The Challenges and Opportunities of LncRNAs in Ovarian Cancer Research and Clinical Use
Abstract
1. Introduction
2. Clinical Relevance of LncRNA in OC: Diagnosis, Prognosis and Treatment Resistance
3. LncRNAs Implicated in OC Development and Progression
4. Regulatory Molecular Mechanisms of LncRNAs in OC
4.1. Transcriptional Regulation
4.2. Post-Transcriptional Regulation
4.3. Post-Translational Regulation
4.4. Regulation of LncRNA Expression
5. Bioinformatics Resources for LncRNA Research
6. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Brett, M.R.; Jennifer, B.P.; Thomas, A.S.; Brett, M.R.; Jennifer, B.P.; Thomas, A.S. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [CrossRef] [PubMed]
- Ueland, F. A Perspective on Ovarian Cancer Biomarkers: Past, Present and Yet-To-Come. Diagnostics 2017, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Cortez, A.J.; Tudrej, P.; Kujawa, K.A.; Lisowska, K.M. Advances in ovarian cancer therapy. Cancer Chemother. Pharmacol. 2018, 81, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, A.; Chen, L. LncRNAs in ovarian cancer. Clin. Chim. Acta 2019, 490, 17–27. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Hu, X.; Bao, J.; Wang, Z.; Zhang, Z.; Gu, P.; Tao, F.; Cui, D.; Jiang, W. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumor Biol. 2016, 37, 3497–3504. [Google Scholar] [CrossRef]
- Zeng, S.; Liu, S.; Feng, J.; Gao, J.; Xue, F. Upregulation of lncRNA AB073614 functions as a predictor of epithelial ovarian cancer prognosis and promotes tumor growth in vitro and in vivo. Cancer Biomark. 2019, 24, 421–428. [Google Scholar] [CrossRef]
- Yang, M.; Zhai, Z.; Zhang, Y.; Wang, Y. Clinical significance and oncogene function of long noncoding RNA HAGLROS overexpression in ovarian cancer. Arch. Gynecol. Obstet. 2019, 300, 703–710. [Google Scholar] [CrossRef]
- Lin, X.; Tang, X.; Zheng, T.; Qiu, J.; Hua, K. Long non-coding RNA NONHSAT076754 promotes invasion and metastasis in epithelial ovarian cancer. J. Cancer 2019, 10, 1930–1940. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.-D.; Chen, X.; Sun, K.-X.; Wang, L.-L.; Chen, S.; Zhao, Y. Role of the lncRNA ABHD11-AS1 in the tumorigenesis and progression of epithelial ovarian cancer through targeted regulation of RhoC. Mol. Cancer 2017, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Xu, Y.; Zhang, D.; Liu, G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem. Biophys. Res. Commun. 2018, 503, 2095–2100. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Song, K.; Hu, H.; Tian, Q.; Wang, W.; Dong, Q.; Yin, X.; Di, W. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J. Exp. Clin. Cancer Res. 2019, 38, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liu, S.; Liu, Y.; Lin, X.; Zheng, T.; Liu, X.; Qiu, J.; Hua, K. Circulating serum exosomal aHIF is a novel prognostic predictor for epithelial ovarian cancer. Onco. Targets. Ther. 2019, 12, 7699–7711. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Li, X.; Zhan, G.; Zhu, Y.; Yu, J.; Wang, J.; Li, L.; Wu, W.; Liu, N.; Guo, X. Long noncoding RNA MAGI1-IT1 promoted invasion and metastasis of epithelial ovarian cancer via the miR-200a/ZEB axis. Cell Cycle 2019, 18, 1393–1406. [Google Scholar] [CrossRef]
- Pei, C.L.; Fei, K.L.; Yuan, X.Y.; Gong, X.J. LncRNA DANCR aggravates the progression of ovarian cancer by downregulating UPF1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10657–10663. [Google Scholar] [CrossRef]
- Worku, T.; Bhattarai, D.; Ayers, D.; Wang, K.; Wang, C.; Rehman, Z.U.; Talpur, H.S.; Yang, L. Long Non-Coding RNAs: The New Horizon of Gene Regulation in Ovarian Cancer. Cell. Physiol. Biochem. 2017, 44, 948–966. [Google Scholar] [CrossRef]
- Sun, D.; Fan, X.H. LncRNA SNHG12 accelerates the progression of ovarian cancer via absorbing miRNA-129 to upregulate SOX4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2345–2352. [Google Scholar] [CrossRef]
- Lu, X.; Wang, F.; Fu, M.; Li, Y.; Wang, L. Long non-coding RNA KCNQ1OT1 accelerates the progression of ovarian cancer via microRNA-212-3/ LCN2 axis. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2019, 28, 135–146. [Google Scholar] [CrossRef]
- Wang, L.-L.; Sun, K.-X.; Wu, D.-D.; Xiu, Y.-L.; Chen, X.; Chen, S.; Zong, Z.-H.; Sang, X.-B.; Liu, Y.; Zhao, Y. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression. J. Cell. Mol. Med. 2017, 21, 3055–3065. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Jiang, H.; Cui, Z.; Wang, X.; Wang, L.; Han, Y. Gene microarray analysis of lncRNA and mRNA expression profiles in patients with high-grade ovarian serous cancer. Int. J. Mol. Med. 2018, 42, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Gao, S.; Zheng, Y.; Yao, M.; Ruan, F. LncRNA CASC15 Functions As An Unfavorable Predictor Of Ovarian Cancer Prognosis And Inhibits Tumor Progression Through Regulation Of miR-221/ARID1A Axis. Onco. Targets. Ther. 2019, 12, 8725–8736. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.B.; Zou, Y.F. Clinical significance of lncRNA FAM83H-AS1 in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4656–4662. [Google Scholar] [CrossRef]
- Wang, A.; Jin, C.; Li, H.; Qin, Q.; Li, L. LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway. Int. J. Biol. Macromol. 2018, 120, 1705–1713. [Google Scholar] [CrossRef]
- Dou, Q.; Xu, Y.; Zhu, Y.; Hu, Y.; Yan, Y.; Yan, H. LncRNA FAM83H-AS1 contributes to the radioresistance, proliferation, and metastasis in ovarian cancer through stabilizing HuR protein. Eur. J. Pharmacol. 2019, 852, 134–141. [Google Scholar] [CrossRef]
- Wang, D.; Dai, J.; Hou, S.; Qian, Y. LncRNA SNHG20 predicts a poor prognosis and promotes cell progression in epithelial ovarian cancer. Biosci. Rep. 2019, 29, 1–9. [Google Scholar] [CrossRef]
- Sun, T.; Yang, P.; Gao, Y. Long non-coding RNA EPB41L4A-AS2 suppresses progression of ovarian cancer by sequestering microRNA-103a to upregulate transcription factor RUNX1T1. Exp. Physiol. 2020, 105, 75–87. [Google Scholar] [CrossRef]
- Xue, Z.; Zhu, X.; Teng, Y. Long non-coding RNA CASC2 inhibits progression and predicts favorable prognosis in epithelial ovarian cancer. Mol. Med. Rep. 2018, 18, 5173–5181. [Google Scholar] [CrossRef]
- Lin, H.; Shen, L.; Lin, Q.; Dong, C.; Maswela, B.; Illahi, G.S.; Wu, X. SNHG5 enhances Paclitaxel sensitivity of ovarian cancer cells through sponging miR-23a. Biomed. Pharmacother. 2020, 123, 109711. [Google Scholar] [CrossRef]
