini, R.; Fox, S.B.; Broggini, M.; Bottini, A.; Zanoni, V.; Bazzola, L.; et al. Triple negative breast cancers have a reduced expression of DNA repair genes. PLoS ONE 2013, 8, e66243. [Google Scholar] [CrossRef] [PubMed]
- Couch, F.J.; Hart, S.N.; Sharma, P.; Toland, A.E.; Wang, X.; Miron, P.; Olson, J.E.; Godwin, A.K.; Pankratz, V.S.; Olswold, C.; et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015, 33, 304–311. [Google Scholar] [CrossRef]
- Bunting, S.F.; Callén, E.; Wong, N.; Chen, H.-T.; Polato, F.; Gunn, A.; Bothmer, A.; Feldhahn, N.; Fernandez-Capetillo, O.; Cao, L.; et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010, 141, 243–254. [Google Scholar] [CrossRef]
- Bouwman, P.; Aly, A.; Escandell, J.M.; Pieterse, M.; Bartkova, J.; van der Gulden, H.; Hiddingh, S.; Thanasoula, M.; Kulkarni, A.; Yang, Q.; et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010, 17, 688–695. [Google Scholar] [CrossRef]
- Escribano-Díaz, C.; Orthwein, A.; Fradet-Turcotte, A.; Xing, M.; Young, J.T.; Tkáč, J.; Cook, M.A.; Rosebrock, A.P.; Munro, M.; Canny, M.D.; et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 2013, 49, 872–883. [Google Scholar] [CrossRef]
- De Gregoriis, G.; Ramos, J.A.; Fernandes, P.V.; Vignal, G.M.; Brianese, R.C.; Carraro, D.M.; Monteiro, A.N.; Struchiner, C.J.; Suarez-Kurtz, G.; Vianna-Jorge, R.; et al. DNA repair genes PAXIP1 and TP53BP1 expression is associated with breast cancer prognosis. Cancer Biol. Ther. 2017, 18, 439–449. [Google Scholar] [CrossRef]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Goncalves, A.; Lee, K.H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef]
- Robson, M.E.; Tung, N.; Conte, P.; Im, S.A.; Senkus, E.; Xu, B.; Masuda, N.; Delaloge, S.; Li, W.; Armstrong, A.; et al. OlympiAD final overall survival and tolerability results: Olaparib versus chemotherapy treatment of physician’s choice in patients with a germline BRCA mutation and HER2-negative metastatic breast cancer. Ann. Oncol. 2019, 30, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; O’Shaughnessy, J.; Loesch, D.; Blum, J.L.; Vahdat, L.T.; Petrakova, K.; Chollet, P.; Manikas, A.; Diéras, V.; Delozier, T.; et al. Eribulin monotherapy versus treatment of physician’s choice in patients with metastatic breast cancer (EMBRACE): A phase 3 open-label randomised study. Lancet 2011, 377, 914–923. [Google Scholar] [CrossRef]
- Pivot, X.; Marmé, F.; Koenigsberg, R.; Guo, M.; Berrak, E.; Wolfer, A. Pooled analyses of eribulin in metastatic breast cancer patients with at least one prior chemotherapy. Ann. Oncol. 2016, 27, 1525–1531. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, A.; Yonemori, K.; Yoshida, M.; Yoshida, T.; Yasojima, H.; Masuda, N.; Aogi, K.; Takahashi, M.; Naito, Y.; Shimizu, S.; et al. Gene Alterations in Triple-Negative Breast Cancer Patients in a Phase I/II Study of Eribulin and Olaparib Combination Therapy. Transl. Oncol. 2019, 12, 1386–1394. [Google Scholar] [CrossRef]
