A Novel Role for the Tumor Suppressor Gene ITF2 in Tumorigenesis and Chemotherapy Response
Abstract
1. Introduction
2. Results
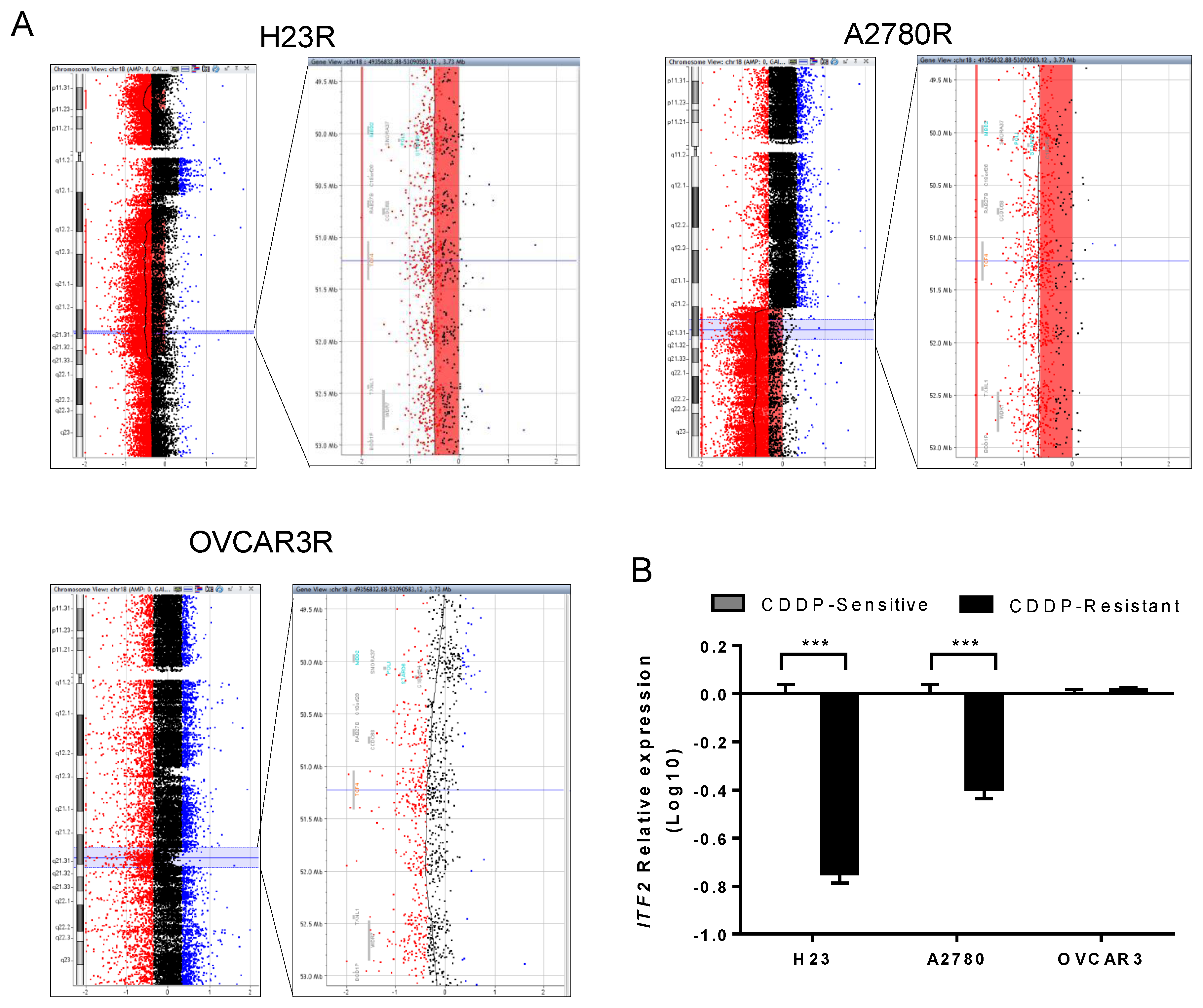
2.1. ITF2 is Frequently Downregulated by Chromosomal Deletion after CDDP Cell Exposure
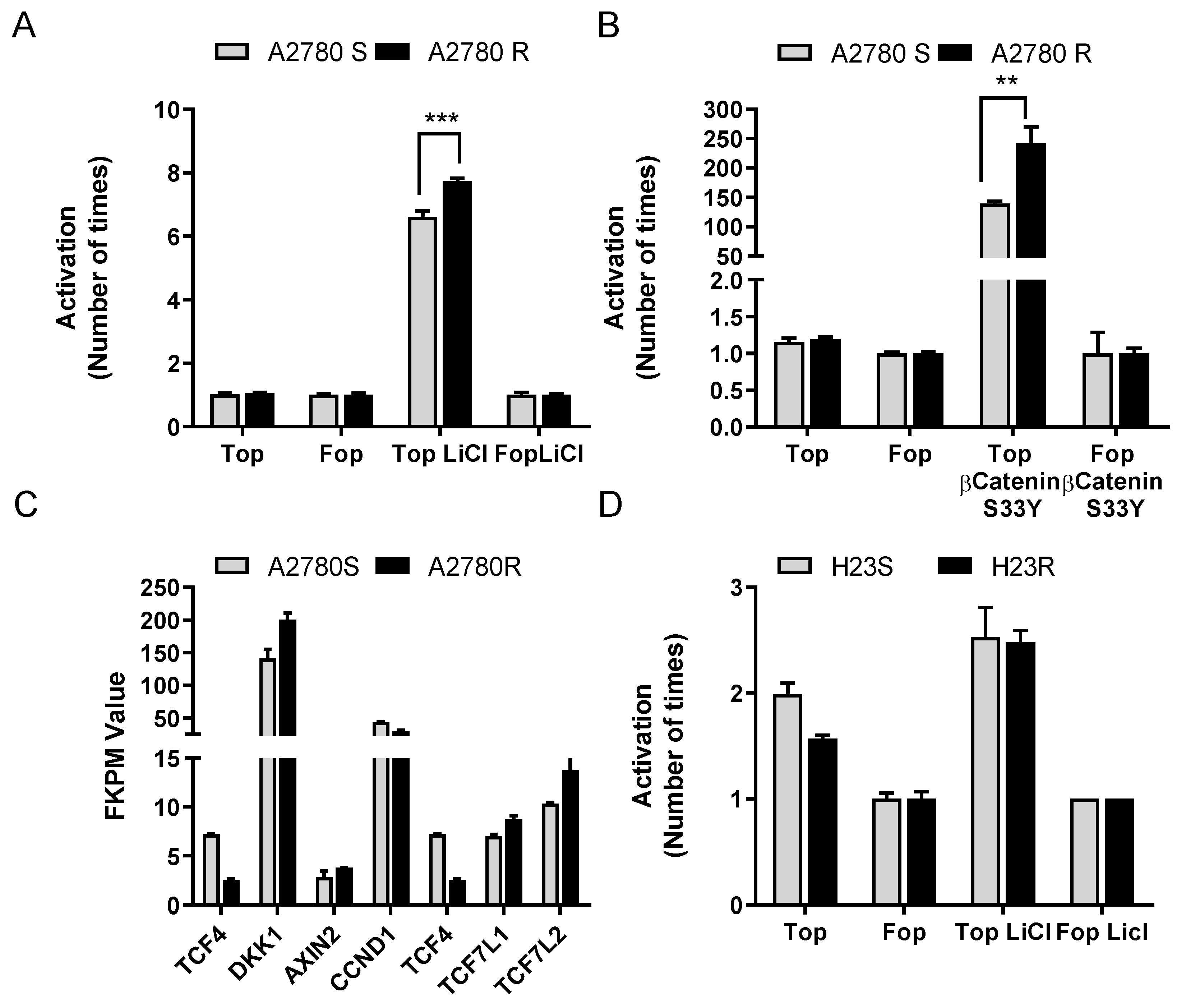
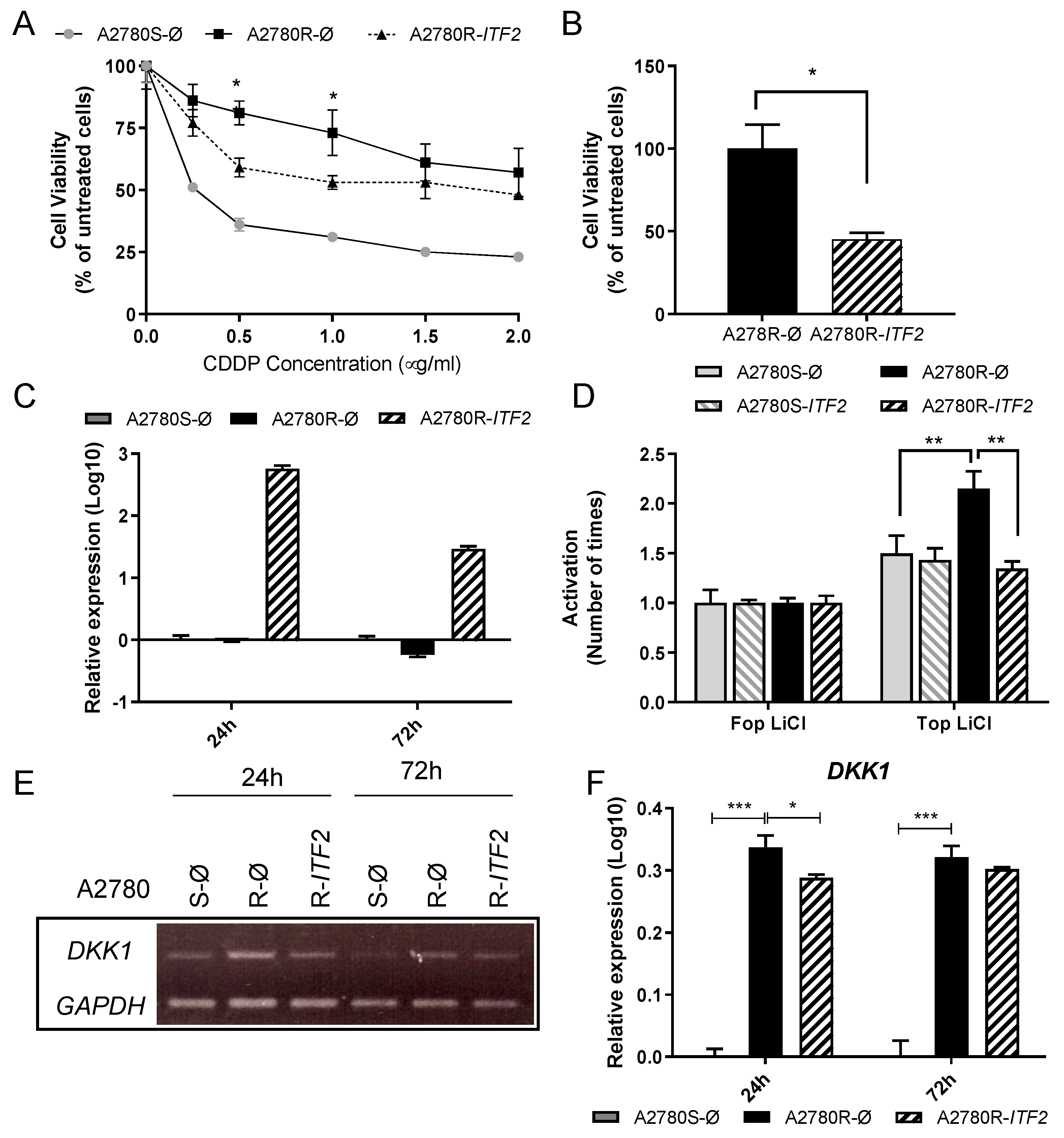
2.2. Restoration of ITF2 Increases the Sensitivity to CDDP and Decreases β-catenin/TCF Transcriptional Activity
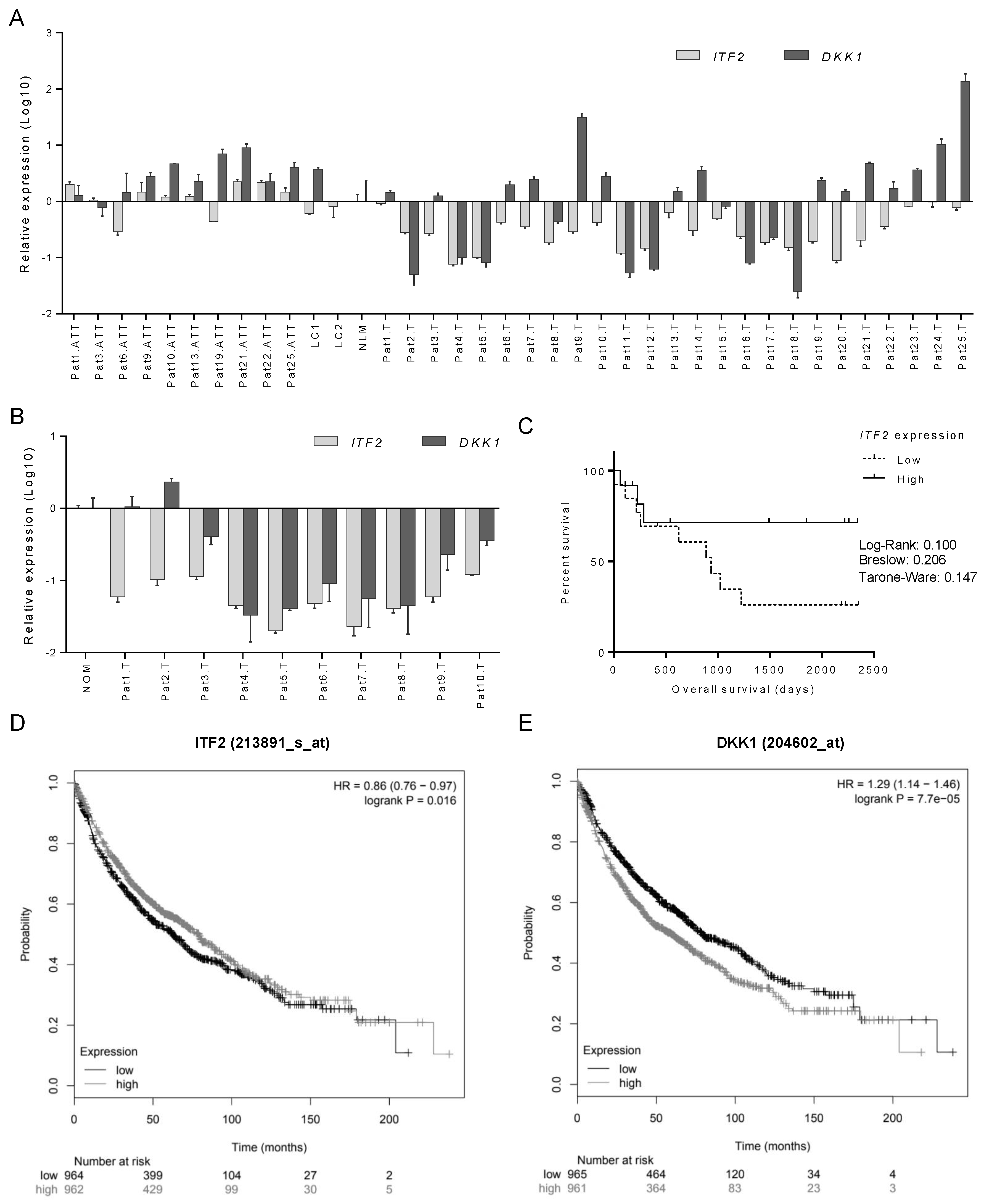
2.3. The Expression of ITF2 is Frequently Downregulated in NSCLC, Ovarian and Other Epithelial Tumors
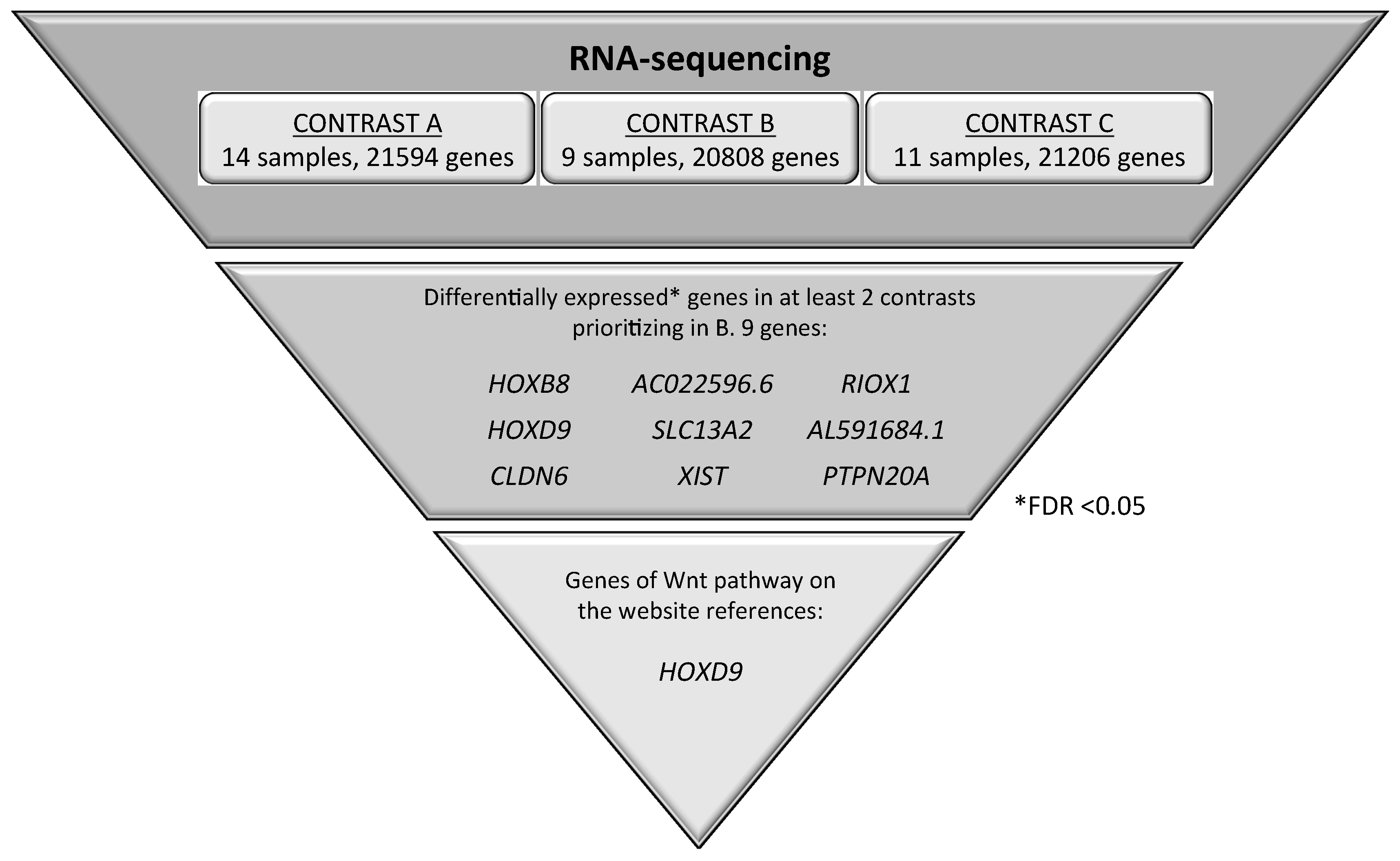
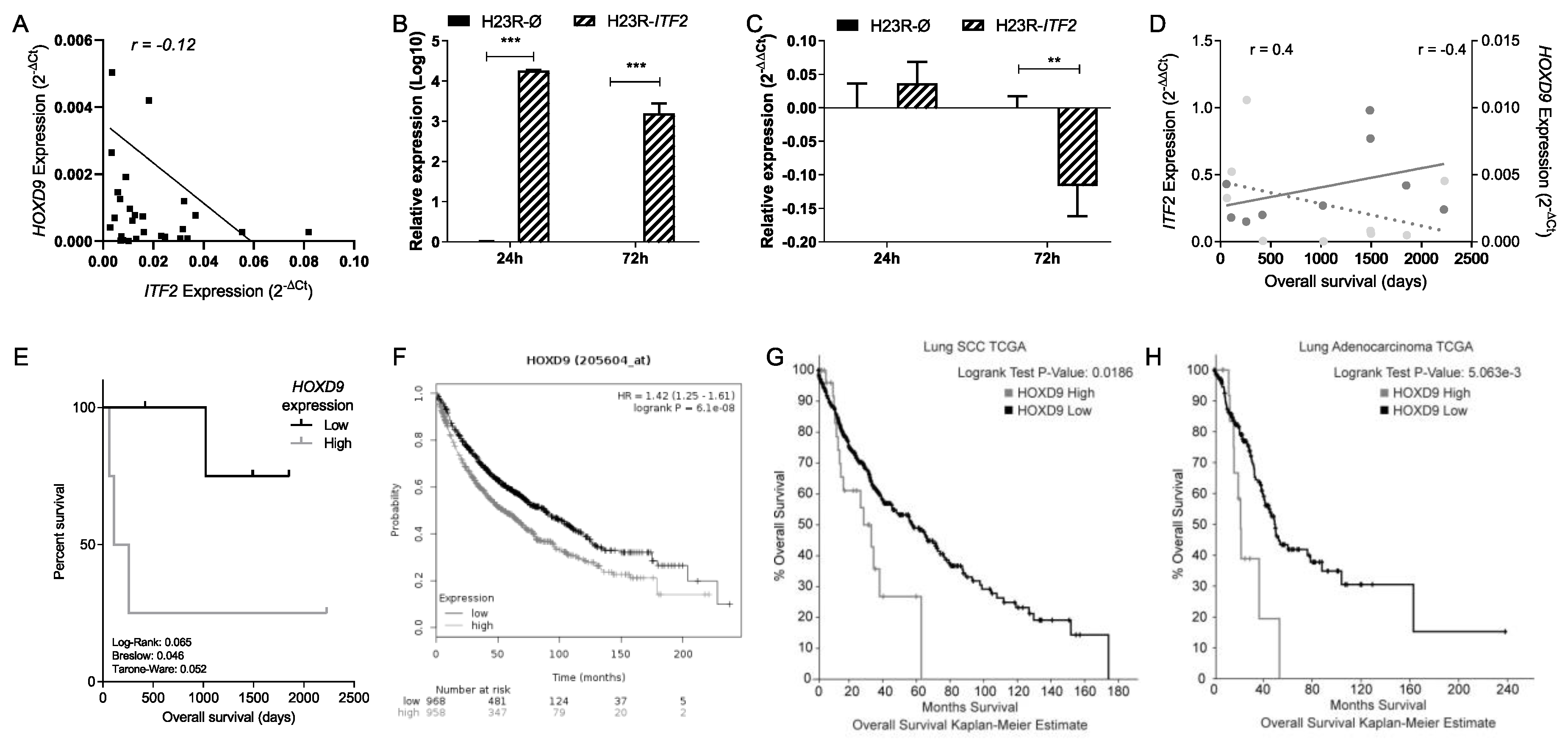
2.4. Identification of Candidate Genes Involved in the Wnt Signaling Pathway through the Analysis of RNA-seq in NSCLC Patients
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Cell-Viability Assays
4.2. Clinical Sample and Data Collection
4.3. DNA Extraction and Array of Comparative Genome Hybridization
4.4. RNA Extraction, RT-PCR, qRT-PCR
4.5. NGS (RNA-seq) and Wnt Signaling Pathways Analysis
4.6. Transfection Assays: Top-fop, B-cat and TCF4 Overexpression
4.7. In Silico Databases: The Cancer Genome Atlas and Kaplan–Meier Plotter
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, S.Y.; Jung, D.K.; Choi, J.E.; Jin, C.C.; Hong, M.J.; Do, S.K.; Kang, H.G.; Lee, W.K.; Seok, Y.; Lee, E.B.; et al. Pd-L1 Polymorphism Can Predict Clinical Outcomes of Non-Small Cell Lung Cancer Patients Treated with First-Line Paclitaxel-Cisplatin Chemotherapy. Sci. Rep. 2016, 6, 25952. [Google Scholar] [CrossRef] [PubMed]
- French, J.D.; Johnatty, S.E.; Lu, Y.; Beesley, J.; Gao, B.; Kalimutho, M.; Henderson, M.J.; Russell, A.J.; Kar, S.; Chen, X.; et al. Germline Polymorphisms in an Enhancer of Psip1 Are Associated with Progression-Free Survival in Epithelial Ovarian Cancer. Oncotarget 2016, 7, 6353–6368. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global Cancer Statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Haslehurst, A.M.; Koti, M.; Dharsee, M.; Nuin, P.; Evans, K.; Geraci, J.; Childs, T.; Chen, J.; Li, J.; Weberpals, J.; et al. Emt Transcription Factors Snail and Slug Directly Contribute to Cisplatin Resistance in Ovarian Cancer. BMC Cancer 2012, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Karaca, B.; Atmaca, H.; Bozkurt, E.; Kisim, A.; Uzunoglu, S.; Karabulut, B.; Sezgin, C.; Sanli, U.A.; Uslu, R. Combination of at-101/Cisplatin Overcomes Chemoresistance by Inducing Apoptosis and Modulating Epigenetics in Human Ovarian Cancer Cells. Mol. Biol. Rep. 2013, 40, 3925–3933. [Google Scholar] [CrossRef] [PubMed]
- Akervall, J.; Guo, X.; Qian, C.N.; Schoumans, J.; Leeser, B.; Kort, E.; Cole, A.; Resau, J.; Bradford, C.; Carey, T.; et al. Genetic and Expression Profiles of Squamous Cell Carcinoma of the Head and Neck Correlate with Cisplatin Sensitivity and Resistance in Cell Lines and Patients. Clin. Cancer Res. 2004, 10, 8204–8213. [Google Scholar]
- Hiorns, L.R.; Seckl, M.J.; Paradinas, F.; Sharp, S.Y.; Skelton, L.A.; Brunstrom, G.; Newlands, E.S.; Kelland, L.R.; Leyland-Jones, B. A Molecular Cytogenetic Approach to Studying Platinum Resistance. J. Inorg. Biochem. 1999, 77, 95–104. [Google Scholar] [CrossRef]
- Leyland-Jones, B.; Kelland, L.R.; Harrap, K.R.; Hiorns, L.R. Genomic Imbalances Associated with Acquired Resistance to Platinum Analogues. Am. J. Pathol. 1999, 155, 77–84. [Google Scholar] [CrossRef][Green Version]
- Yasui, K.; Mihara, S.; Zhao, C.; Okamoto, H.; Saito-Ohara, F.; Tomida, A.; Funato, T.; Yokomizo, A.; Naito, S.; Imoto, I.; et al. Alteration in Copy Numbers of Genes as a Mechanism for Acquired Drug Resistance. Cancer Res. 2004, 64, 1403–1410. [Google Scholar] [CrossRef]
- Vera, O.; Jimenez, J.; Pernia, O.; Rodriguez-Antolin, C.; Rodriguez, C.; Sanchez Cabo, F.; Soto, J.; Rosas, R.; Lopez-Magallon, S.; Esteban Rodriguez, I.; et al. DNA Methylation of Mir-7 Is a Mechanism Involved in Platinum Response through Mafg Overexpression in Cancer Cells. Theranostics 2017, 7, 4118–4134. [Google Scholar] [CrossRef]