- Miao, J.-T.; Gao, J.-H.; Chen, Y.-Q.; Chen, H.; Meng, H.-Y.; Lou, G. LncRNA ANRIL affects the sensitivity of ovarian cancer to cisplatin via regulation of let-7a/HMGA2 axis. Biosci. Rep. 2019, 39, BSR20182101. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, H.R. Down-regulation of long noncoding RNA DLX6-AS1 defines good prognosis and inhibits proliferation and metastasis in human epithelial ovarian cancer cells via Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3243–3252. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.H.; Du, X.; Sun, F.D.; Wang, P.C.; Li, M. Cisplatin suppresses tumor proliferation by inhibiting autophagy in ovarian cancer via long non-coding RNA RP11-135L22.1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Chen, W.; Wu, D.; Wang, Y. Upregulation of SNHG3 expression associated with poor prognosis and enhances malignant progression of ovarian cancer. Cancer Biomark. 2018, 22, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Gao, S.; Xuan, L.; Liu, X. Long non-coding RNA FEZF1-AS1 induced progression of ovarian cancer via regulating miR-130a-5p/SOX4 axis. J. Cell. Mol. Med. 2020. In print. [Google Scholar] [CrossRef] [PubMed]
- Zuo, K.; Zhao, Y.; Zheng, Y.; Chen, D.; Liu, X.; Du, S.; Liu, Q. Long non-coding RNA XIST promotes malignant behavior of epithelial ovarian cancer. Onco. Targets. Ther. 2019, 12, 7261–7267. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, M.K.; Doxtater, K.; Keramatnia, F.; Zacheaus, C.; Yallapu, M.M.; Jaggi, M.; Chauhan, S.C. Role of lncRNAs in ovarian cancer: Defining new biomarkers for therapeutic purposes. Drug Discov. Today 2018, 23, 1635–1643. [Google Scholar] [CrossRef]
- Yan, H.; Li, H.; Silva, M.A.; Guan, Y.; Yang, L.; Zhu, L.; Zhang, Z.; Li, G.; Ren, C. LncRNA FLVCR1-AS1 mediates miR-513/YAP1 signaling to promote cell progression, migration, invasion and EMT process in ovarian cancer. J. Exp. Clin. Cancer Res. 2019, 38, 356. [Google Scholar] [CrossRef]
- Zhao, H.; Yu, H.; Zheng, J.; Ning, N.; Tang, F.; Yang, Y.; Wang, Y. Lowly-expressed lncRNA GAS5 facilitates progression of ovarian cancer through targeting miR-196-5p and thereby regulating HOXA5. Gynecol. Oncol. 2018, 151, 345–355. [Google Scholar] [CrossRef]
- Pan, L.; Meng, Q.; Li, H.; Liang, K.; Li, B. LINC00339 promotes cell proliferation, migration, and invasion of ovarian cancer cells via miR-148a-3p/ROCK1 axes. Biomed. Pharmacother. 2019, 120, 109423. [Google Scholar] [CrossRef]
- Chu, Z.P.; Dai, J.; Jia, L.G.; Li, J.; Zhang, Y.; Zhang, Z.Y.; Yan, P. Increased expression of long noncoding RNA HMMR-AS1 in epithelial ovarian cancer: An independent prognostic factor. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8145–8150. [Google Scholar] [CrossRef]
- Gong, M.; Luo, C.; Meng, H.; Li, S.; Nie, S.; Jiang, Y.; Wan, Y.; Li, H.; Cheng, W. Upregulated LINC00565 Accelerates Ovarian Cancer Progression By Targeting GAS6. Onco. Targets. Ther. 2019, 12, 10011–10022. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, H. Long non-coding RNA GEHT1 promoted the proliferation of ovarian cancer cells via modulating the protein stability of HIF1α. Biosci. Rep. 2019, 39, 1–10. [Google Scholar] [CrossRef]
- Ni, M.-W.; Zhou, J.; Zhang, Y.-L.; Zhou, G.-M.; Zhang, S.-J.; Feng, J.-G.; Xu, Q.; Zhou, Y.; Mou, H.-Z.; Zheng, Z.-G. Downregulation of LINC00515 in high-grade serous ovarian cancer and its relationship with platinum resistance. Biomark. Med. 2019, 13, 535–543. [Google Scholar] [CrossRef]
- Li, J.; Feng, L.; Tian, C.; Tang, Y.L.; Tang, Y.; Hu, F.Q. Long noncoding RNA-JPX predicts the poor prognosis of ovarian cancer patients and promotes tumor cell proliferation, invasion and migration by the PI3K/Akt/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8135–8144. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shi, L.; Zhou, C.; Wang, Z.; Yu, T.; Zhou, J.; Yang, Y. Long non-coding RNA MCM3AP-AS1 inhibits cell viability and promotes apoptosis in ovarian cancer cells by targeting miR-28-5p. Int. J. Clin. Exp. Med. 2019, 12, 2939–2951. [Google Scholar] [CrossRef]
- Guo, Q.; Wang, L.; Zhu, L.; Lu, X.; Song, Y.; Sun, J.; Wu, Z.; Shi, J.; Wang, Z.; Zhou, X. The clinical significance and biological function of lncRNA SOCAR in serous ovarian carcinoma. Gene 2019, 713, 143969. [Google Scholar] [CrossRef] [PubMed]
- Oncul, S.; Amero, P.; Rodriguez-Aguayo, C.; Calin, G.A.; Sood, A.K.; Lopez-Berestein, G. Long non-coding RNAs in ovarian cancer: Expression profile and functional spectrum. Rna Biol. 2019. In print. [Google Scholar] [CrossRef]
- He, S.; Zhao, Y.; Wang, X.; Deng, Y.; Wan, Z.; Yao, S.; Shen, H. Up-regulation of long non-coding RNA SNHG20 promotes ovarian cancer progression via Wnt/β-catenin signaling. Biosci. Rep. 2018, 38, 1–8. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Wen, J.; Qi, T.; Wang, Y. Long non-coding RNA TTN-AS1 promotes tumorigenesis of ovarian cancer through modulating the miR-139-5p/ROCK2 axis. Biomed. Pharmacother. 2020, 125, 109882. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Zhang, Y.; Luo, C.; Zhu, T.; Zhang, R.; Yao, R. LINC01210 accelerates proliferation, invasion and migration in ovarian cancer through epigenetically downregulating KLF4. Biomed. Pharmacother. 2019, 119, 109431. [Google Scholar] [CrossRef]
- Li, R.; Wang, Y.; Xu, Y.; He, X.; Li, Y. Silencing the long noncoding RNA, TINCR, a molecular sponge of miR-335, inhibits the malignant phenotype of epithelial ovarian cancer via FGF2 suppression. Int. J. Oncol. 2019, 55, 1110–1124. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Yao, D.; Cui, D. LncSOX4 serves an oncogenic role in the tumorigenesis of epithelial ovarian cancer by promoting cell proliferation and inhibiting apoptosis. Mol. Med. Rep. 2018, 17, 8282–8288. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Wei, R.; Rodríguez, R.A.; Requena, M. LncRNA PCAT-1 plays an oncogenic role in epithelial ovarian cancer by modulating cyclinD1/CDK4 expression. Int. J. Clin. Exp. Pathol. 2019, 12, 2148–2156. [Google Scholar] [PubMed]
- Wang, L.; Yu, M.; Zhao, S. lncRNA MEG3 modified epithelial-mesenchymal transition of ovarian cancer cells by sponging miR-219a-5p and regulating EGFR. J. Cell. Biochem. 2019, 120, 17709–17722. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Wang, P.L.; Gao, Y.; Liang, W.T. Long noncoding RNA HOTTIP is a significant indicator of ovarian cancer prognosis and enhances cell proliferation and invasion. Cancer Biomark. 2019, 25, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Gao, H.; Li, X.; Zhu, Y.; Peng, S.; Yu, J.; Zhan, G.; Wang, J.; Liu, N.; Guo, X. LncRNA TPT1-AS1 promotes tumorigenesis and metastasis in epithelial ovarian cancer by inducing TPT1 expression. Cancer Sci. 2019, 110, 1587–1598. [Google Scholar] [CrossRef]
- Wang, L.; Ye, T.Y.; Wu, H.; Chen, S.Y.; Weng, J.R.; Xi, X.W. LINC00702 accelerates the progression of ovarian cancer through interacting with EZH2 to inhibit the transcription of KLF2. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 201–208. [Google Scholar] [CrossRef]
