DNA Damage/Repair Management in Cancers
Abstract
1. Introduction
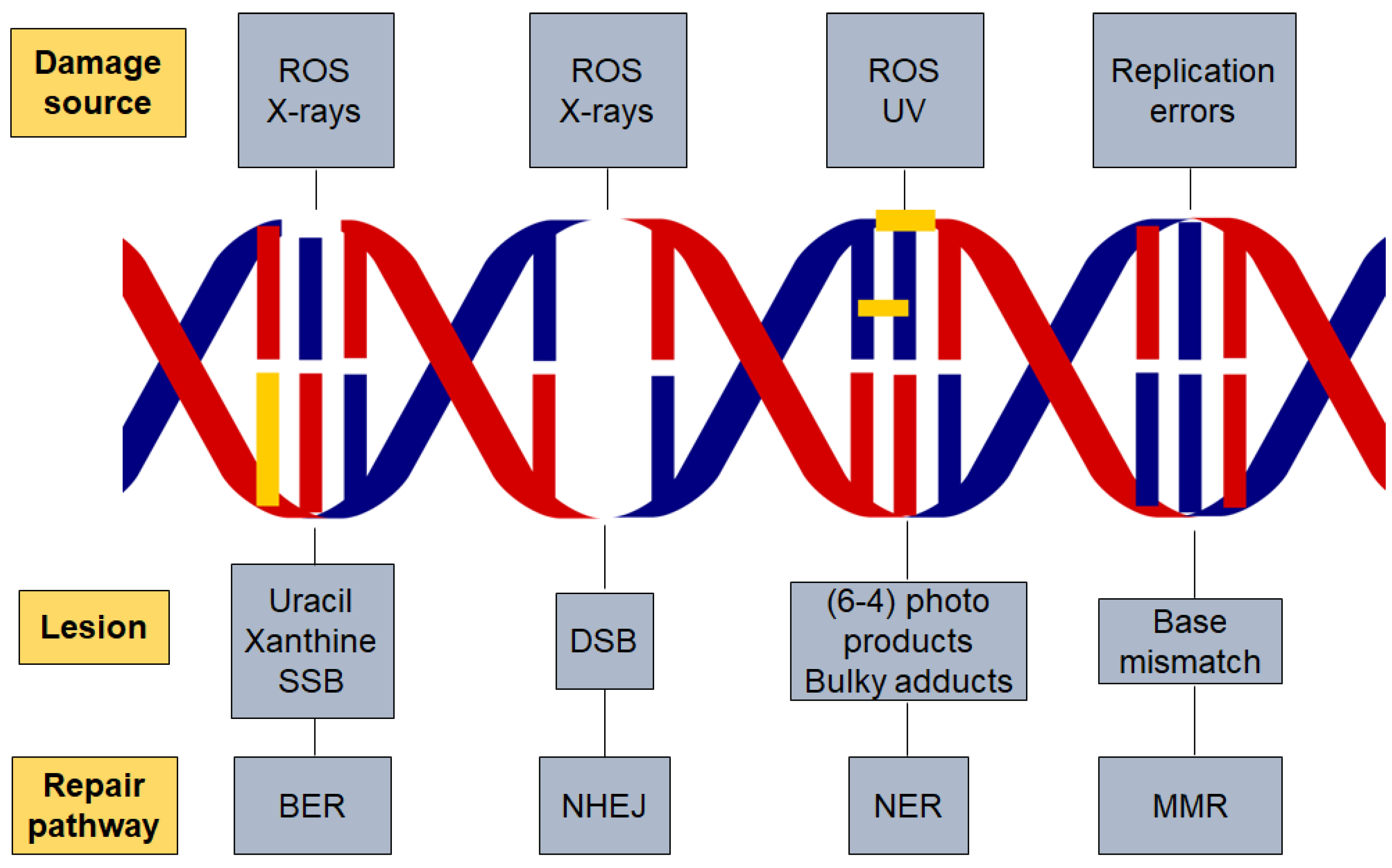
2. Types of DNA Damage
3. DNA Damage Response
4. Components of the DNA Damage Response
5. DDR and Disease Treatment
6. DNA Repair Pathways
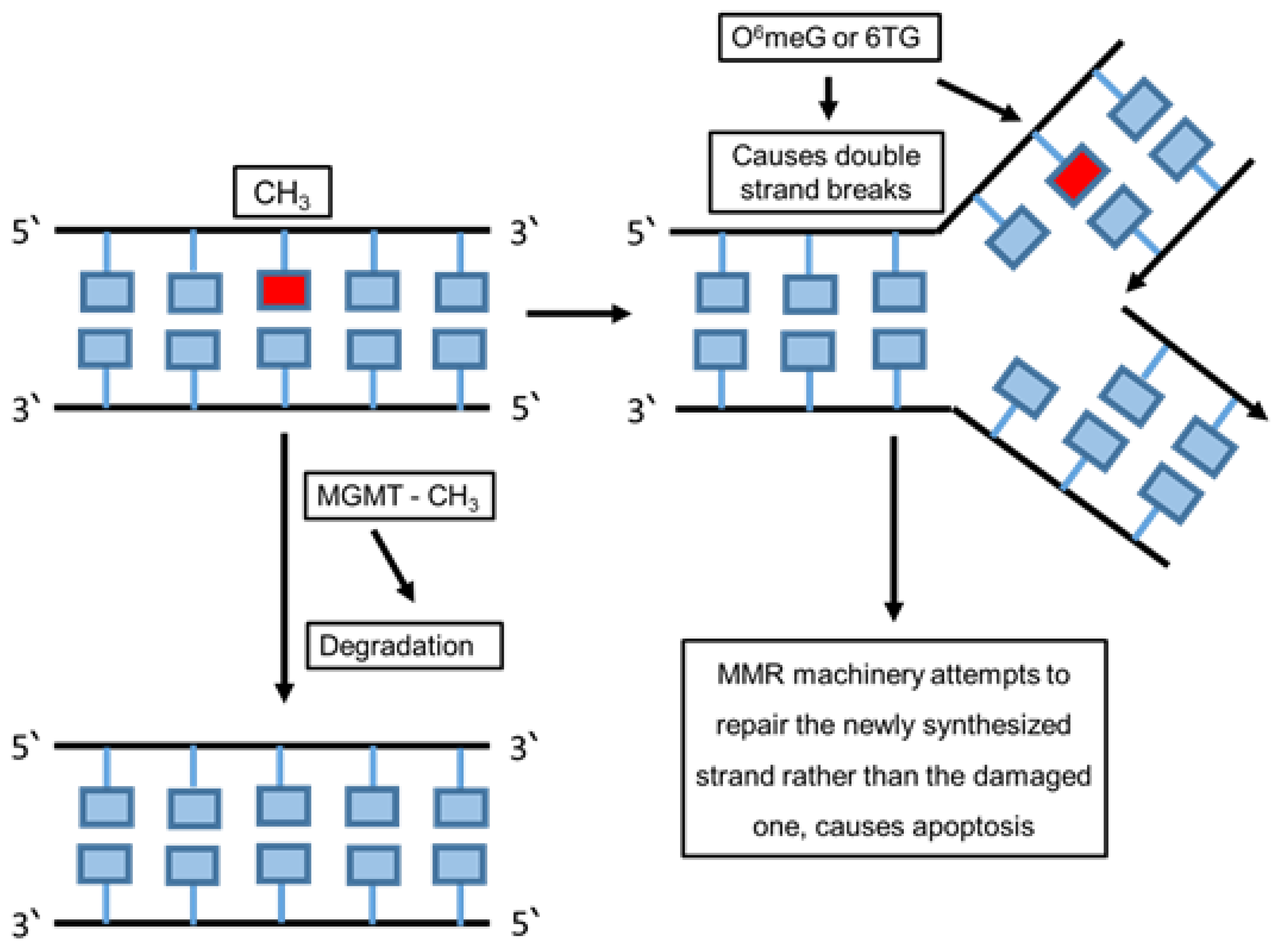
6.1. Direct Repair
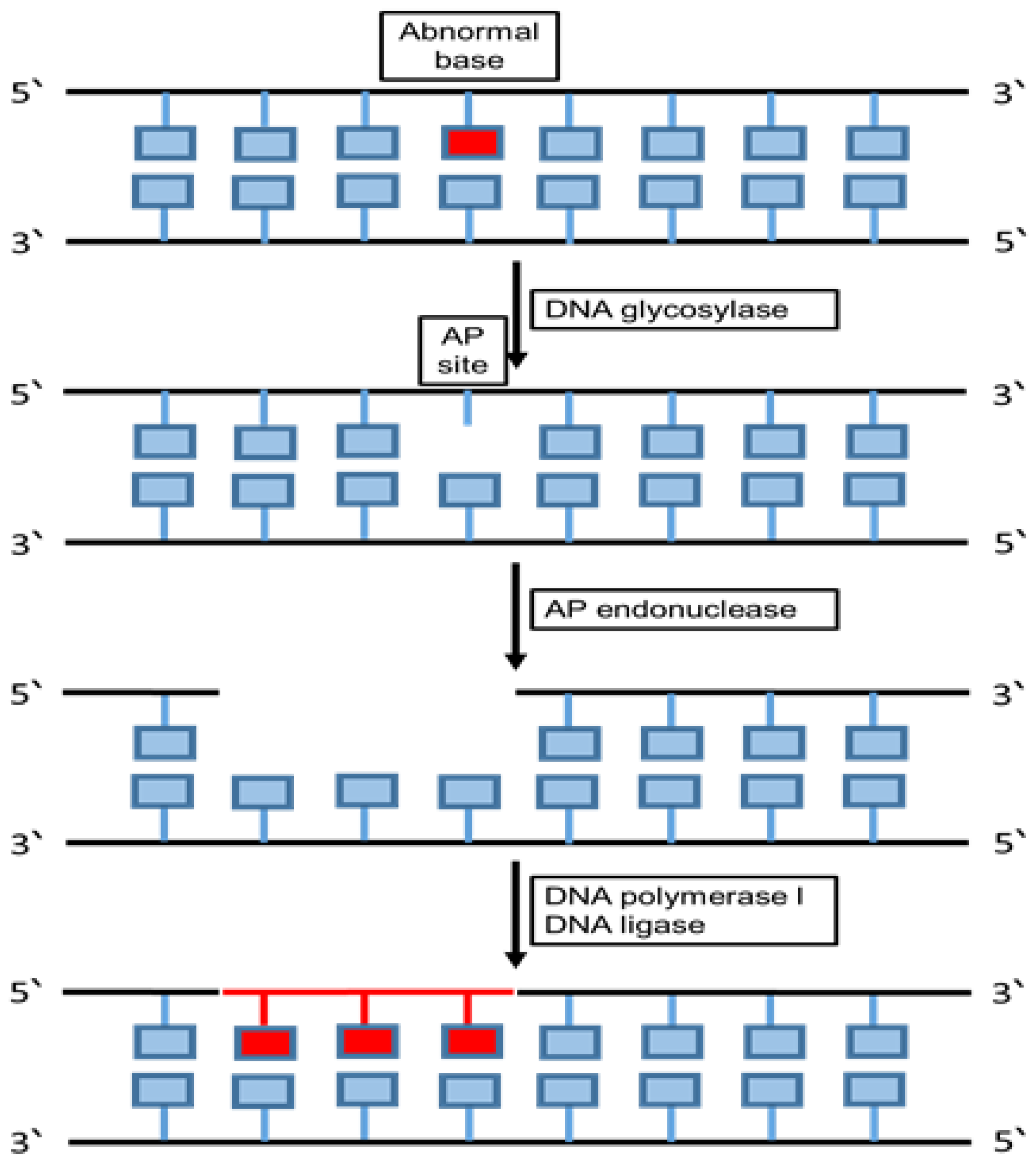
6.2. Base Excision Repair (BER)
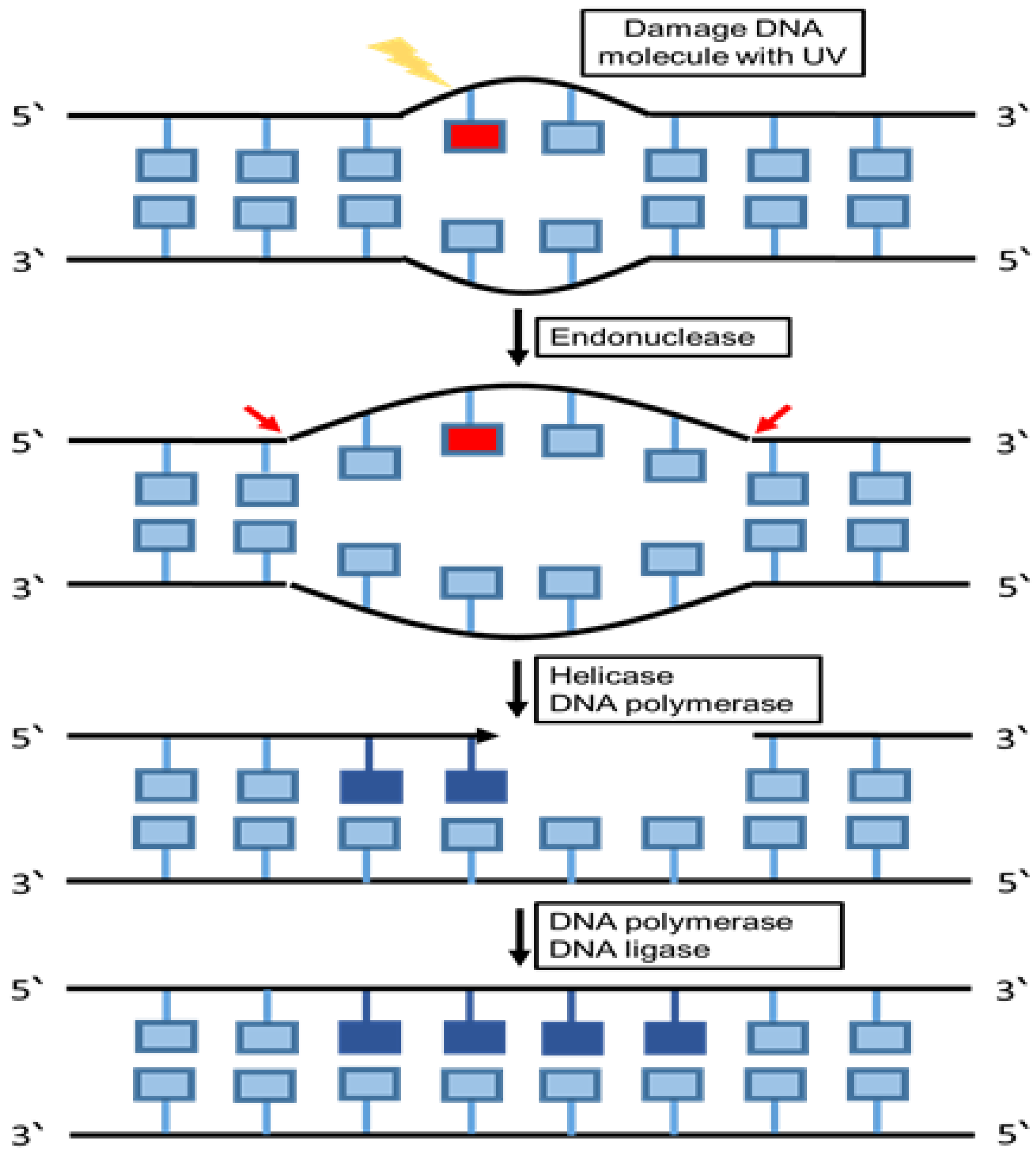
6.3. Nucleotide Excision Repair (NER)
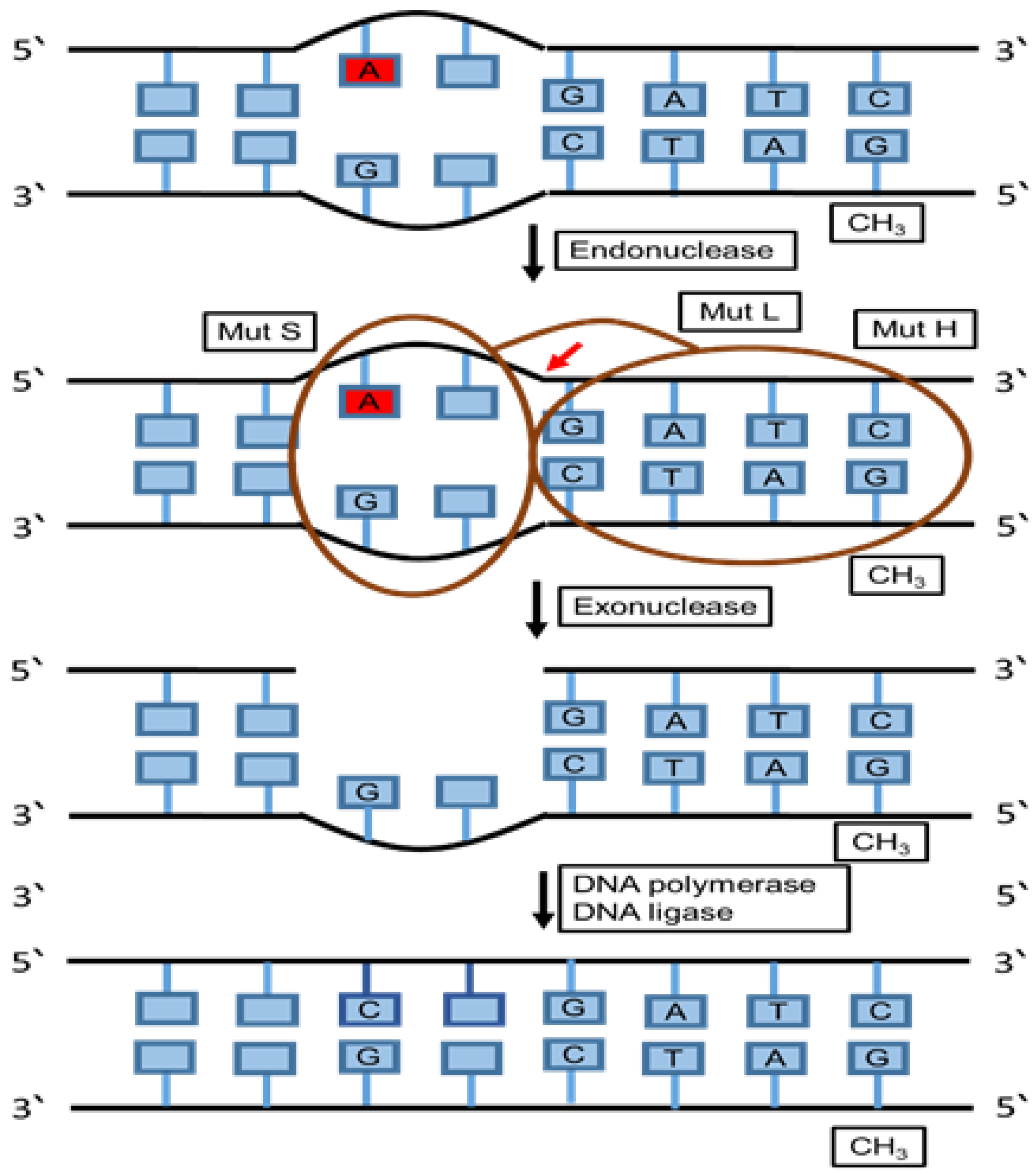
6.4. Mismatch Repair (MMR)
6.5. Non-Homologous End Joining (NHEJ) and Homologous Recombination (HRR)
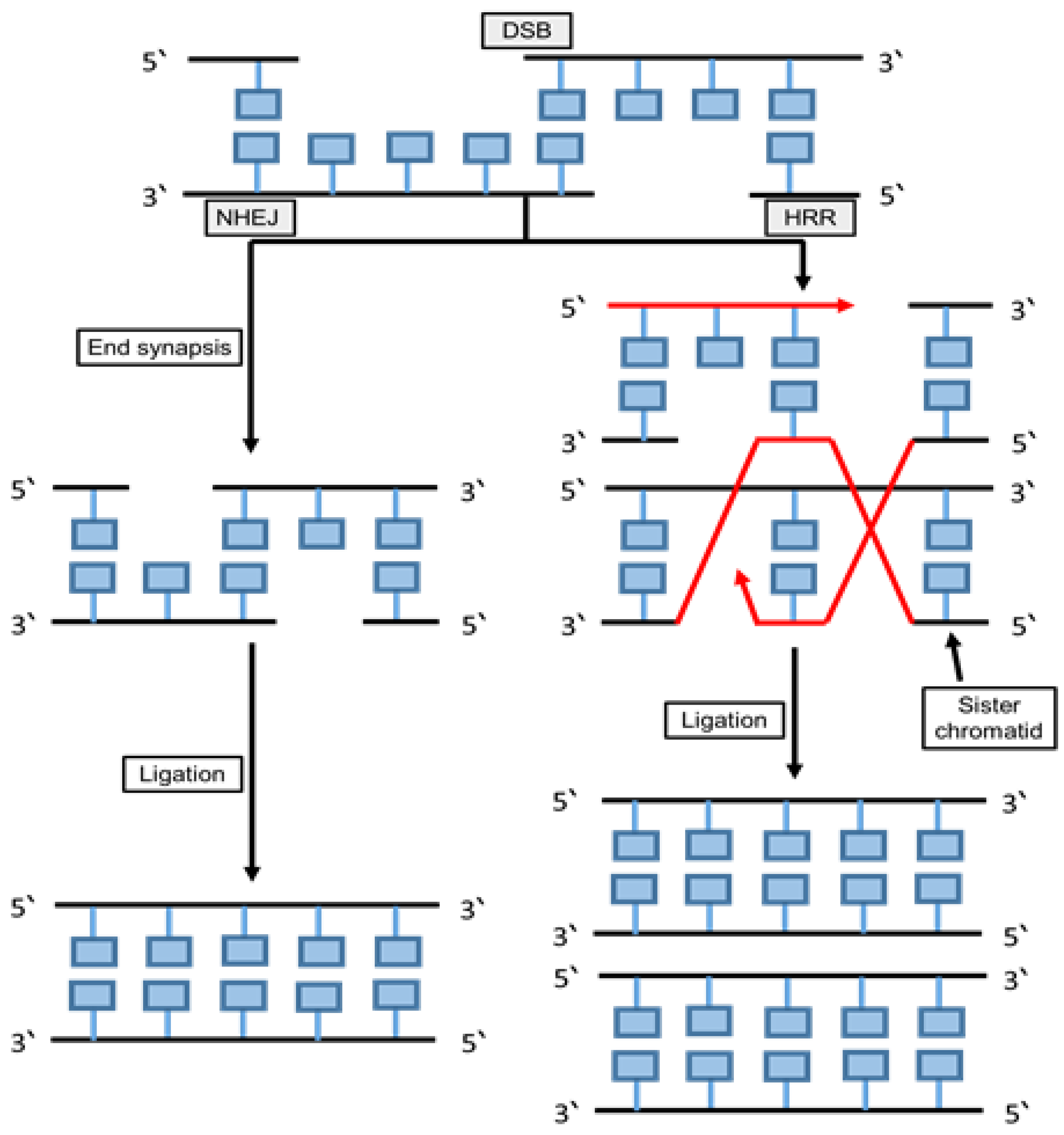
6.5.1. Non-Homologous End Joining (NHEJ)
6.5.2. Homologous Recombination Repair (HRR)
7. Cell Cycle as a Checkpoint in DNA Damage
8. Effects of Chemotherapy or Radiation in Cancer Treatments
9. Potential Biomarkers of Chromosomal Abnormalities
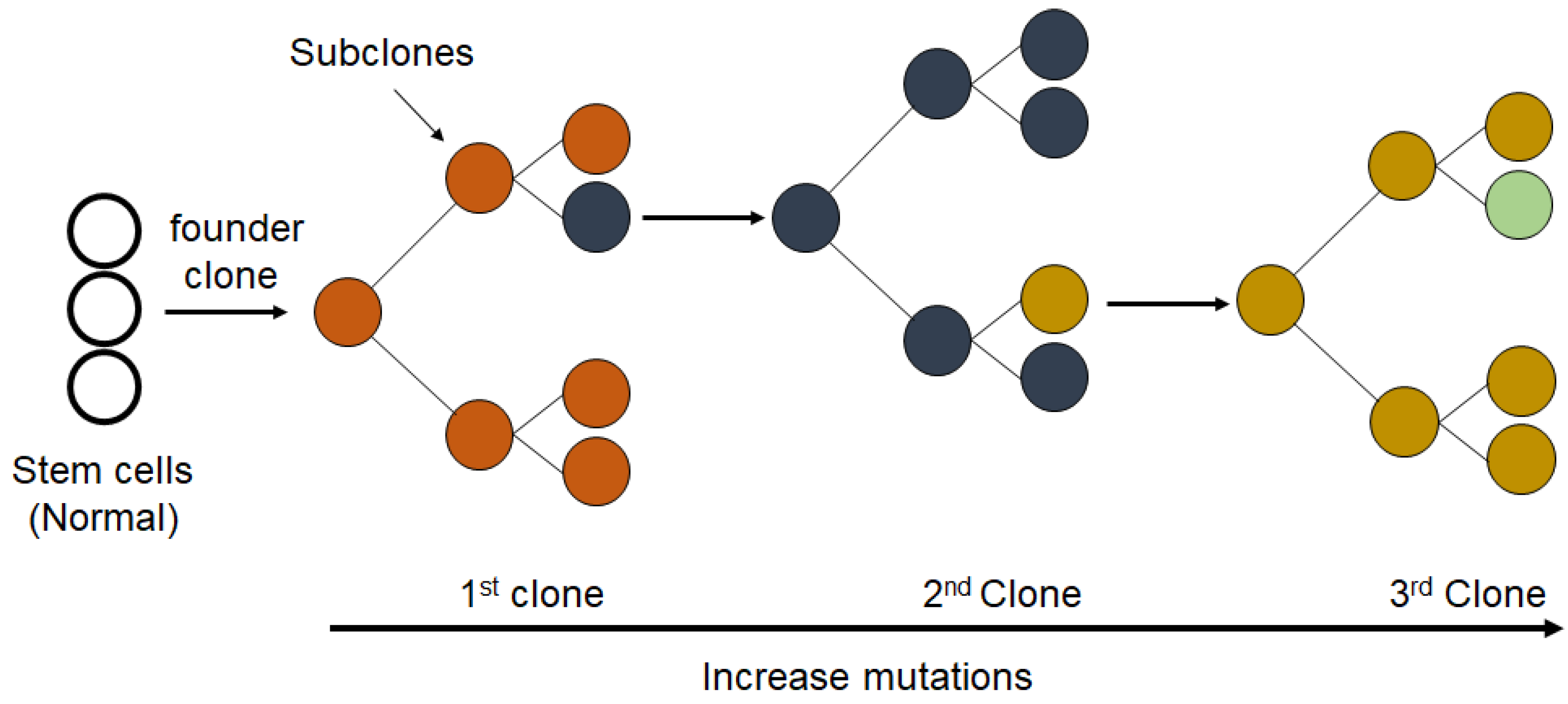
10. Clonal Evolution in Cancer
11. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Srinivas, U.S.; Tan, B.W.; Vellayappan, B.; Jeyasekharan, A. ROS and the DNA damage response in cancer. Redox Boil. 2019. [Google Scholar] [CrossRef]
- Terradas, M.; Martin, M.; Tusell, L.; Genescà, A. Genetic activities in micronuclei: Is the DNA entrapped in micronuclei lost for the cell? Mutat. Res. Mutat. Res. 2010. [Google Scholar] [CrossRef]
- Cannan, W.; Pederson, D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell. Physiol. 2016. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009. [Google Scholar] [CrossRef]
- Pucci, B.; Kasten, M.; Giordano, A. Cell Cycle and Apoptosis. Neoplasia 2000. [Google Scholar] [CrossRef]
- Basu, A.K. DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef]
- Cooper, G.M. The Cell: A Molecular Approach, 2nd ed.; ASM Press: Washington, DC, USA, 2000. [Google Scholar]
- Arjunan, K.P.; Sharma, V.K.; Ptasinska, S. Effects of Atmospheric Pressure Plasmas on Isolated and Cellular DNA—A Review. Int. J. Mol. Sci. 2015, 16, 2971–3016. [Google Scholar] [CrossRef]
- Friedberg, E.C. Fixing Your Damaged and Incorrect Genes; World Scientific: Singapore, 2019. [Google Scholar]
- Vignard, J.; Mirey, G.; Salles, B. Ionizing-radiation induced DNA double-strand breaks: A direct and indirect lighting up. Radiother. Oncol. 2013. [Google Scholar] [CrossRef]
- Abrahamson, S. Adverse Reproductive Outcomes in Families of Atomic Veterans: The Feasibility of Epidemiologic Studies. Radiat. Res. 1995. [Google Scholar] [CrossRef]
- Talty, J.J. Principles of Ionizing Radiation. In Industrial Hygiene Engineering; Elsevier: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Lomax, M.; Folkes, L.; O’Neill, P. Biological Consequences of Radiation-induced DNA Damage: Relevance to Radiotherapy. Clin. Oncol. 2013. [Google Scholar] [CrossRef]
- Baskar, R.; Dai, J.; Wenlong, N.; Yeo, R.; Yeoh, K.-W. Biological response of cancer cells to radiation treatment. Front. Mol. Biosci. 2014. [Google Scholar] [CrossRef]
- Smith, T.A.; Kirkpatrick, D.R.; Smith, S.; Smith, T.K.; Pearson, T.; Kailasam, A.; Herrmann, K.Z.; Schubert, J.; Agrawal, D.K. Radioprotective agents to prevent cellular damage due to ionizing radiation. J. Transl. Med. 2017. [Google Scholar] [CrossRef]
- Borek, C. Antioxidants and radiation therapy. J. Nutr. 2004. [Google Scholar] [CrossRef]
- Fuchs, Y.; Steller, H. Programmed cell death in animal development and disease. Cell 2011. [Google Scholar] [CrossRef]