- Kolligs, F.T.; Nieman, M.T.; Winer, I.; Hu, G.; Van Mater, D.; Feng, Y.; Smith, I.M.; Wu, R.; Zhai, Y.; Cho, K.R.; et al. Itf-2, a Downstream Target of the Wnt/Tcf Pathway, Is Activated in Human Cancers with Beta-Catenin Defects and Promotes Neoplastic Transformation. Cancer Cell 2002, 1, 145–155. [Google Scholar]
- Shin, H.W.; Choi, H.; So, D.; Kim, Y.I.; Cho, K.; Chung, H.J.; Lee, K.H.; Chun, Y.S.; Cho, C.H.; Kang, G.H.; et al. Itf2 Prevents Activation of the Beta-Catenin-Tcf4 Complex in Colon Cancer Cells and Levels Decrease with Tumor Progression. Gastroenterology 2014, 147, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Chikazawa, N.; Tanaka, H.; Tasaka, T.; Nakamura, M.; Tanaka, M.; Onishi, H.; Katano, M. Inhibition of Wnt Signaling Pathway Decreases Chemotherapy-Resistant Side-Population Colon Cancer Cells. Anticancer Res. 2010, 30, 2041–2048. [Google Scholar] [PubMed]
- Nagaraj, A.B.; Joseph, P.; Kovalenko, O.; Singh, S.; Armstrong, A.; Redline, R.; Resnick, K.; Zanotti, K.; Waggoner, S.; DiFeo, A. Critical Role of Wnt/Beta-Catenin Signaling in Driving Epithelial Ovarian Cancer Platinum Resistance. Oncotarget 2015, 6, 23720–23734. [Google Scholar] [CrossRef]
- Zhang, Q.; Meng, X.K.; Wang, W.X.; Zhang, R.M.; Zhang, T.; Ren, J.J. The Wnt/Beta-Catenin Signaling Pathway Mechanism for Pancreatic Cancer Chemoresistance in a Three-Dimensional Cancer Microenvironment. Am. J. Transl. Res. 2016, 8, 4490–4498. [Google Scholar]
- Bosquet, J.G.; Marchion, D.C.; Chon, H.; Lancaster, J.M.; Chanock, S. Analysis of Chemotherapeutic Response in Ovarian Cancers Using Publicly Available High-Throughput Data. Cancer Res. 2014, 74, 3902–3912. [Google Scholar] [CrossRef]
- Kohno, T.; Otsuka, A.; Girard, L.; Sato, M.; Iwakawa, R.; Ogiwara, H.; Sanchez-Cespedes, M.; Minna, J.D.; Yokota, J. A Catalog of Genes Homozygously Deleted in Human Lung Cancer and the Candidacy of Ptprd as a Tumor Suppressor Gene. Genes Chromosom. Cancer 2010, 49, 342–352. [Google Scholar]