- Yan, H.; Li, H.; Li, P.; Li, X.; Lin, J.; Zhu, L.; Silva, M.A.; Wang, X.; Wang, P.; Zhang, Z. Long noncoding RNA MLK7-AS1 promotes ovarian cancer cells progression by modulating miR-375/YAP1 axis. J. Exp. Clin. Cancer Res. 2018, 37, 237. [Google Scholar] [CrossRef]
- Zhan, L.; Li, J.; Wei, B. Long non-coding RNAs in ovarian cancer. J. Exp. Clin. Cancer Res. 2018, 37, 120. [Google Scholar] [CrossRef]
- Chen, X.; Wu, W.; Cao, X.; Zhao, X.; Li, W.; Deng, C.; Huang, Z. lncRNA mortal obligate RNA transcript was downregulated in ovarian carcinoma and inhibits cancer cell proliferation by downregulating miRNA-21. J. Cell. Biochem. 2019, 120, 11949–11954. [Google Scholar] [CrossRef]
- Kong, F.R.; Lv, Y.H.; Yao, H.M.; Zhang, H.Y.; Zhou, Y.; Liu, S.E. LncRNA PCAT6 promotes occurrence and development of ovarian cancer by inhibiting PTEN. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8230–8238. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Jiang, H.; Cui, Z.; Wang, L.; Wang, X.; Tian, T. Linc-ROR induces epithelial-to-mesenchymal transition in ovarian cancer by increasing Wnt/β-catenin signaling. Oncotarget 2017, 8, 69983–69994. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fei, J.; Yu, S.; Shen, J.; Zhu, X.; Sadhukhan, A.; Lu, W.; Zhou, J. LINC01088 inhibits tumorigenesis of ovarian epithelial cells by targeting miR-24-1-5p. Sci. Rep. 2018, 8, 2876. [Google Scholar] [CrossRef]
- Zhang, S.; Li, J.; Wu, L.; Pei, M. Interaction between LncRNA-ROR and miR-145 contributes to epithelial-mesenchymal transition of ovarian cancer cells. Gen. Physiol. Biophys. 2019, 38, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhai, Z.; Guo, S.; Li, X.; Zhu, Y.; Wang, Y. Long non-coding RNA FLJ33360 participates in ovarian cancer progression by sponging miR-30b-3p. Onco. Targets. Ther. 2019, 12, 4469–4480. [Google Scholar] [CrossRef]
- Qu, C.; Dai, C.; Guo, Y.; Qin, R.; Liu, J. Long noncoding RNA SNHG15 serves as an oncogene and predicts poor prognosis in epithelial ovarian cancer. Onco. Targets. Ther. 2018, 12, 101–111. [Google Scholar] [CrossRef]
- Prat, J. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int. J. Gynecol. Obstet. 2014, 124, 1–5. [Google Scholar] [CrossRef]
- Chen, J.; Peng, X.; Dai, Y. The long non-coding RNA (lncRNA) AGAP2-AS1 is upregulated in ovarian carcinoma and negatively regulates lncRNA MEG3. Med. Sci. Monit. 2019, 25, 4699–4704. [Google Scholar] [CrossRef]
- Jiang, J.N.; Hong, Q.Y.; Gao, H.J.; Lai, B.L.; Lan, J.F.; Yang, Q. Lnc00908 promotes the development of ovarian cancer by regulating microRNA-49. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1388–1396. [Google Scholar] [CrossRef]
- Zhao, X.; Cheng, Z.; Wang, J. Long noncoding RNA FEZF1-AS1 promotes proliferation and inhibits apoptosis in ovarian cancer by activation of JAK-STAT3 pathway. Med. Sci. Monit. 2018, 24, 8088–8095. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Xu, P.; Xu, S. LncRNA RP11-597D13.9 expression and clinical significance in serous Ovarian Cancer based on TCGA database. Cancer Cell Res. 2018, 19, 484–488. [Google Scholar]
- Hou, B.; Hou, X.; Ni, H. Long non-coding RNA LNC01133 promotes the tumorigenesis of ovarian cancer by sponging miR-126. Int. J. Clin. Exp. Pathol. 2018, 11, 5809–5819. [Google Scholar] [PubMed]
- Ma, S.Y.; Wei, P.; Qu, F. KCNMA1-AS1 attenuates apoptosis of epithelial ovarian cancer cells and serves as a risk factor for poor prognosis of epithelial ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4629–4641. [Google Scholar] [CrossRef]
- Wang, J.; Tian, Y.; Zheng, H.; Ding, Y.; Wang, X. An integrated analysis reveals the oncogenic function of lncRNA LINC00511 in human ovarian cancer. Cancer Med. 2019, 8, 3026–3035. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, T.; Wang, C.; Wang, F. Sinomenine hydrochloride exerts antitumor outcome in ovarian cancer cells by inhibition of long non-coding RNA HOST2 expression. Artif. Cellsnanomedicinebiotechnol. 2019, 47, 4131–4138. [Google Scholar] [CrossRef]
- Qiu, J.; Lin, X.; Tang, X.; Zheng, T.; Zhang, X.; Hua, K. Long noncoding RNA TC0101441 induces epithelial–mesenchymal transition in epithelial ovarian cancer metastasis by downregulating KiSS1. Int. J. Cancer 2020, 146, 2588–2598. [Google Scholar] [CrossRef]
- Li, L.; Zhang, R.; Li, S.J. Long noncoding RNA SNHG14 promotes ovarian cancer cell proliferation and metastasis via sponging miR-219a-5p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4136–4142. [Google Scholar] [CrossRef]
- Guo, J.; Pan, H. Long noncoding RNA LINC01125 enhances cisplatin sensitivity of ovarian cancer via miR-1972. Med. Sci. Monit. 2019, 25, 9844–9854. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, C.; Liu, Y.; Zhang, G. Long noncoding RNA SNHG14 enhances migration and invasion of ovarian cancer by upregulating DGCR8. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10226–10233. [Google Scholar] [CrossRef]
- Liu, H.; Chen, R.; Kang, F.; Lai, H.; Wang, Y. KCNQ1OT1 promotes ovarian cancer progression via modulating MIR-142-5p / CAPN10 axis. Mol. Genet. Genom. Med. 2020, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.L.; Huang, Y.M. LncSNHG14 promotes ovarian cancer by targeting microRNA-125a-5p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3235–3242. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fang, Q.; Li, Y.; Wang, Y. Amplification of lncRNA PVT1 promotes ovarian cancer proliferation by binding to miR-140. Mamm. Genome 2019, 30, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Guan, W.; Ren, W.; Zhang, L.; Zhang, J.; Xu, G. MALAT1 affects ovarian cancer cell behavior and patient survival. Oncol. Rep. 2018, 39, 2644–2652. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiao, Y.; Hao, J.; Xing, H.; Li, C. Long noncoding RNA TP73-AS1 accelerates the epithelial ovarian cancer via epigenetically repressing p21. Am. J. Transl. Res. 2019, 11, 2447–2454. [Google Scholar]
- Zhang, Y.; Ruan, F. LncRNA LEF1-AS1 Promotes Ovarian Cancer Development Through Interacting with miR-1285-3p. Cancer Manag. Res. 2020, 12, 687–694. [Google Scholar] [CrossRef]
- Yao, N.; Yu, L.; Zhu, B.; Gan, H.Y.; Guo, B.Q. LncRNA GIHCG promotes development of ovarian cancer by regulating microRNA-429. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8127–8134. [Google Scholar] [CrossRef]
- Du, W.; Feng, Z.; Sun, Q. LncRNA LINC00319 accelerates ovarian cancer progression through miR-423-5p/NACC1 pathway. Biochem. Biophys. Res. Commun. 2018, 507, 198–202. [Google Scholar] [CrossRef]