- Müller, V.; Riethdorf, S.; Rack, B.; Janni, W.; Fasching, P.A.; Solomayer, E.; Aktas, B.; Kasimir-Bauer, S.; Pantel, K.; Fehm, T. DETECT study group; Prognostic impact of circulating tumor cells assessed with the CellSearch SystemTM and AdnaTest BreastTM in metastatic breast cancer patients: The DETECT study. Breast Cancer Res. 2012, 14, R118. [Google Scholar]
- Bidard, F.C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Madic, J.; Kiialainen, A.; Bidard, F.C.; Birzele, F.; Ramey, G.; Leroy, Q.; Rio Frio, T.; Vaucher, I.; Raynal, V.; Bernard, V.; et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int. J. Cancer 2015, 136, 2158–2165. [Google Scholar] [CrossRef]
- Pierga, J.Y.; Hajage, D.; Bachelot, T.; Delaloge, S.; Brain, E.; Campone, M.; Diéras, V.; Rolland, E.; Mignot, L.; Mathiot, C.; et al. High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann. Oncol. 2012, 23, 618–624. [Google Scholar] [CrossRef]
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J. Clin. Oncol. 2014, 32, 3483–3489. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Pierga, J.Y.; Reuben, J.; Rademaker, A.; Davis, A.A.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit. Rev. Oncol. Hematol. 2019, 134, 39–45. [Google Scholar] [CrossRef]
- Banys-Paluchowski, M.; Krawczyk, N.; Fehm, T. Potential Role of Circulating Tumor Cell Detection and Monitoring in Breast Cancer: A Review of Current Evidence. Front. Oncol. 2016, 6, 255. [Google Scholar] [CrossRef] [PubMed]
- Schramm, A.; Friedl, T.W.; Schochter, F.; Scholz, C.; de Gregorio, N.; Huober, J.; Rack, B.; Trapp, E.; Alunni-Fabbroni, M.; Müller, V.; et al. Therapeutic intervention based on circulating tumor cell phenotype in metastatic breast cancer: Concept of the DETECT study program. Arch. Gynecol. Obstet. 2016, 293, 271–281. [Google Scholar] [CrossRef]
- Jaeger, B.A.S.; Neugebauer, J.; Andergassen, U.; Melcher, C.; Schochter, F.; Mouarrawy, D.; Ziemendorff, G.; Clemens, M.; V Abel, E.; Heinrich, G.; et al. The HER2 phenotype of circulating tumor cells in HER2-positive early breast cancer: A translational research project of a prospective randomized phase III trial. PLoS ONE 2017, 12, e0173593. [Google Scholar] [CrossRef]
- Obermeier, K.; Sachsenweger, J.; Friedl, T.W.P.; Pospiech, H.; Winqvist, R.; Wiesmüller, L. Heterozygous PALB2, c. 1592delT mutation channels DNA double-strand break repair into error-prone pathways in breast cancer patients. Oncogene 2016, 35, 3796–3806. [Google Scholar] [CrossRef] [PubMed]
- Grotsky, D.A.; Gonzalez-Suarez, I.; Novell, A.; Neumann, M.A.; Yaddanapudi, S.C.; Croke, M.; Martinez-Alonso, M.; Redwood, A.B.; Ortega-Martinez, S.; Feng, Z.; et al. BRCA1 loss activates cathepsin L-mediated degradation of 53BP1 in breast cancer cells. J. Cell Biol. 2013, 200, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Nagelkerke, A.; van Kuijk, S.J.; Sweep, F.C.; Nagtegaal, I.D.; Hoogerbrugge, N.; Martens, J.W.; Timmermans, M.A.; van Laarhoven, H.W.; Bussink, J.; Span, P.N. Constitutive expression of γ-H2AX has prognostic relevance in triple negative breast cancer. Radiother. Oncol. 2011, 101, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Caparica, R.; Lambertini, M.; de Azambuja, E. How I treat metastatic triple-negative breast cancer. ESMO Open 2019, 4 (Suppl. 2), e00050. [Google Scholar] [CrossRef] [PubMed]