- Cho, E.K. Array-Based Comparative Genomic Hybridization and Its Application to Cancer Genomes and Human Genetics. J. Lung Cancer 2011, 10, 77–86. [Google Scholar] [CrossRef]
- Brown, J.; Bothma, H.; Veale, R.; Willem, P. Genomic Imbalances in Esophageal Carcinoma Cell Lines Involve Wnt Pathway Genes. World J. Gastroenterol. 2011, 17, 2909–2923. [Google Scholar] [CrossRef]
- Ni, S.; Hu, J.; Duan, Y.; Shi, S.; Li, R.; Wu, H.; Qu, Y.; Li, Y. Down Expression of Lrp1b Promotes Cell Migration Via Rhoa/Cdc42 Pathway and Actin Cytoskeleton Remodeling in Renal Cell Cancer. Cancer Sci. 2013, 104, 817–825. [Google Scholar] [CrossRef]
- Liu, C.X.; Musco, S.; Lisitsina, N.M.; Forgacs, E.; Minna, J.D.; Lisitsyn, N.A. Lrp-Dit, a Putative Endocytic Receptor Gene, Is Frequently Inactivated in Non-Small Cell Lung Cancer Cell Lines. Cancer Res. 2000, 60, 1961–1967. [Google Scholar] [PubMed]
- Ruiz de Garibay, G.; Mateo, F.; Stradella, A.; Valdes-Mas, R.; Palomero, L.; Serra-Musach, J.; Puente, D.A.; Diaz-Navarro, A.; Vargas-Parra, G.; Tornero, E.; et al. Tumor Xenograft Modeling Identifies an Association between Tcf4 Loss and Breast Cancer Chemoresistance. Dis. Model. Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Murre, C. Helix-Loop-Helix Proteins and Lymphocyte Development. Nat. Immunol. 2005, 6, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Sepp, M.; Kannike, K.; Eesmaa, A.; Urb, M.; Timmusk, T. Functional Diversity of Human Basic Helix-Loop-Helix Transcription Factor Tcf4 Isoforms Generated by Alternative 5’ Exon Usage and Splicing. PLoS ONE 2011, 7, e22138. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, K.; Pedroso, I.; De Jong, S.; Stefansson, H.; Steinberg, S.; Stefansson, K.; Ophoff, R.A.; Schalkwyk, L.C.; Collier, D.A. Tcf4 (E2-2; Itf2): A Schizophrenia-Associated Gene with Pleiotropic Effects on Human Disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013, 162B, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Akiri, G.; Cherian, M.M.; Vijayakumar, S.; Liu, G.; Bafico, A.; Aaronson, S.A. Wnt Pathway Aberrations Including Autocrine Wnt Activation Occur at High Frequency in Human Non-Small-Cell Lung Carcinoma. Oncogene 2009, 28, 2163–2172. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. Rna-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Katz, Y.; Wang, E.T.; Airoldi, E.M.; Burge, C.B. Analysis and Design of Rna Sequencing Experiments for Identifying Isoform Regulation. Nat. Methods 2010, 7, 1009–1015. [Google Scholar] [CrossRef]
- Plowright, L.; Harrington, K.J.; Pandha, H.S.; Morgan, R. Hox Transcription Factors Are Potential Therapeutic Targets in Non-Small-Cell Lung Cancer (Targeting Hox Genes in Lung Cancer). Br. J. Cancer 2009, 100, 470–475. [Google Scholar] [CrossRef]
- Tabuse, M.; Ohta, S.; Ohashi, Y.; Fukaya, R.; Misawa, A.; Yoshida, K.; Kawase, T.; Saya, H.; Thirant, C.; Chneiweiss, H.; et al. Functional Analysis of Hoxd9 in Human Gliomas and Glioma Cancer Stem Cells. Mol. Cancer 2011, 10, 60. [Google Scholar] [CrossRef]
- Lv, X.; Li, L.; Lv, L.; Qu, X.; Jin, S.; Li, K.; Deng, X.; Cheng, L.; He, H.; Dong, L. Hoxd9 Promotes Epithelial-Mesenchymal Transition and Cancer Metastasis by Zeb1 Regulation in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 133. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human Hox Loci by Noncoding Rnas. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.J.; Kingston, R.E. Hotair Lifts Noncoding Rnas to New Levels. Cell 2007, 129, 1257–1279. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, M.; Lu, K.; Liu, J.; Zhang, M.; Wu, W.; De, W.; Wang, Z.; Wang, R. The Long Noncoding Rna Hotair Contributes to Cisplatin Resistance of Human Lung Adenocarcinoma Cells Via Downregualtion of P21(Waf1/Cip1) Expression. PLoS ONE 2013, 8, e77293. [Google Scholar]