- Shang, A.; Wang, W.; Gu, C.; Chen, C.; Zeng, B.; Yang, Y.; Ji, P.; Sun, J.; Wu, J.; Lu, W.; et al. Long non-coding RNA HOTTIP enhances IL-6 expression to potentiate immune escape of ovarian cancer cells by upregulating the expression of PD-L1 in neutrophils. J. Exp. Clin. Cancer Res. 2019, 38, 411. [Google Scholar] [CrossRef]
- Liu, Y.; He, X.; Chen, Y.; Cao, D. Long non-coding RNA LINC00504 regulates the Warburg effect in ovarian cancer through inhibition of miR-1244. Mol. Cell. Biochem. 2020, 464, 39–50. [Google Scholar] [CrossRef]
- Jing, L.; Gong, M.; Lu, X.; Jiang, Y.; Li, H.; Cheng, W. LINC01127 promotes the development of ovarian tumors by regulating the cell cycle. Am. J. Transl. Res. 2019, 11, 406–417. [Google Scholar] [PubMed]
- Fan, Y.; Wang, L.; Han, X.-C.; Ma, H.-Y.; Zhang, N.; Zhe, L. LncRNA MIF-AS1 aggravates the progression of ovarian cancer by sponging miRNA-31-5p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2248–2255. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, W.; Xu, Y.; Tao, E.; Mo, M.; Xu, W.; Cai, X.; Chen, X.; Yuan, J.; Wu, X. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany NY) 2020, 12, 4558–4572. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhan, X.; Zhan, X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol. Oncol. 2018, 150, 343–354. [Google Scholar] [CrossRef]
- Wang, S.; Li, G. LncRNA XIST inhibits ovarian cancer cell growth and metastasis via regulating miR-150-5p/PDCD4 signaling pathway. Naunyn. Schmiedebergs. Arch. Pharmacol. 2020. [Google Scholar] [CrossRef]
- Shen, X.; Zhu, W. Long non-coding RNA LINC01627 is a prognostic risk factor for epithelial ovarian cancer. Oncol. Lett. 2019, 18, 2861–2868. [Google Scholar] [CrossRef]
- Tong, L.; Wang, Y.; Ao, Y.; Sun, X. CREB1 induced lncRNA HAS2-AS1 promotes epithelial ovarian cancer proliferation and invasion via the miR-466/RUNX2 axis. Biomed. Pharmacother. 2019, 115, 108891. [Google Scholar] [CrossRef]
- Xue, F.; Xu, Y.H.; Shen, C.C.; Qin, Z.L.; Zhou, H. Bin Non-coding RNA LOXL1-AS1 exhibits oncogenic activity in ovarian cancer via regulation of miR-18b-5p/VMA21 axis. Biomed. Pharmacother. 2020, 125, 109568. [Google Scholar] [CrossRef]
- Zhu, D.; Huang, X.; Liang, F.; Zhao, L. LncRNA miR503HG interacts with miR-31-5p through multiple ways to regulate cancer cell invasion and migration in ovarian cancer. J. Ovarian Res. 2020, 13, 3. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, Y.; Fan, X.; Liu, Y.; Feng, Q. Upregulation of long non-coding RNA CCEPR is associated with poor prognosis and contributes to the progression of ovarian cancer through regulating the Wnt/β-catenin signaling pathway. Mol. Med. Rep. 2020, 21, 1950–1958. [Google Scholar] [CrossRef]
- Han, S.; Li, D.; Xiao, M. LncRNA ZFAS1 serves as a prognostic biomarker to predict the survival of patients with ovarian cancer. Exp. Ther. Med. 2019, 18, 4673–4681. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-Y.; Li, N.; Cui, Y.-L. Long non-coding RNA CCAT1 sponges miR-454 to promote chemoresistance of ovarian cancer cells to cisplatin by regulation of surviving. Cancer Res. Treat. 2020. In print. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Li, B.; Zhang, X.; Meng, X. NCK1-AS1 promotes NCK1 expression to facilitate tumorigenesis and chemo-resistance in ovarian cancer. Biochem. Biophys. Res. Commun. 2020, 522, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wu, J.; Fu, C.; Teng, F.; Liu, S.; Dai, C.; Shen, R.; Jia, X. Multidrug resistant lncRNA profile in chemotherapeutic sensitive and resistant ovarian cancer cells. J. Cell. Physiol. 2018, 233, 5034–5043. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Han, X.; Pan, Y.; Lan, Y.; Ning, J.; Zhao, Y.; Zhang, L. Long non-coding RNA CCAT2 regulates proliferation, drug sensitivity and metastasis of ovarian cancer. Arch. Med. Sci. 2019, 1–6. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, S.; Fu, C.; Wang, X.; Zhang, M.; Liu, G.; Dai, C.; Gong, Z.; Xu, H.; Fu, Z.; et al. LncRNA KB-1471A8.2 overexpression suppresses cell proliferation and migration and antagonizes the paclitaxel resistance of ovarian cancer cells. Cancer Biother. Radiopharm. 2019, 34, 316–324. [Google Scholar] [CrossRef]
- Wambecke, A.; Ahmad, M.; Lambert, B.; Joly, F.; Poulain, L.; Denoyelle, C.; Meryet-Figuiere, M. The influence of long non-coding RNAs on the response to chemotherapy in ovarian cancer. Gynecol. Oncol. 2020, 156, 726–733. [Google Scholar] [CrossRef]
- Xu, M.; Zhou, K.; Wu, Y.; Wang, L.; Lu, S. Linc00161 regulated the drug resistance of ovarian cancer by sponging microRNA-128 and modulating MAPK1. Mol. Carcinog. 2019, 58, 577–587. [Google Scholar] [CrossRef]
- Qu, J.; Kamal, M.A.; Yuan, C. The Regulatory Roles of Long Non-Coding RNA in the Chemoresistance Process of Ovarian cancer. Curr. Pharm. Des. 2019, 25, 856–861. [Google Scholar] [CrossRef]
- Li, Z.Y.; Wang, X.L.; Dang, Y.; Zhu, X.Z.; Zhang, Y.H.; Cai, B.X.; Zheng, L. Long non-coding RNA UCA1 promotes the progression of paclitaxel resistance in ovarian cancer by regulating the miR-654-5p/SIK2 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 591–603. [Google Scholar] [CrossRef]
- Zou, H.; Li, H. Knockdown of long non-coding RNA LINC00152 increases cisplatin sensitivity in ovarian cancer cells. Exp. Ther. Med. 2019, 18, 4510–4516. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, Y.; He, J.; Sun, H.; Jin, Z. Long non-coding RNA H19 mediates ovarian cancer cell cisplatin-resistance and migration during EMT. Int. J. Clin. Exp. Pathol. 2019, 12, 2506–2515. [Google Scholar] [PubMed]
- Özeş, A.R.; Wang, Y.; Zong, X.; Fang, F.; Pilrose, J.; Nephew, K.P. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci. Rep. 2017, 7, 894. [Google Scholar] [CrossRef] [PubMed]
- Nikpayam, E.; Tasharrofi, B.; Sarrafzadeh, S.; Ghafouri-Fard, S. The role of long non-coding RNAs in ovarian cancer. Iran. Biomed. J. 2017, 21, 3–15. [Google Scholar] [CrossRef]
- Tong, L.; Ao, Y.; Zhang, H.; Wang, K.; Wang, Y.; Ma, Q. Long noncoding RNA NORAD is upregulated in epithelial ovarian cancer and its downregulation suppressed cancer cell functions by competing with miR-155-5p. Cancer Med. 2019, 8, 4782–4791. [Google Scholar] [CrossRef]
- Yang, X.; Yan, Y.; Chen, Y.; Li, J.; Yang, J. Involvement of NORAD/miR-608/STAT3 axis in carcinostasis effects of physcion 8-O-β-glucopyranoside on ovarian cancer cells. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2855–2865. [Google Scholar] [CrossRef]
- Park, S.-A.; Kim, L.K.; Kim, H.J. Long non-coding RNA E2F4as promotes tumor progression and predicts patient prognosis in human ovarian cancer. Ann. Oncol. 2019, 30, v775. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03738319 (accessed on 20 November 2019).
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03742856 (accessed on 21 November 2019).