- Tang, H.L.; Tang, H.M.; Mak, K.H.; Hu, S.; Wang, S.S.; Wong, K.M.; Wong, C.S.T.; Wu, H.Y.; Law, H.T.; Liu, K.; et al. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol. Boil. Cell 2012. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000. [Google Scholar] [CrossRef]
- Cooper, G.M.; Hausman, R.E. The Cell: A Molecular Approach, 4th ed.; ASM Press: Washington, DC, USA, 2007. [Google Scholar]
- Harrison, J.C.; Haber, J.E. Surviving the Breakup: The DNA Damage Checkpoint. Annu. Rev. Genet. 2006. [Google Scholar] [CrossRef]
- McGowan, C.H.; Russell, P. The DNA damage response: Sensing and signaling. Curr. Opin. Cell Biol. 2004. [Google Scholar] [CrossRef]
- Blow, J.J.; Lukas, J.; Bartkova, J. DNA Damage Response as an Anti-Cancer Barrier: Damage Threshold and the Concept of ‘Conditional Haploinsufficiency’. Cell Cycle 2007. [Google Scholar] [CrossRef]
- Maréchal, A.; Zou, L. DNA Damage Sensing by the ATM and ATR Kinases. Cold Spring Harb. Perspect. Boil. 2013. [Google Scholar] [CrossRef]
- Giglia-Mari, A.; Zotter, A.; Vermeulen, W. DNA Damage Response. Cold Spring Harb. Perspect. Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Gorgoulis, V.G.; Vassiliou, L.-V.F.; Karakaidos, P.; Zacharatos, P.; Kotsinas, A.; Liloglou, T.; Venere, M.; DiTullio, R.A.; Kastrinakis, N.G.; Levy, B.; et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 2005. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011. [Google Scholar] [CrossRef] [PubMed]
- Bristow, R.G.; Hill, R.P. Hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer 2008. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M. Chromosomal biomarkers of genomic instability relevant to cancer. Drug Discov. Today 2002. [Google Scholar] [CrossRef]
- Bartek, J. DNA damage response, genetic instability and cancer: From mechanistic insights to personalized treatment. Mol. Oncol. 2011. [Google Scholar] [CrossRef]
- Dai, Y.Y.W. Genomic Instability and Cancer. J. Carcinog. Mutagen. 2014. [Google Scholar] [CrossRef]
- Al-Tassan, N.A.; Chmiel, N.H.; Maynard, J.; Fleming, N.; Livingston, A.L.; Williams, G.T.; Hodges, A.; Davies, D.R.; David, S.S.; Sampson, J.R.; et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 2002. [Google Scholar] [CrossRef]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An Oncogene-Induced DNA Damage Model for Cancer Development. Science 2008. [Google Scholar] [CrossRef]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 2010. [Google Scholar] [CrossRef]
- McKinnon, P.J. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 2009. [Google Scholar] [CrossRef]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Boil. 2008. [Google Scholar] [CrossRef]
- Shiloh, Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 2003. [Google Scholar] [CrossRef] [PubMed]
- Riley, T.; Sontag, E.D.; Chen, P.A.; Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Huen, M.S.; Chen, J. The DNA damage response pathways: At the crossroad of protein modifications. Cell Res. 2007. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007. [Google Scholar] [CrossRef]
- Campisi, J.; Daddadifagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007. [Google Scholar] [CrossRef]
- Misteli, T.; Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 2009. [Google Scholar] [CrossRef]
- Ziv, Y.; Bielopolski, D.; Galanty, Y.; Lukas, C.; Taya, Y.; Schultz, D.C.; Lukas, J.; Bekker-Jensen, S.; Bartek, J.; Shiloh, Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature 2006. [Google Scholar] [CrossRef]