- Ibanez de Caceres, I.; Cortes-Sempere, M.; Moratilla, C.; Machado-Pinilla, R.; Rodriguez-Fanjul, V.; Manguan-Garcia, C.; Cejas, P.; Lopez-Rios, F.; Paz-Ares, L.; de CastroCarpeno, J.; et al. Igfbp-3 Hypermethylation-Derived Deficiency Mediates Cisplatin Resistance in Non-Small-Cell Lung Cancer. Oncogene 2010, 29, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Machado-Pinilla, R.; Manguan-Garcia, C.; Belda-Iniesta, C.; Moratilla, C.; Cejas, P.; Fresno-Vara, J.A.; de Castro-Carpeno, J.; Casado, E.; Nistal, M.; et al. Mkp1/Cl100 Controls Tumor Growth and Sensitivity to Cisplatin in Non-Small-Cell Lung Cancer. Oncogene 2006, 25, 3335–3345. [Google Scholar] [CrossRef] [PubMed]
- Ibanez de Caceres, I.; Dulaimi, E.; Hoffman, A.M.; Al-Saleem, T.; Uzzo, R.G.; Cairns, P. Identification of Novel Target Genes by an Epigenetic Reactivation Screen of Renal Cancer. Cancer Res. 2006, 66, 5021–5028. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. Rsem: Accurate Transcript Quantification from Rna-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A Scaling Normalization Method for Differential Expression Analysis of Rna-Seq Data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Gyorffy, B.; Surowiak, P.; Budczies, J.; Lanczky, A. Online Survival Analysis Software to Assess the Prognostic Value of Biomarkers Using Transcriptomic Data in Non-Small-Cell Lung Cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef] [PubMed]
| Patient | Histology | Sex | Stage | Chemotherapy | Relapse | Status | OS (days) | PFS (days) | TCF4 | DKK1 | HOXD9 | RIOX1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (2−ΔCt) | (2−ΔCt) | (2−ΔCt) | (2−ΔCt) | |||||||||
| Pat1.T | Adenocarcinoma | Female | IA | No | Yes | Alive | 2220 | 1490 | 0.03364 | 0.00089 | 0.00008 | 0.00872 |
| Pat2.T | Adenocarcinoma | Male | NA | No | Yes | Alive | 2352 | 1860 | 0.01041 | 0.00003 | 0.04277 | 0.01605 |
| Pat3.T | Epidermoid | Male | IB | No | Yes | Exitus | 1022 | 825 | 0.01012 | 0.00075 | 0.00001 | 0.00114 |
| Pat4.T | Adenocarcinoma | Male | IB | No | No | Exitus | 3 | 3 | 0.00286 | 0.00006 | 0.00041 | 0.00624 |