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef]
- DiStefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. In Disease Gene Identification. Methods in Molecular Biology; Springer: Berlin, Germany, 2018; pp. 91–110. [Google Scholar] [CrossRef]
- Wang, J.; Xu, W.; He, Y.; Xia, Q.; Liu, S. LncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN. Inflamm. Res. 2018, 67, 927–936. [Google Scholar] [CrossRef]
- Ye, W.; Ni, Z.; Yicheng, S.; Pan, H.; Huang, Y.; Xiong, Y.; Liu, T. Anisomycin inhibits angiogenesis in ovarian cancer by attenuating the molecular sponge effect of the lncRNA-Meg3/miR-421/PDGFRA axis. Int. J. Oncol. 2019, 55, 1296–1312. [Google Scholar] [CrossRef]
- Miao, S.; Wang, J.; Xuan, L.; Liu, X. LncRNA TTN-AS1 acts as sponge for miR-15b-5p to regulate FBXW7 expression in ovarian cancer. BioFactors 2020. In print. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Li, L.; Li, L.; Wang, D. Long non-coding RNA HAL suppresses the migration and invasion of serous ovarian cancer by inhibiting EMT signaling pathway. Biosci. Rep. 2020, 40, 28–29. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Zhang, X.; Zhao, Y.; Qian, H.; Wang, H.; He, C.; Liu, X.; Guo, T.; Lin, M.; Yu, H.; et al. Role of lncRNA-ATB in ovarian cancer and its mechanisms of action. Exp. Ther. Med. 2019, 19, 965–971. [Google Scholar] [CrossRef]
- Li, J.; Yang, C.; Li, Y.; Chen, A.; Li, L.; You, Z. LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci. Rep. 2018, 38, 1–11. [Google Scholar] [CrossRef]
- Yun, C.; Lee, S. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest. 2009, 119, 1429–1437. [Google Scholar] [CrossRef]
- Lu, Y.; Hu, Z.; Mangala, L.S.; Stine, Z.E.; Hu, X.; Jiang, D.; Xiang, Y.; Zhang, Y.; Pradeep, S.; Rodriguez-Aguayo, C.; et al. MYC Targeted Long Noncoding RNA DANCR Promotes Cancer in Part by Reducing p21 Levels. Cancer Res. 2018, 78, 64–74. [Google Scholar] [CrossRef]
- Zheng, X.; Zhou, Y.; Chen, W.; Chen, L.; Lu, J.; He, F.; Li, X.; Zhao, L. Ginsenoside 20(S)-Rg3 Prevents PKM2-Targeting miR-324-5p from H19 Sponging to Antagonize the Warburg Effect in Ovarian Cancer Cells. Cell. Physiol. Biochem. 2018, 51, 1340–1353. [Google Scholar] [CrossRef]
- Lin, X.; Yang, F.; Qi, X.; Li, Q.; Wang, D.; Yi, T.; Yin, R.; Zhao, X.; Zhong, X.; Bian, C. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol. Carcinog. 2019, 58, 2286–2296. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, J.; Pan, J.; Qing, Q.; Li, D.; Liao, J.; Sun, C.; Zhou, H. LncRNA HNF1A-AS1 promotes ovarian cancer growth by countering miR-214-mediated suppression of the sema 4D/plexin B1 pathway. Lancet 2019, 30. [Google Scholar] [CrossRef]
- Charbonneau, B.; Block, M.S.; Bamlet, W.R.; Vierkant, R.A.; Kalli, K.R.; Fogarty, Z.; Rider, D.N.; Sellers, T.A.; Tworoger, S.S.; Poole, E.; et al. Risk of Ovarian Cancer and the NF- B Pathway: Genetic Association with IL1A and TNFSF10. Cancer Res. 2014, 74, 852–861. [Google Scholar] [CrossRef]
- Zeng, X.; Jiang, X.-Y.; Yong, J.-H.; Xie, H.; Yuan, J.; Zeng, D.; Dou, Y.-Y.; Xiao, S.-S. lncRNA ABHD11-AS1, regulated by the EGFR pathway, contributes to the ovarian cancer tumorigenesis by epigenetically suppressing TIMP2. Cancer Med. 2019, 8, 7074–7085. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Su, H.; Tan, Y. UNC5B-AS1 promoted ovarian cancer progression by regulating the H3K27me on NDRG2 via EZH2. Cell Biol. Int. 2020, 44, 1028–1036. [Google Scholar] [CrossRef]
- Guo, L.; Wang, S. Downregulated Long Noncoding RNA GAS5 Fails to Function as Decoy of CEBPB, Resulting in Increased GDF15 Expression and Rapid Ovarian Cancer Cell Proliferation. Cancer Biother. Radiopharm. 2019, 34, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Niu, J.; Feng, Y. Knockdown of long non-coding RNA LINC00176 suppresses ovarian cancer progression by BCL3-mediated down-regulation of ceruloplasmin. J. Cell. Mol. Med. 2020, 24, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Drak Alsibai, K.; Vacher, S.; Meseure, D.; Nicolas, A.; Lae, M.; Schnitzler, A.; Chemlali, W.; Cros, J.; Longchampt, E.; Cacheux, W.; et al. High Positive Correlations between ANRIL and p16-CDKN2A/p15-CDKN2B/p14-ARF Gene Cluster Overexpression in Multi-Tumor Types Suggest Deregulated Activation of an ANRIL–ARF Bidirectional Promoter. Non-Coding Rna 2019, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Leng, T.; Zhang, Q.; Zhao, Q.; Nie, X.; Yang, L. Sanguinarine inhibits epithelial ovarian cancer development via regulating long non-coding RNA CASC2-EIF4A3 axis and/or inhibiting NF-κB signaling or PI3K/AKT/mTOR pathway. Biomed. Pharmacother. 2018, 102, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Qi, Y.; Wang, X.; Gu, J.; Shi, T. Down-regulation of lncRNA BLACAT1 inhibits ovarian cancer progression by suppressing the Wnt/β-catenin signaling pathway via regulating miR-519d-3p. Mol. Cell. Biochem. 2020, 467, 95–105. [Google Scholar] [CrossRef]