- Xiao, A.; Li, H.; Shechter, D.; Ahn, S.H.; Fabrizio, L.A.; Erdjument-Bromage, H.; Ishibe-Murakami, S.; Wang, B.; Tempst, P.; Hofmann, K.; et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 2008. [Google Scholar] [CrossRef]
- Cook, P.J.; Ju, B.G.; Telese, F.; Wang, X.; Glass, C.K.; Rosenfeld, M.G. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 2009. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006. [Google Scholar] [CrossRef] [PubMed]
- Helleday, T.; Petermann, E.; Lundin, C.; Hodgson, B.; Sharma, R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 2008. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.; Lord, C.J.; Ashworth, A. DNA repair deficiency as a therapeutic target in cancer. Curr. Opin. Genet. Dev. 2008. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Reinhardt, H.C.; Bartkova, J.; Tommiska, J.; Blomqvist, C.; Nevanlinna, H.; Bartek, J.; Yaffe, M.B.; Hemann, M.T. The combined status of ATM and p53 link tumor development with therapeutic response. Genome Res. 2009. [Google Scholar] [CrossRef] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005. [Google Scholar] [CrossRef]
- Hosoya, N.; Miyagawa, K. Targeting DNA damage response in cancer therapy. Cancer Sci. 2014. [Google Scholar] [CrossRef]
- Ashworth, A. A Synthetic Lethal Therapeutic Approach: Poly(ADP) Ribose Polymerase Inhibitors for the Treatment of Cancers Deficient in DNA Double-Strand Break Repair. J. Clin. Oncol. 2008. [Google Scholar] [CrossRef]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’Connor, M.J.; et al. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors fromBRCAMutation Carriers. N. Engl. J. Med. 2009. [Google Scholar] [CrossRef] [PubMed]
- Audeh, M.W.; Carmichael, J.; Penson, R.; Friedlander, M.L.; Powell, B.; Bell-McGuinn, K.M.; Scott, C.; Weitzel, J.N.; Oaknin, A.; Loman, N.; et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet 2010. [Google Scholar] [CrossRef]
- Rouleau, M.; Patel, A.; Hendzel, M.J.; Kaufmann, S.H.; Poirier, G.G. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 2010. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xiao, Z.; Gu, W.-Z.; Xue, J.; Bui, M.H.; Kovár, P.; Li, G.; Wang, G.; Tao, Z.-F.; Tong, Y.; et al. Selective Chk1 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int. J. Cancer 2006. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, W. CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation. Cancer Lett. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Sandhu, S.; Carden, C.P.; De Bono, J.S. Poly(ADP-Ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA Cancer J. Clin. 2011. [Google Scholar] [CrossRef]
- Slade, D. PARP and PARG inhibitors in cancer treatment. Genome Res. 2020. [Google Scholar] [CrossRef]
- Fang, P.; De Souza, C.; Minn, K.; Chien, J. Genome-scale CRISPR knockout screen identifies TIGAR as a modifier of PARP inhibitor sensitivity. Commun. Boil. 2019. [Google Scholar] [CrossRef]
- Aleskandarany, M.A.; Caracappa, D.; Nolan, C.C.; Macmillan, R.D.; Ellis, I.O.; Rakha, E.A.; Green, A.R. DNA damage response markers are differentially expressed in BRCA-mutated breast cancers. Breast Cancer Res. Treat. 2015. [Google Scholar] [CrossRef]
- Garcia-Cao, I.; Garcia-Cao, M.; Martín-Caballero, J.; Criado, L.M.; Klatt, P.; Flores, J.M.; Weill, J.C.; Blasco, M.A.; Serrano, M. ‘Super p53’ mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002. [Google Scholar] [CrossRef]