| Pat5.T | Adenocarcinoma | Male | NA | No | No | Exitus | 626 | 626 | 0.00365 | 0.00005 | 0.00503 | 0.00932 |
| Pat6.T | Large cell | Male | IIB | No | No | Exitus | 62 | 62 | 0.01584 | 0.00119 | 0.00074 | 0.00216 |
| Pat7.T | Adenocarcinoma | Male | IIIA | Otro | Yes | Exitus | 228 | 138 | 0.01278 | 0.00150 | 0.00078 | 0.12070 |
| Pat8.T | Epidermoid | Female | IIIB | CDDP + Others | No | Exitus | 109 | 109 | 0.00675 | 0.00026 | 0.00126 | 0.00196 |
| Pat9.T | Adenocarcinoma | Female | IIA | CDDP + Others | Yes | Alive | 2260 | 2260 | 0.01070 | 0.01826 | 0.00097 | 0.04697 |
| Pat10.T | Epidermoid | Male | IB | No | No | Alive | 1853 | 1853 | 0.01626 | 0.00162 | 0.00027 | 0.00723 |
| Pat11.T | Adenocarcinoma | Male | IA | No | No | Exitus | 216 | 216 | 0.00456 | 0.00003 | 0.00070 | 0.01399 |
| Pat12.T | Adenocarcinoma | Female | IIIA | CBDCA + Others | Yes | Alive | 2192 | 2192 | 0.00580 | 0.00004 | 0.00146 | 0.01059 |
| Pat13.T | Epidermoid | Male | IB | CDDP + Others | No | Alive | 2341 | 2341 | 0.02466 | 0.00090 | 0.00013 | 0.14752 |
| Pat14.T | Epidermoid | Male | IIA | No | ND | Exitus | 289 | 289 | 0.01179 | 0.00206 | 0.00061 | 0.02547 |
| Pat15.T | Epidermoid | Male | IIA | No | ND | ND | 109 | 109 | 0.01833 | 0.01370 | 0.00420 | 0.07619 |
| Pat16.T | Adenocarcinoma | Female | IIIA | CDDP + Others | ND | Alive | 2228 | 2228 | 0.00907 | 0.00005 | 0.00191 | 0.00814 |
| Pat17.T | Adenocarcinoma | Male | IIB | Otro | Yes | Exitus | 888 | 443 | 0.00704 | 0.00014 | 0.00002 | 0.00833 |
| Pat18.T | Epidermoid | Male | IIB | CBDCA + Others | No | Exitus | 259 | 259 | 0.00563 | 0.00001 | 0.01455 | |
| Pat19.T | Adenocarcinoma | Female | IB | CDDP + Others | Yes | Exitus | 936 | 428 | 0.00738 | 0.00137 | 0.00014 | 0.01572 |
| Pat20.T | Epidermoid | Male | IIB | CDDP + Others | Yes | Exitus | 1224 | 637 | 0.00346 | 0.00089 | 0.00264 | 0.00422 |
| Pat21.T | Adenocarcinoma | Male | IIIA | CDDP + Others | No | ND | 421 | 421 | 0.00792 | 0.00279 | 0.00003 | 0.00509 |