- Huang, K.; Fan, W.S.; Fu, X.Y.; Li, Y.L.; Meng, Y.G. Long noncoding RNA DARS-AS1 acts as an oncogene by targeting miR-532-3p in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2353–2359. [Google Scholar] [CrossRef]
- You, Q.; Shi, H.-Y.; Gong, C.-F.; Tian, X.-Y.; Li, S. Long non-coding RNA DLX6-AS1 acts as an oncogene by targeting miR-613 in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6429–6435. [Google Scholar] [CrossRef] [PubMed]
- Yiwei, T.; Hua, H.; Hui, G.; Mao, M.; Xiang, L. HOTAIR Interacting with MAPK1 Regulates Ovarian Cancer skov3 Cell Proliferation, Migration, and Invasion. Med. Sci. Monit. 2015, 21, 1856–1863. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, H.; Zhang, T.; Chu, Y.; Chen, D.; Zuo, J. miR-200c overexpression inhibits the invasion and tumorigenicity of epithelial ovarian cancer cells by suppressing lncRNA HOTAIR in mice. J. Cell. Biochem. 2020, 121, 1514–1523. [Google Scholar] [CrossRef]
- Dong, L.; Hui, L. HOTAIR promotes proliferation, migration, and invasion of ovarian cancer SKOV3 cells through regulating PIK3R3. Med. Sci. Monit. 2016, 22, 325–331. [Google Scholar] [CrossRef]
- Liu, H.; Liu, G.; Pang, W.; Zhang, H.; Zeng, Z.; Wang, H. LncRNA LUCAT1 promotes proliferation of ovarian cancer cells by regulating miR-199a-5p expression. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1682–1687. [Google Scholar] [CrossRef] [PubMed]
- Gokulnath, P.; De Cristofaro, T.; Manipur, I.; Di Palma, T.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long Non-Coding RNA MAGI2-AS3 is a New Player with a Tumor Suppressive Role in High Grade Serous Ovarian Carcinoma. Cancers 2019, 11, 2008. [Google Scholar] [CrossRef]
- Sun, Q.; Li, Q.; Xie, F. LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. Onco. Targets. Ther. 2019, 12, 6297–6307. [Google Scholar] [CrossRef]
- Zhou, S.; Xu, A.; Song, T.; Gao, F.; Sun, H.; Kong, X. lncRNA MIAT Regulates Cell Growth, Migration, and Invasion Through Sponging miR-150-5p in Ovarian Cancer. Cancer Biother. Radiopharm. 2020. In print. [Google Scholar] [CrossRef]
- Xu, C.; Zhu, L.-X.; Sun, D.-M.; Yao, H.; Han, D.-X. Regulatory mechanism of lncRNA NORAD on proliferation and invasion of ovarian cancer cells through miR-199a-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1672–1681. [Google Scholar] [CrossRef]
- Liang, H.; Yu, M.; Yang, R.; Zhang, L.; Zhang, L.; Zhu, D.; Luo, H.; Hong, Y.; Yu, T.; Sun, J.; et al. A PTAL-miR-101-FN1 Axis Promotes EMT and Invasion-Metastasis in Serous Ovarian Cancer. Mol. Ther. Oncolytics 2020, 16, 53–62. [Google Scholar] [CrossRef]
- Liang, H.; Yu, T.; Han, Y.; Jiang, H.; Wang, C.; You, T.; Zhao, X.; Shan, H.; Yang, R.; Yang, L.; et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol. Cancer 2018, 17, 119. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xin, N.; Qu, H.; Wei, L.; Han, Z. Long noncoding RNA TUG1 facilitates cell ovarian cancer progression through targeting MiR-29b-3p/MDM2 axis. Anat. Rec. 2020. In print. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, S.; Bai, X.; Pan, W.; Ai, L.; Tan, W. Long noncoding RNA WDFY3-AS2 suppresses tumor progression by acting as a competing endogenous RNA of microRNA-18a in ovarian cancer. J. Cell. Physiol. 2020, 235, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Spindler, T.J.; De Souza Fonseca, M.A.; Corona, R.I.; Seo, J.H.; Dezem, F.S.; Li, L.; Lee, J.M.; Long, H.W.; Sellers, T.A.; et al. Super-Enhancer-Associated LncRNA UCA1 Interacts Directly with AMOT to Activate YAP Target Genes in Epithelial Ovarian Cancer. iScience 2019, 17, 242–255. [Google Scholar] [CrossRef]
- Qiu, J.; Lin, X.; Zheng, T.; Tang, X.; Hua, K. Natural antisense transcript of hypoxia-inducible factor 1 regulates hypoxic cell apoptosis in epithelial ovarian cancer. Onco. Targets. Ther. 2018, 11, 9101–9110. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Li, J.; Han, L.; Roebuck, P.; Diao, L.; Liu, L.; Yuan, Y.; Weinstein, J.N.; Liang, H. TANRIC: An interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015, 75, 3728–3737. [Google Scholar] [CrossRef]
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef]
- Hou, M.; Tang, X.; Tian, F.; Shi, F.; Liu, F.; Gao, G. AnnoLnc: A web server for systematically annotating novel human lncRNAs. BMC Genom. 2016, 17, 931. [Google Scholar] [CrossRef] [PubMed]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.V.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015, 43, D168–D173. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Deb, A.; Maji, R.K.; Saha, S.; Ghosh, Z. LncRBase: An enriched resource for lncRNA information. PLoS ONE 2014, 9, e108010. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, T.; Cui, T.; Wang, Z.; Zhang, Y.; Tan, P.; Huang, Y.; Yu, J.; Wang, D. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2020, 48, D189–D197. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef] [PubMed]