- Wolters, S.; Schumacher, B. Genome maintenance and transcription integrity in aging and disease. Front. Genet. 2013. [Google Scholar] [CrossRef] [PubMed]
- Broustas, C.; Lieberman, H.B. DNA Damage Response Genes and the Development of Cancer Metastasis. Radiat. Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Kelley, M.R.; Logsdon, D.; Fishel, M. Targeting DNA repair pathways for cancer treatment: What’s new? Future Oncol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Loeb, K.R.; Loeb, L.A. Significance of multiple mutations in cancer. Carcinogenesis 2000. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J. DNA Damage, Aging, and Cancer. N. Engl. J. Med. 2009. [Google Scholar] [CrossRef]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001. [Google Scholar] [CrossRef]
- Eker, A.P.M.; Quayle, C.; Chaves, I.; Van Der Horst, G. DNA Repair in Mammalian Cells. Cell. Mol. Life Sci. 2009. [Google Scholar] [CrossRef]
- Yi, C.; He, C. DNA Repair by Reversal of DNA Damage. Cold Spring Harb. Perspect. Biol. 2013. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Richa; Kumar, A.; Tyagi, M.B.; Sinha, R.P. Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair. J. Nucleic Acids 2010. [Google Scholar] [CrossRef]
- Campbell, N.; Reece, J. Biology, 8th ed.; Pearson Education: New York, NY, USA, 2008. [Google Scholar]
- Morita, R.; Nakane, S.; Shimada, A.; Inoue, M.; Iino, H.; Wakamatsu, T.; Fukui, K.; Nakagawa, N.; Masui, R.; Kuramitsu, S. Molecular Mechanisms of the Whole DNA Repair System: A Comparison of Bacterial and Eukaryotic Systems. J. Nucleic Acids 2010. [Google Scholar] [CrossRef]
- Pegg, A.E. Multifaceted Roles of Alkyltransferase and Related Proteins in DNA Repair, DNA Damage, Resistance to Chemotherapy, and Research Tools. Chem. Res. Toxicol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Christmann, M.; Kaina, B. O6-methylguanine-DNA methyltransferase (MGMT): Impact on cancer risk in response to tobacco smoke. Mutat. Res. Mol. Mech. Mutagen. 2012. [Google Scholar] [CrossRef] [PubMed]
- Christmann, M.; Verbeek, B.; Roos, W.; Kaina, B. O6-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: Enzyme activity, promoter methylation and immunohistochemistry. Biochim. Biophys. Acta Rev. Cancer 2011. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, B.; Bates, P.A.; Paik, J.; Jacobs, S.; Lindahl, T. Repair of alkylated DNA: Recent advances. DNA Repair 2007. [Google Scholar] [CrossRef]
- Kavli, B.; Otterlei, M.; Slupphaug, G.; Krokan, H. Uracil in DNA—General mutagen, but normal intermediate in acquired immunity. DNA Repair 2007. [Google Scholar] [CrossRef]
- Frosina, G.; Fortini, P.; Rossi, O.; Carrozzino, F.; Raspaglio, G.; Cox, L.S.; Lane, D.P.; Abbondandolo, A.; Dogliotti, E. Two Pathways for Base Excision Repair in Mammalian Cells. J. Boil. Chem. 1996. [Google Scholar] [CrossRef]
- Robertson, A.B.; Klungland, A.; Rognes, T.; Leiros, I. DNA Repair in Mammalian Cells. Cell. Mol. Life Sci. 2009. [Google Scholar] [CrossRef]
- Schormann, N.; Ricciardi, R.; Chattopadhyay, D. Uracil-DNA glycosylases—Structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 2014. [Google Scholar] [CrossRef] [PubMed]
- Slupphaug, G.; Eftedal, I.; Kavli, B.; Bharati, S.; Helle, N.M.; Haug, T.; Levine, D.W.; Krokan, H.E. Properties of a Recombinant Human Uracil-DNA Glycosylase from the UNG Gene and Evidence that UNG Encodes the Major Uracil-DNA Glycosylase. Biochemistry 1995. [Google Scholar] [CrossRef]