- Le Du, F.; Eckhardt, B.L.; Lim, B.; Litton, J.K.; Moulder, S.; Meric-Bernstamm, F.; Gonzalez-Angulo, A.M.; Ueno, N.T. Is the future of personalized therapy in triple-negative breast cancer based on molecular subtype? Oncotarget 2015, 6, 12890–12908. [Google Scholar] [CrossRef]
- Yap, T.A.; Lorente, D.; Omlin, A.; Olmos, D.; de Bono, J.S. Circulating tumor cells: A multifunctional biomarker. Clin. Cancer Res. 2014, 20, 2553–2568. [Google Scholar] [CrossRef]
- Easton, D.F.; Pharoah, P.D.; Antoniou, A.C.; Tischkowitz, M.; Tavtigian, S.V.; Nathanson, K.L.; Devilee, P.; Meindl, A.; Couch, F.J.; Southey, M.; et al. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 2015, 372, 2243–2257. [Google Scholar] [CrossRef]
- Plo, I.; Laulie, C.; Gauthier, L.; Lebrun, F.; Calvo, F.; Lopez, B.S. AKT1 inhibits homologous recombination by inducing cytoplasmic retention of BRCA1 and RAD51. Cancer Res. 2009, 68, 9404–9412. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.L.; Adams, D.K.; He, J.; Kalhor, N.; Zhang, M.; Xu, T.; Gao, H.; Reuben, J.M.; Qiao, Y.; Komaki, R.; et al. Sequential Tracking of PD-L1 Expression and RAD50 Induction in Circulating Tumor and Stromal Cells of Lung Cancer Patients Undergoing Radiotherapy. Clin. Cancer Res. 2017, 23, 5948–5958. [Google Scholar] [CrossRef] [PubMed]
- Sedelnikova, O.A.; Bonner, W.M. GammaH2AX in cancer cells: A potential biomarker for cancer diagnostics, prediction and recurrence. Cell Cycle 2006, 5, 2909–2913. [Google Scholar]
- Cortes, J.; Schöffski, P.; Littlefield, B.A. Multiple modes of action of eribulin mesylate: Emerging data and clinical implications. Cancer Treat. Rev. 2018, 70, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Poruchynsky, M.S.; Komlodi-Pasztor, E.; Trostel, S.; Wilkerson, J.; Regairaz, M.; Pommier, Y.; Zhang, X.; Kumar Maity, T.; Robey, R.; Burotto, M.; et al. Microtubule-targeting agents augment the toxicity of DNA-damaging agents by disrupting intracellular trafficking of DNA repair proteins. Proc. Natl. Acad. Sci. USA 2015, 112, 1571–1576. [Google Scholar] [CrossRef]
- Lottersberger, F.; Karssemeijer, R.A.; Dimitrova, N.; de Lange, T. 53BP1 and the LINC Complex Promote Microtubule-Dependent DSB Mobility and DNA Repair. Cell 2015, 163, 880–893. [Google Scholar] [CrossRef]
- Linn, S.; Van ’t Veer, L. Clinical relevance of the triple-negative breast cancer concept: Genetic basis and clinical utility of the concept. Eur. J. Cancer 2009, 45 (Suppl. 1), 11–26. [Google Scholar] [CrossRef]
- Hassan, S.; Esch, A.; Liby, T.; Gray, J.W.; Heiser, L.M. Pathway-Enriched Gene Signature Associated with 53BP1 Response to PARP Inhibition in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2017, 16, 2892–2901. [Google Scholar] [CrossRef]