| Pat22.T | Adenocarcinoma | Female | IIB | CDDP + Others | ND | ND | 184 | 184 | 0.01319 | 0.00101 | 0.00007 | 0.01480 |
| Pat23.T | Epidermoid | Male | IIIA | CDDP + Others | ND | ND | 542 | 542 | 0.03221 | 0.00214 | 0.00120 | 0.01741 |
| Pat24.T | Adenocarcinoma | Male | IIA | CBDCA + Others | No | Alive | 1491 | 1491 | 0.03680 | 0.00614 | 0.00077 | 0.01088 |
| Pat25.T | Adenocarcinoma | Male | IIA | CDDP + Others | No | Alive | 1496 | 1496 | 0.00008 | 0.00872 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pernía, O.; Sastre-Perona, A.; Rodriguez-Antolín, C.; García-Guede, A.; Palomares-Bralo, M.; Rosas, R.; Sanchez-Cabrero, D.; Cruz, P.; Rodriguez, C.; Diestro, M.; et al. A Novel Role for the Tumor Suppressor Gene ITF2 in Tumorigenesis and Chemotherapy Response. Cancers 2020, 12, 786. https://doi.org/10.3390/cancers12040786
Pernía O, Sastre-Perona A, Rodriguez-Antolín C, García-Guede A, Palomares-Bralo M, Rosas R, Sanchez-Cabrero D, Cruz P, Rodriguez C, Diestro M, et al. A Novel Role for the Tumor Suppressor Gene ITF2 in Tumorigenesis and Chemotherapy Response. Cancers. 2020; 12(4):786. https://doi.org/10.3390/cancers12040786
Chicago/Turabian StylePernía, Olga, Ana Sastre-Perona, Carlos Rodriguez-Antolín, Alvaro García-Guede, María Palomares-Bralo, Rocío Rosas, Darío Sanchez-Cabrero, Patricia Cruz, Carmen Rodriguez, MDolores Diestro, and et al. 2020. "A Novel Role for the Tumor Suppressor Gene ITF2 in Tumorigenesis and Chemotherapy Response" Cancers 12, no. 4: 786. https://doi.org/10.3390/cancers12040786
APA StylePernía, O., Sastre-Perona, A., Rodriguez-Antolín, C., García-Guede, A., Palomares-Bralo, M., Rosas, R., Sanchez-Cabrero, D., Cruz, P., Rodriguez, C., Diestro, M., Martín-Arenas, R., Pulido, V., Santisteban, P., de Castro, J., Vera, O., & Ibáñez de Cáceres, I. (2020). A Novel Role for the Tumor Suppressor Gene ITF2 in Tumorigenesis and Chemotherapy Response. Cancers, 12(4), 786. https://doi.org/10.3390/cancers12040786