- Mjelle, R.; Hegre, S.A.; Aas, P.A.; Slupphaug, G.; Drabløs, F.; Sætrom, P.; Krokan, H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair 2015. [Google Scholar] [CrossRef]
- Cabelof, D.C.; Raffoul, J.J.; Yanamadala, S.; Guo, Z.; Heydari, A.R. Induction of DNA polymerase beta-dependent base excision repair in response to oxidative stress in vivo. Carcinogesis 2002. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schär, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2011. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Iii, D.M.W. Overview of Base Excision Repair Biochemistry. Curr. Mol. Pharmacol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Schärer, O.D.; Jiricny, J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. BioEssays 2001. [Google Scholar] [CrossRef]
- Freudenthal, B.D. Base excision repair of oxidative DNA damage from mechanism to disease. Front. Biosci. 2017. [Google Scholar] [CrossRef]
- Eide, L.; Luna, L.; Gustad, E.C.; Henderson, P.T.; Essigmann, J.M.; Demple, B.; Seeberg, E. Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry 2001. [Google Scholar] [CrossRef]
- Zahn, K.E.; Averill, A.; Wallace, S.S.; Doublié, S. The Miscoding Potential of 5-Hydroxycytosine Arises Due to Template Instability in the Replicative Polymerase Active Site. Biochemistry 2011. [Google Scholar] [CrossRef]
- Petruseva, I.O.; Evdokimov, A.N.; Lavrik, O. Molecular Mechanism of Global Genome Nucleotide Excision Repair. Acta Nat. 2014. [Google Scholar] [CrossRef]
- Lehmann, A.R. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 2003. [Google Scholar] [CrossRef]
- Hanawalt, P.C.; Spivak, G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008. [Google Scholar] [CrossRef]
- Friedberg, E.C. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 2001. [Google Scholar] [CrossRef] [PubMed]
- Sugasawa, K. Regulation of damage recognition in mammalian global genomic nucleotide excision repair. Mutat. Res. Mol. Mech. Mutagen. 2010. [Google Scholar] [CrossRef] [PubMed]
- Svilar, D.; Goellner, E.M.; Almeida, K.H.; Sobol, R.W. Base Excision Repair and Lesion-Dependent Subpathways for Repair of Oxidative DNA Damage. Antioxidants Redox Signal. 2011. [Google Scholar] [CrossRef]
- Puumalainen, M.-R.; Rüthemann, P.; Min, J.-H.; Naegeli, H. Xeroderma pigmentosum group C sensor: Unprecedented recognition strategy and tight spatiotemporal regulation. Cell. Mol. Life Sci. 2015. [Google Scholar] [CrossRef] [PubMed]
- Sugasawa, K.; Shimizu, Y.; Iwai, S.; Hanaoka, F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair 2002. [Google Scholar] [CrossRef]
- Missura, M.; Buterin, T.; Hindges, R.; Hübscher, U.; Kasparkova, J.; Brabec, V.; Naegeli, H. Double-check probing of DNA bending and unwinding by XPA–RPA: An architectural function in DNA repair. EMBO J. 2001. [Google Scholar] [CrossRef] [PubMed]
- Volker, M.; Moné, M.J.; Karmakar, P.; Van Hoffen, A.; Schul, W.; Vermeulen, W.; Hoeijmakers, J.H.; Van Driel, R.; Van Zeeland, A.A.; Mullenders, L.H. Sequential Assembly of the Nucleotide Excision Repair Factors In Vivo. Mol. Cell 2001. [Google Scholar] [CrossRef]
- Kunkel, T.; Erie, R.A. DNA mismatch repair. Annu. Rev. Biochem. 2005. [Google Scholar] [CrossRef]
- Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2007. [Google Scholar] [CrossRef]