- Perez-Garcia, J.M.; Cortes, J. The safety of eribulin for the treatment of metastatic breast cancer. Exp. Opin. Drug Saf. 2019, 18, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Fritsche, H.; Müller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Jänicke, F.; et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the CellSearch system. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, T.M.; Riethdorf, S.; Nees, J.; Hartkopf, A.D.; Schönfisch, B.; Domschke, C.; Sprick, M.R.; Schütz, F.; Brucker, S.Y.; Stefanovic, S.; et al. Impact of apoptotic circulating tumor cells (aCTC) in metastatic breast cancer. Breast Cancer Res. Treat. 2016, 160, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Müller, V.; Zhang, L.; Rau, T.; Loibl, S.; Komor, M.; Roller, M.; Huober, J.; Fehm, T.; Schrader, I.; et al. Detection and HER2 expression of circulating tumor cells: Prospective monitoring in breast cancer patients treated in the neoadjuvant Gepar-Quattro trial. Clin. Cancer Res. 2010, 16, 2634–2645. [Google Scholar] [CrossRef] [PubMed]
- Polzer, B.; Medoro, G.; Pasch, S.; Fontana, F.; Zorzino, L.; Pestka, A.; Andergassen, U.; Meier-Stiegen, F.; Czyz, Z.T.; Alberter, B.; et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 2014, 6, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Concin, N.; Zeillinger, C.; Tong, D.; Stimpfl, M.; König, M.; Printz, D.; Stonek, F.; Schneeberger, C.; Hefler, L.; Kainz, C.; et al. Comparison of p53 mutational status with mRNA and protein expression in a panel of 24 human breast carcinoma cell lines. Breast Cancer Res. Treat. 2003, 79, 37–46. [Google Scholar] [CrossRef]
- Keimling, M.; Wiesmüller, L. DNA double-strand break repair activities in mammary epithelial cells--influence of endogenous p53 variants. Carcinogenesis 2009, 30, 1260–1268. [Google Scholar] [CrossRef]
- Ireno, I.C.; Wiehe, R.S.; Stahl, A.I.; Hampp, S.; Aydin, S.; Troester, M.A.; Selivanova, G.; Wiesmüller, L. Modulation of the poly (ADP-ribose) polymerase inhibitor response and DNA recombination in breast cancer cells by drugs affecting endogenous wild-type p53. Carcinogenesis 2014, 35, 2273–2282. [Google Scholar] [CrossRef]
- Deniz, M.; Kaufmann, J.; Stahl, A.; Gundelach, T.; Janni, W.; Hoffmann, I.; Keimling, M.; Hampp, S.; Ihle, M.; Wiesmüller, L. In vitro model for DNA double-strand break repair analysis in breast cancer reveals cell type-specific associations with age and prognosis. FASEB J. 2016, 30, 3786–3799. [Google Scholar] [CrossRef]
| Immunophenotype | Primary Tumor (N = 63) | Metastases (N = 43) | ||||
|---|---|---|---|---|---|---|
| Triple-Negative N (%) | Luminal-Like (HR+) N (%) | P-Value 2 | Triple-Negative N (%) | Luminal-Like (HR+) N (%) | P-Value 2 | |
| 15 (23.8%) | 48 (76.2%) | 14 (32.6%) | 29 (67.4%) | |||
| Age (years) | Median (Range) | 0.272 3 | Median (Range) | 0.613 3 | ||
| 65 (43–78) | 62 (34–81) | 64 (43–78) | 63 (39–75) | |||
| BMI (kg/m2) | Median (Range) | 0.116 3 | Median (Range) | 1.000 3 | ||
| 23.3 (18.0–39.7) | 26.4 (18.4–40.6) | 24.6 (18.6–39.7) | 24.0 (19.4–35.3) | |||
| ECOG | N (%) | 0.004 4 | N (%) | 0.035 4 | ||
| 0 | 4 (26.7%) | 35 (72.9%) | 5 (35.7%) | 22 (75.9%) | ||
| 1 | 9 (60.0%) | 12 (25.0%) | 7 (50.0%) | 6 (20.7%) | ||