- Vafaee, F.; Krycer, J.R.; Ma, X.; Burykin, T.; James, D.E.; Kuncic, Z. ORTI: An Open-access Repository of Transcriptional Interactions for interrogating mammalian gene expression data. PLoS ONE 2016, 11, e0164535. [Google Scholar] [CrossRef]
- Xiong, Y.; Wei, Y.; Gu, Y.; Zhang, S.; Lyu, J.; Zhang, B.; Chen, C.; Zhu, J.; Wang, Y.; Liu, H.; et al. DiseaseMeth version 2.0: A major expansion and update of the human disease methylation database. Nucleic Acids Res. 2017, 45, D888–D895. [Google Scholar] [CrossRef]
- Zhi, H.; Li, X.; Wang, P.; Gao, Y.; Gao, B.; Zhou, D.; Zhang, Y.; Guo, M.; Yue, M.; Shen, W.; et al. Lnc2Meth: A manually curated database of regulatory relationships between long non-coding RNAs and DNA methylation associated with human disease. Nucleic Acids Res. 2018, 46, D133–D138. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, P.; Tian, R.; Wang, S.; Guo, Q.; Luo, M.; Zhou, W.; Liu, G.; Jiang, H.; Jiang, Q. LncRNA2Target v2.0: A comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019, 47, D140–D144. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, G.; Li, J.; Zhang, X.; Huang, S.; Xiang, S.; Hu, X.; Liu, C. CRISPRlnc: A manually curated database of validated sgRNAs for lncRNAs. Nucleic Acids Res. 2019, 47, D63–D68. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Zhang, L.; Niu, Y.; Cai, T.; Luo, J.; He, S.; Zhang, B.; Zhang, D.; Qin, Y.; Yang, F.; et al. SmProt: A database of small proteins encoded by annotated coding and non-coding RNA loci. Brief. Bioinform. 2018, 19, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shi, J.; Zhang, Y.; Xie, A.; Yu, L.; Zhang, C.; Lei, J.; Xu, H.; Leng, Z.; Li, T.; et al. LncTarD: A manually-curated database of experimentally-supported functional lncRNA–target regulations in human diseases. Nucleic Acids Res. 2019, 48, D118–D126. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, L.; Wang, Z.; Pan, T.; Sahni, N.; Jin, X.; Wang, G.; Li, J.; Zheng, X.; Zhang, Y.; et al. LncMAP: Pan-cancer atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Res. 2018, 46, 1113–1123. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, P.; Wang, Y.; Ma, X.; Zhi, H.; Zhou, D.; Li, X.; Fang, Y.; Shen, W.; Xu, Y.; et al. Lnc2Cancer v2.0: Updated database of experimentally supported long non-coding RNAs in human cancers. Nucleic Acids Res. 2019, 47, D1028–D1033. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Chen, W.; Li, J.; Liu, C. CRlncRNA: A manually curated database of cancer-related long non-coding RNAs with experimental proof of functions on clinicopathological and molecular features. BMC Med. Genom. 2018, 11, 114. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhou, W.; Han, Y.; Peng, F.; Wang, R.; Yu, R.; Wang, C.; Liang, H.; Guo, Z.; Gu, Y. EMT-Regulome: A database for EMT-related regulatory interactions, motifs and network. Cell Death Dis. 2017, 8, e2872. [Google Scholar] [CrossRef]
- Wang, C.; Yang, F.; Chen, T.; Dong, Q.; Zhao, Z.; Liu, Y.; Chen, B.; Liang, H.; Yang, H.; Gu, Y. RHPCG: A database of the Regulation of the Hippo Pathway in Cancer Genome. Database 2019, 2019, 1–10. [Google Scholar] [CrossRef]
- Muppirala, U.K.; Honavar, V.G.; Dobbs, D. Predicting RNA-Protein Interactions Using Only Sequence Information. BMC Bioinform. 2011, 12, 489. [Google Scholar] [CrossRef]
- Suresh, V.; Liu, L.; Adjeroh, D.; Zhou, X. RPI-Pred: Predicting ncRNA-protein interaction using sequence and structural information. Nucleic Acids Res. 2015, 43, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Ren, S.; Lu, M.; Zhang, Y.; Zhu, D.; Zhang, X.; Li, T. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genom. 2013, 14, 651. [Google Scholar] [CrossRef] [PubMed]
- Agostini, F.; Zanzoni, A.; Klus, P.; Marchese, D.; Cirillo, D.; Tartaglia, G.G. catRAPID omics: A web server for large-scale prediction of protein-RNA interactions. Bioinformatics 2013, 29, 2928–2930. [Google Scholar] [CrossRef]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.-C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, T.; Iwakiri, J.; Ono, Y.; Hamada, M. LncRRIsearch: A Web Server for lncRNA-RNA Interaction Prediction Integrated With Tissue-Specific Expression and Subcellular Localization Data. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhang, H.; Liu, H.; Zhu, H. LongTarget: A tool to predict lncRNA DNA-binding motifs and binding sites via Hoogsteen base-pairing analysis. Bioinformatics 2015, 31, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Lou, U.K.; Wong, C.H.; Chen, Y. A simple and rapid colorimetric detection of serum lncRNA biomarkers for diagnosis of pancreatic cancer. RSC Adv. 2020, 10, 8087–8092. [Google Scholar] [CrossRef]
- Islam, M.N.; Moriam, S.; Umer, M.; Phan, H.-P.; Salomon, C.; Kline, R.; Nguyen, N.-T.; Shiddiky, M.J.A. Naked-eye and electrochemical detection of isothermally amplified HOTAIR long non-coding RNA. Analyst 2018, 143, 3021–3028. [Google Scholar] [CrossRef]