- Aaltonen, L.; Peltomaki, P.; Leach, F.; Sistonen, P.; Pylkkanen, L.; Mecklin, J.; Jarvinen, H.; Powell, S.; Jen, J. Clues to the pathogenesis of familial colorectal cancer. Science 1993. [Google Scholar] [CrossRef]
- Cortés-Ciriano, I.; Lee, S.; Park, W.-Y.; Kim, T.-M.; Park, P. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 2017. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.V.; Yamada, H.Y.; Yao, Y.; Dai, W. Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: A perspective from genetic studies in mice. Carcinogenesis 2009. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Amon, A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 2015. [Google Scholar] [CrossRef]
- Orans, J.; McSweeney, E.A.; Iyer, R.R.; Hast, M.A.; Hellinga, H.W.; Modrich, P.; Beese, L.S. Structures of Human Exonuclease 1 DNA Complexes Suggest a Unified Mechanism for Nuclease Family. Cell 2011. [Google Scholar] [CrossRef]
- Vallur, A.C.; Maizels, N. Distinct Activities of Exonuclease 1 and Flap Endonuclease 1 at Telomeric G4 DNA. PLoS ONE 2010. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010. [Google Scholar] [CrossRef]
- Schipler, A.; Iliakis, G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013. [Google Scholar] [CrossRef]
- Vilenchik, M.M.; Knudson, A. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA 2003. [Google Scholar] [CrossRef]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2007. [Google Scholar] [CrossRef]
- Mahaney, B.L.; Meek, K.; Lees-Miller, S.P. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009. [Google Scholar] [CrossRef]
- McClendon, A.K.; Osheroff, N. DNA topoisomerase II, genotoxicity, and cancer. Mutat. Res. Mol. Mech. Mutagen. 2007. [Google Scholar] [CrossRef] [PubMed]
- Lamarche, B.J.; Orazio, N.I.; Weitzman, M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010. [Google Scholar] [CrossRef] [PubMed]
- Lempiäinen, H.; Halazonetis, T.D. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 2009. [Google Scholar] [CrossRef] [PubMed]
- Boulton, S.; Kyle, S.; Durkacz, B. Mechanisms of enhancement of cytotoxicity in etoposide and ionising radiation-treated cells by the protein kinase inhibitor wortmannin. Eur. J. Cancer 2000. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017. [Google Scholar] [CrossRef]
- Saleh-Gohari, N.; Bryant, H.E.; Schultz, N.; Parker, K.M.; Cassel, T.N.; Helleday, T. Spontaneous Homologous Recombination Is Induced by Collapsed Replication Forks That Are Caused by Endogenous DNA Single-Strand Breaks. Mol. Cell. Boil. 2005. [Google Scholar] [CrossRef]
- Deans, A.J.; West, S. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011. [Google Scholar] [CrossRef]
- Curtin, N.J. DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 2012. [Google Scholar] [CrossRef]
- Aloyz, R.; Grzywacz, K.; Xu, Z.-Y.; Loignon, M.; Alaoui-Jamali, M.; Panasci, L. Imatinib sensitizes CLL lymphocytes to chlorambucil. Leukemia 2003. [Google Scholar] [CrossRef][Green Version]
- Choudhury, A.; Zhao, H.; Jalali, F.; Al Rashid, S.; Ran, J.; Supiot, S.; Kiltie, A.E.; Bristow, R.G. Targeting homologous recombination using imatinib results in enhanced tumor cell chemosensitivity and radiosensitivity. Mol. Cancer Ther. 2009. [Google Scholar] [CrossRef]
- Popp, H.D.; Naumann, N.; Brendel, S.; Henzler, T.; Weiss, C.; Hofmann, W.-K.; Fabarius, A. Increase of DNA damage and alteration of the DNA damage response in myelodysplastic syndromes and acute myeloid leukemias. Leuk. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Nowak, B.J.; Keating, M.J.; Plunkett, W. DNA repair initiated in chronic lymphocytic leukemia lymphocytes by 4-hydroperoxycyclophosphamide is inhibited by fludarabine and clofarabine. Clin. Cancer Res. 2001, 7, 3580–3589. [Google Scholar] [PubMed]