| 2 | 2 (13.3%) | 1 (2.1%) | 2 (14.3%) | 1 (3.4%) | ||
| Histotype | N (%) | 0.549 4 | N (%) | 0.537 4 | ||
| Ductal | 11 (73.3%) | 37 (77.1%) | 9 (64.3%) | 23 (79.3%) | ||
| Lobular | 1 (6.7%) | 6 (12.5%) | 2 (14.3%) | 3 (10.3%) | ||
| Mixed and Other | 3 (20.0%) | 5 (10.4%) | 3 (21.4%) | 3 (10.3%) | ||
| Grading | N (%) | 0.004 4 | N (%) | 0.041 4 | ||
| 1 | 0 (0.0%) | 1 (2.1%) | 0 (0.0%) | 0 (0.0%) | ||
| 2 | 4 (26.7%) | 31 (64.6%) | 4 (28.6%) | 17 (58.6%) | ||
| 3 | 11 (73.3%) | 11 (22.9%) | 9 (64.3%) | 9 (31.0%) | ||
| Unknown | 0 (0.0%) | 5 (10.4%) | 1 (7.1%) | 3 (10.3%) | ||
| Metastatic site—locally advanced 1 | N (%) | 0.028 5 | N (%) | 0.252 5 | ||
| Yes | 6 (40.0%) | 6 (12.5%) | 5 (35.7%) | 5 (17.2%) | ||
| No | 9 (60.0%) | 42 (87.5%) | 9 (64.3%) | 24 (82.8%) | ||
| Metastatic site—bone 1 | N (%) | 0.012 4 | N (%) | 0.507 5 | ||
| Yes | 6 (40.0%) | 36 (75.0%) | 8 (57.1%) | 20 (69.0%) | ||
| No | 9 (60.0%) | 12 (25.0%) | 6 (42.9%) | 9 (31.0%) | ||
| Metastatic site—visceral 1 | N (%) | 0.291 5 | N (%) | 0.055 5 | ||
| Yes | 10 (66.7%) | 39 (81.3%) | 8 (57.1%) | 25 (86.2%) | ||
| No | 5 (33.3%) | 9 (18.8%) | 6 (42.9%) | 4 (13.8%) | ||
| Metastatic site—CNS 1 | N (%) | 0.039 5 | N (%) | 0.094 5 | ||
| Yes | 3 (20.0%) | 1 (2.1%) | 3 (21.4%) | 1 (3.4%) | ||
| No | 12 (80.0%) | 47 (97.9%) | 11 (78.6%) | 28 (96.6%) | ||
| Line of chemotherapeutical treatment (in metastatic setting) | N (%) | 0.741 4 | N (%) | 0.899 4 | ||
| 1 | 6 (40.0%) | 27 (56.3%) | 8 (57.1%) | 15 (51.7%) | ||
| 2 | 5 (33.3%) | 13 (27.1%) | 4 (28.6%) | 8 (27.6%) | ||
| 3 or more | 3 (20.0%) | 8 (16.7%) | 2 (14.3%) | 6 (20.7%) | ||
| Unknown | 1 (6.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schochter, F.; Werner, K.; Köstler, C.; Faul, A.; Tzschaschel, M.; Alberter, B.; Müller, V.; Neubauer, H.; Fehm, T.; Friedl, T.W.P.; et al. 53BP1 Accumulation in Circulating Tumor Cells Identifies Chemotherapy-Responsive Metastatic Breast Cancer Patients. Cancers 2020, 12, 930. https://doi.org/10.3390/cancers12040930
Schochter F, Werner K, Köstler C, Faul A, Tzschaschel M, Alberter B, Müller V, Neubauer H, Fehm T, Friedl TWP, et al. 53BP1 Accumulation in Circulating Tumor Cells Identifies Chemotherapy-Responsive Metastatic Breast Cancer Patients. Cancers. 2020; 12(4):930. https://doi.org/10.3390/cancers12040930
Chicago/Turabian StyleSchochter, Fabienne, Kim Werner, Cäcilia Köstler, Anke Faul, Marie Tzschaschel, Barbara Alberter, Volkmar Müller, Hans Neubauer, Tanja Fehm, Thomas W.P. Friedl, and et al. 2020. "53BP1 Accumulation in Circulating Tumor Cells Identifies Chemotherapy-Responsive Metastatic Breast Cancer Patients" Cancers 12, no. 4: 930. https://doi.org/10.3390/cancers12040930
APA StyleSchochter, F., Werner, K., Köstler, C., Faul, A., Tzschaschel, M., Alberter, B., Müller, V., Neubauer, H., Fehm, T., Friedl, T. W. P., Polzer, B., Janni, W., Rack, B., & Wiesmüller, L. (2020). 53BP1 Accumulation in Circulating Tumor Cells Identifies Chemotherapy-Responsive Metastatic Breast Cancer Patients. Cancers, 12(4), 930. https://doi.org/10.3390/cancers12040930