- Barnum, K.J.; O’Connell, M.J. Cell cycle regulation by checkpoints. Breast Cancer 2014. [Google Scholar] [CrossRef]
- Telser, A. Molecular Biology of the Cell, 4th Edition. Shock 2002. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008. [Google Scholar] [CrossRef]
- Bouwman, P.; Jonkers, J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer 2012. [Google Scholar] [CrossRef]
- Vacchelli, E.; Galluzzi, L.; Fridman, W.H.; Galon, J.; Sautès-Fridman, C.; Tartour, E.; Kroemer, G. Trial watch. OncoImmunology 2012. [Google Scholar] [CrossRef]
- Meng, Y.; Efimova, E.V.; Hamzeh, K.W.; Darga, T.; Mauceri, H.J.; Fu, Y.-X.; Kron, S.J.; Weichselbaum, R.R. Radiation-inducible Immunotherapy for Cancer: Senescent Tumor Cells as a Cancer Vaccine. Mol. Ther. 2012. [Google Scholar] [CrossRef]
- Mayer, E.L. Early and Late Long-Term Effects of Adjuvant Chemotherapy. Am. Soc. Clin. Oncol. Educ. Book 2013. [Google Scholar] [CrossRef]
- Landau, D.A.; Sun, C.; Rosebrock, D.; Herman, S.E.M.; Fein, J.A.; Sivina, M.; Underbayev, C.; Liu, D.; Hoellenriegel, J.; Ravichandran, S.; et al. The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nat. Commun. 2017. [Google Scholar] [CrossRef]
- Barcellos-Hoff, M.H.; Park, C.; Wright, E.G. Radiation and the microenvironment—Tumorigenesis and therapy. Nat. Rev. Cancer 2005. [Google Scholar] [CrossRef]
- Krem, M.M.; Press, O.; Horwitz, M.S.; Tidwell, T. Mechanisms and clinical applications of chromosomal instability in lymphoid malignancy. Br. J. Haematol. 2015. [Google Scholar] [CrossRef] [PubMed]
- El-Zein, R.A.; Schabath, M.B.; Etzel, C.J.; Lopez, M.S.; Franklin, J.D.; Spitz, M.R. Cytokinesis-Blocked Micronucleus Assay as a Novel Biomarker for Lung Cancer Risk. Cancer Res. 2006. [Google Scholar] [CrossRef] [PubMed]
- Kirsch-Volders, M.; Plas, G.; Elhajouji, A.; Lukamowicz, M.; Gonzalez, L.; Loock, K.V.; Decordier, I. The in vitro MN assay in 2011: Origin and fate, biological significance, protocols, high throughput methodologies and toxicological relevance. Arch. Toxicol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Makowski, M.; Archer, K.J. Generalized Monotone Incremental Forward Stagewise Method for Modeling Count Data: Application Predicting Micronuclei Frequency. Cancer Inf. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kirsch-Volders, M.; Vanhauwaert, A.; Eichenlaub-Ritter, U.; Decordier, I. Indirect mechanisms of genotoxicity. Toxicol. Lett. 2003. [Google Scholar] [CrossRef]
- Iarmarcovai, G.; Bonassi, S.; Botta, A.; Baan, R.; Orsière, T. Genetic polymorphisms and micronucleus formation: A review of the literature. Mutat. Res. Mutat. Res. 2008. [Google Scholar] [CrossRef]
- Alkan, O.; Schoeberl, B.; Shah, M.; Koshkaryev, A.; Heinemann, T.; Drummond, D.C.; Yaffe, M.B.; Raue, A. Modeling chemotherapy-induced stress to identify rational combination therapies in the DNA damage response pathway. Sci. Signal. 2018. [Google Scholar] [CrossRef]
- Driessens, G.; Harsan, L.; Robaye, B.; Waroquier, D.; Browaeys, P.; Giannakopoulos, X.; Velu, T.; Bruyns, C. Micronuclei to detect in vivo chemotherapy damage in a p53 mutated solid tumour. Br. J. Cancer 2003. [Google Scholar] [CrossRef]
- Gashi, G.; Mahovlić, V.; Manxhuka-Kerliu, S.; Podrimaj-Bytyqi, A.; Gashi, L.; Elezaj, I.R. The association between micronucleus, nucleoplasmic bridges, and nuclear buds frequency and the degree of uterine cervical lesions. Biomarkers 2018. [Google Scholar] [CrossRef]
- Bitgen, N.; Altuntas, H.D.; Bayram, F.; Cakir, I.; Hamurcu, Z.; Diri, H.; Baskol, G.; Senol, S.; Durak, A. Increased micronucleus, nucleoplasmic bridge, nuclear bud frequency and oxidative DNA damage associated with prolactin levels and pituitary adenoma diameters in patients with prolactinoma. Biotech. Histochem. 2015. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Brenes, I.; Wodarz, D. Preventing clonal evolutionary processes in cancer: Insights from mathematical models. Proc. Natl. Acad. Sci. USA 2015. [Google Scholar] [CrossRef] [PubMed]
- Ibragimova, M.; Tsyganov, M.M.; Litviakov, N.V. Natural and chemotherapy-induced clonal evolution of tumors. Biochemistry 2017. [Google Scholar] [CrossRef] [PubMed]
- Greaves, M.; Mailey, C.C. Clonal evolution in cancer. Nature 2012. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017. [Google Scholar] [CrossRef]
- Carina, L.; Susana, V.; Elisabete, C.; Manuel, G.; Miguel, B. Correlation between the genotoxicity endpoints measured by two different genotoxicity assays: Comet assay and CBMN assay. Front. Genet. 2015. [Google Scholar] [CrossRef]
- Yap, T.A.; Plummer, R.; Azad, N.S.; Helleday, T. The DNA Damaging Revolution: PARP Inhibitors and Beyond. Am. Soc. Clin. Oncol. Educ. Book 2019. [Google Scholar] [CrossRef]
- Yao, Y.; Dai, W. Genomic Instability and Cancer Yixin. J. Carcinog. Mutagen. 2014. [Google Scholar] [CrossRef]
- Faraoni, I.; Graziani, G. Role of BRCA Mutations in Cancer Treatment with Poly(ADP-ribose) Polymerase (PARP) Inhibitors. Cancers 2018, 10, 487. [Google Scholar] [CrossRef]
- Williams, D.T.; Staples, C. Approaches for Identifying Novel Targets in Precision Medicine: Lessons from DNA Repair. In Advances in Experimental Medicine and Biology; Springer: Berlin, Germany, 2017. [Google Scholar]
- Beggs, R.; Yang, E.S. Targeting DNA repair in precision medicine. In Advances in Protein Chemistry and Structural Biology; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Ivashkevich, A.; Redon, C.E.; Nakamura, A.J.; Martin, R.F.; Martin, O.A. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2011. [Google Scholar] [CrossRef]
- Lippi, G.; Plebani, M. Personalized medicine: Moving from simple theory to daily practice. Clin. Chem. Lab. Med. 2015. [Google Scholar] [CrossRef] [PubMed]
- Reddig, A.; Rübe, C.E.; Rödiger, S.; Schierack, P.; Reinhold, D.; Roggenbuck, D. DNA damage assessment and potential applications in laboratory diagnostics and precision medicine. J. Lab. Precis. Med. 2018. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.-E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. https://doi.org/10.3390/cancers12041050
Alhmoud JF, Woolley JF, Al Moustafa A-E, Malki MI. DNA Damage/Repair Management in Cancers. Cancers. 2020; 12(4):1050. https://doi.org/10.3390/cancers12041050
Chicago/Turabian StyleAlhmoud, Jehad F., John F. Woolley, Ala-Eddin Al Moustafa, and Mohammed Imad Malki. 2020. "DNA Damage/Repair Management in Cancers" Cancers 12, no. 4: 1050. https://doi.org/10.3390/cancers12041050
APA StyleAlhmoud, J. F., Woolley, J. F., Al Moustafa, A.-E., & Malki, M. I. (2020). DNA Damage/Repair Management in Cancers. Cancers, 12(4), 1050. https://doi.org/10.3390/cancers12041050