- Guo, B.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Wang, Y.; Zhang, S.; Wu, R.; Lu, J.; et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J. 2020, 39, e102190. [Google Scholar] [CrossRef]
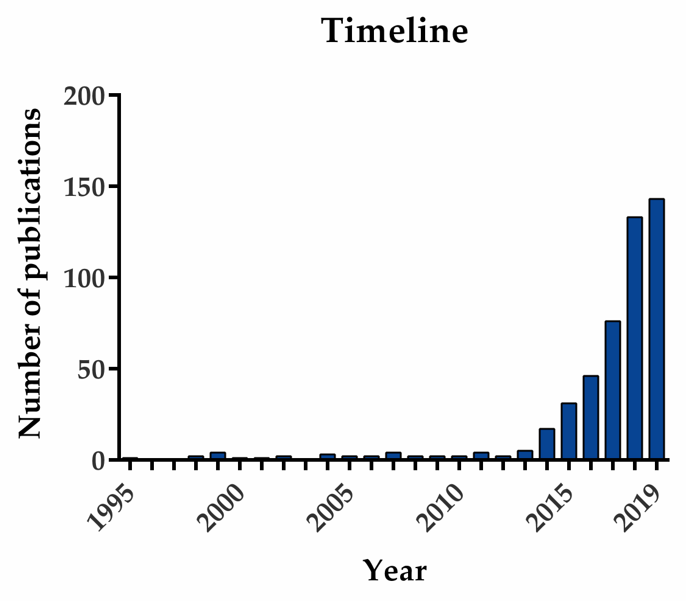
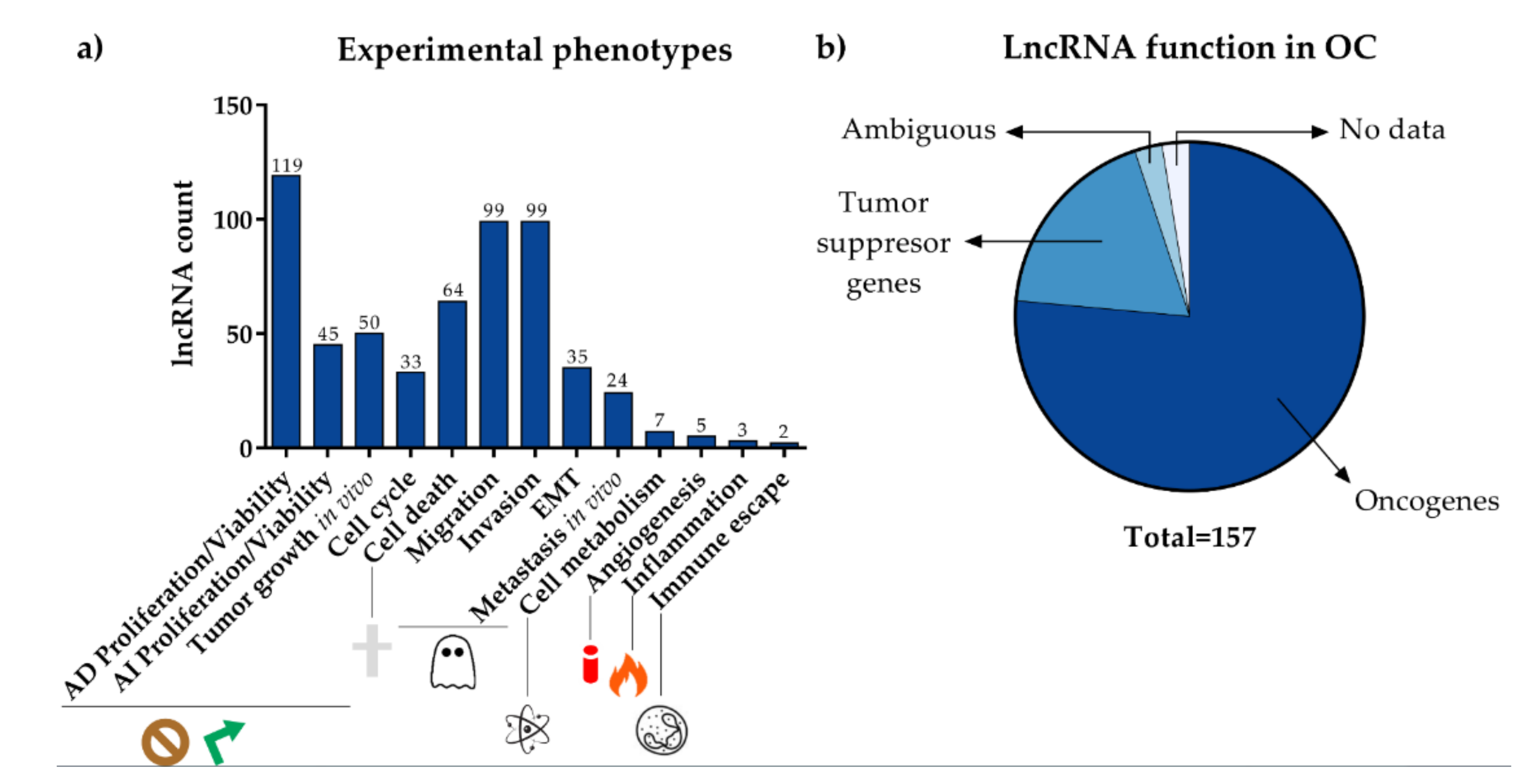
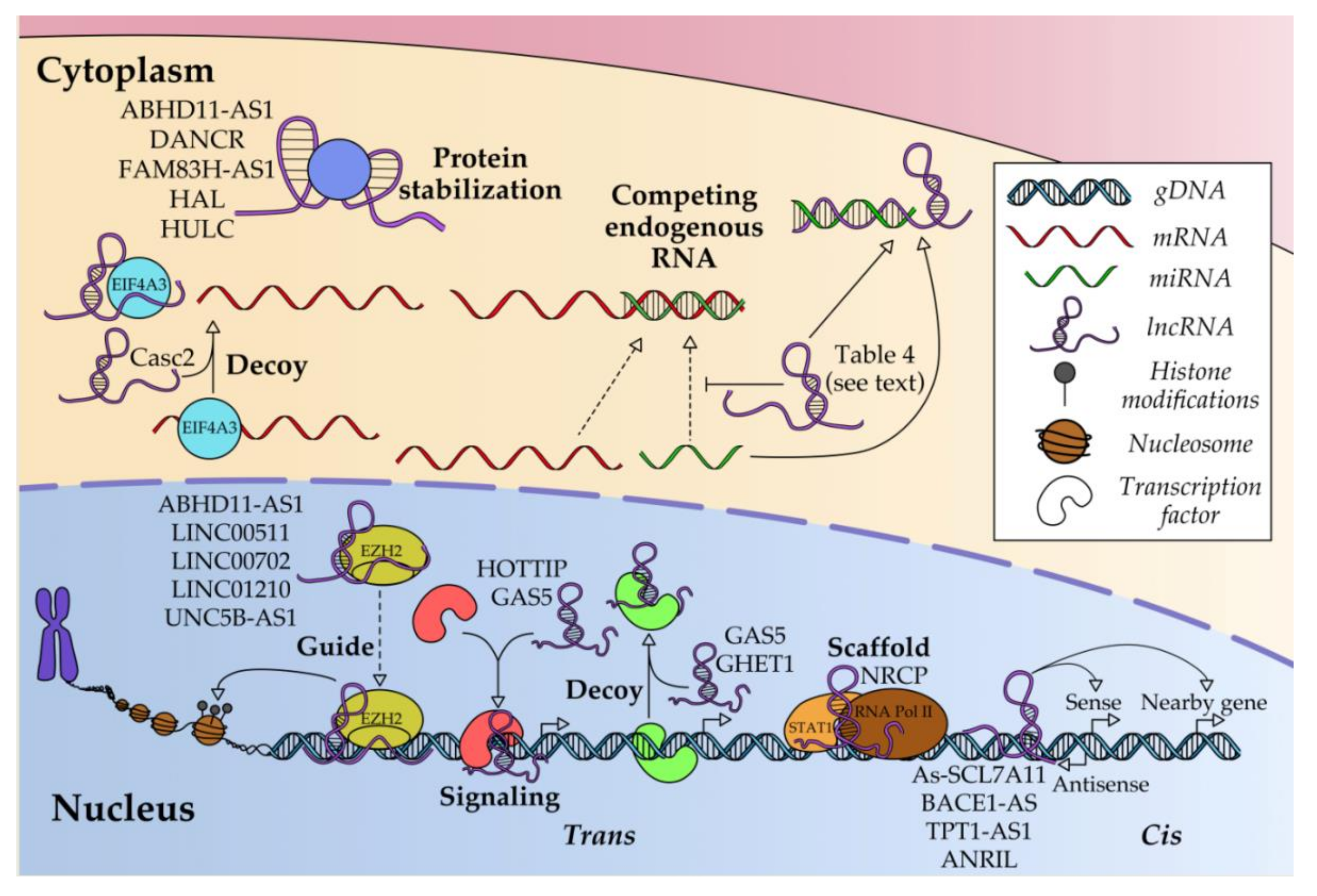
| Tumor Size | Histological Dedifferentiation Grade | Lymph Node Metastasis | Distant Metastasis | ||||
|---|---|---|---|---|---|---|---|
| LncRNA | Ref. | LncRNA | Ref. | LncRNA | Ref. | LncRNA | Ref. |
| AB073614 | [9] | ABHD11-AS1 | [10] | AB073614 | [9] | DANCR | [11] |
| Casc15 | [12] | aHIF | [13] | AC093818.1 | [14] | FAM83H-AS1 | [15,16] |
| Casc2 | [17] | ANRIL | [6,18] | ADAMTS9-AS2 | [19] | FEZF1-AS1 | [20] |
| DUXAP10 | [6] | ASAP1-IT1 | [21] | AK130076 | [14] | GAS5 | [22] |
| EPB41L4A-AS2 | [23] | Casc15 | [12] | ANRIL | [6] | GHET1 | [24] |
| FAM83H-AS1 | [15] | Casc2 | [17] | Casc2 | [17] | JPX | [25] |
| GAS5 | [22,26] | CCAT1 | [6] | CCAT1 | [6] | LINC01210 | [27] |
| GHET1 | [24] | CCAT2 | [6] | CCAT2 | [6] | LncSOX4 | [28] |
| HAGLROS | [29] | CPS1-IT1 | [6] | CPS1-IT1 | [6] | LUCAT1 | [30] |
| HOTAIR | [31] | DLEU1 | [32] | DLX6-AS1 | [33] | MAGI1-IT1 | [34] |
| JPX | [25] | DUXAP10 | [6] | EIBC | [6] | MALAT1 | [6] |
| KCNQ1OT1 | [35] | EIBC | [6] | FAM83H-AS1 | [15] | MCM3AP-AS1 | [36] |
| LINC00565 | [37] | EPB41L4A-AS2 | [23] | FAS-AS1 | [14] | MEG3 | [38] |
| LINC00702 | [39] | FAM215A | [21] | FEZF1-AS1 | [20] | MLK7-AS1 | [40] |
| LncSOX4 | [28] | FAM83H-AS1 | [16] | FLVCR1-AS1 | [41] | NEAT1 | [42] |
| MORT | [43] | FEZF1-AS1 | [20] | GAS5 | [6] | PCAT6 | [44] |
| PCAT-1 | [45] | GAS5 | [6,46] | GTSE1-AS1 | [14] | SNHG12 | [47] |
| RP11-135L22.1 | [48] | HOTAIR | [21] | HMMR-AS1 | [49] | SNHG20 | [50] |
| TINCR | [51] | HOXA11-AS | [42] | HOTTIP | [52] | TPT1-AS1 | [53] |
| TPT1-AS1 | [53] | KCNQ1OT1 | [35] | HOXD-AS1 | [6] | XIST | [54] |
| LINC00339 | [55] | LINC00515 | [56] | ||||
| LINC00472 | [42] | Linc-ROR | [57] | ||||
| LINC01088 | [58] | lncARSR | [6] | ||||
| Linc-ROR | [59] | lncBRM | [6] | ||||
| lncARSR | [6] | Lnc-OC1 | [6] | ||||
| lncBRM | [6] | MCM3AP-AS1 | [36] | ||||
| Lnc-OC1 | [6] | MIR22HG | [6] | ||||
| MEG3 | [38] | MLK7-AS1 | [40] | ||||
| MIR22HG | [6] | MNX1-AS1 | [6] | ||||
| MNX1-AS1 | [6] | NBAT-1 | [6] | ||||
| NBAT-1 | [6] | NEAT1 | [6] | ||||
| NEAT1 | [31] | NONHSAT076754 | [60] | ||||
| NONHSAT076754 | [60] | PCAT6 | [44] | ||||
| SNHG20 | [61] | RP11-199F11.2 | [14] | ||||
| SNHG5 | [62] | SNHG20 | [61] | ||||
| SOX2OT | [6] | SNHG3 | [63] | ||||
| SPRY4-IT1 | [6] | SNHG5 | [62] | ||||
| TC0100223 | [21] | SOCAR | [64] | ||||
| TC0101441 | [21] | SOX2OT | [6] | ||||
| TPT1-AS1 | [53] | SPRY4-IT1 | [6] | ||||
| TUBA4B | [6] | TC0100223 | [42] | ||||
| XIST | [54] | TC0101441 | [42] | ||||
| TINCR | [51] | ||||||
| TTN-AS1 | [65] | ||||||
| TUBA4B | [6] | ||||||
| UCA1 | [21] | ||||||
| Overall Survival | |||||||
|---|---|---|---|---|---|---|---|
| LncRNA | Ref. | LncRNA | Ref. | LncRNA | Ref. | LncRNA | Ref. |
| AB073614 | [6,9] | FAM83H-AS1 | [15,16] | LINC00504 | [70] | MLK7-AS1 | [40] |
| AC104699.1.1 | [42] | FEZF1-AS1 | [20,71] | LINC00511 | [72] | MNX1-AS1 | [6] |
| ADAMTS9-AS2 | [19] | FLVCR1-AS1 | [41] | LINC00565 | [37] | NEAT1 | [6] |
| aHIF | [13] | GAS5 | [22,31,46] | LINC01125 | [73] | PVT1 | [74] |
| AK021924 | [42] | GIHCG | [75] | LINC01127 | [76] | RHPN1-AS1 | [77] |
| AK094536 | [42] | H19 | [42] | LINC01210 | [27] | RP11-135L22.1 | [48] |
| ANRIL | [6] | HAGLROS | [29] | LINC01627 | [78] | RP11-284N8.3.1 | [21] |
| ASAP1-IT1 | [42] | HAS2-AS1 | [79] | LNC00908 | [80] | RP11-597D13.9 | [81] |
| BC004123 | [42] | HMMR-AS1 | [49] | LNC01133 | [82] | RUNX1-IT1 | [42] |
| BC007937 | [42] | HOST2 | [83] | lncARSR | [6] | SNHG12 | [47] |
| BC037530 | [42] | HOTAIR | [21] | lncBRM | [6] | SNHG14 | [84,85,86] |
| BC062365 | [42] | HOTAIRM1 | [42] | lnc-HRCT1-1 | [42] | SNHG15 | [67] |
| Casc15 | [12] | HOTTIP | [52,87] | Lnc-OC1 | [6] | SNHG20 | [61] |
| Casc2 | [17] | HOXA11-AS | [6] | lnc-SERTAD2-3 | [42] | SNHG3 | [63,88] |
| CCAT1 | [6] | HOXD-AS1 | [6] | LOC100190986 | [42] | SNHG5 | [62] |
| CCAT2 | [6] | JPX | [25] | LOXL1-AS1 | [89] | SPRY4-IT1 | [42] |
| CCEPR | [90] | KCNMA1-AS1 | [91] | LUCAT1 | [30] | TC0101441 | [42,92] |
| DLX6-AS1 | [33] | KCNQ1OT1 | [35,93] | MALAT1 | [94] | TP73-AS1 | [95] |
| DUXAP10 | [42] | LEF1-AS1 | [96] | MEG3 | [38] | TTN-AS1 | [65] |
| EIBC | [6] | LINC00319 | [97] | MIF-AS1 | [98] | UCA1 | [26] |
| EPB41L4A-AS2 | [23] | LINC00339 | [55] | MIR22HG | [6] | XIST | [54,99] |
| FAM215A | [42] | LINC00472 | [21] | miR503HG | [100] | ZFAS1 | [101] |
| Disease-Free Survival | Progression-Free Survival | ||||
|---|---|---|---|---|---|
| LncRNA | Ref. | LncRNA | Ref. | LncRNA | Ref. |
| DLX6-AS1 | [33] | ANRIL | [6] | lnc-HRCT1-1 | [42] |
| GAS5 | [6,46] | Casc15 | [12] | Lnc-OC1 | [6] |
| H19 | [42] | Casc2 | [17] | lnc-SERTAD2-3 | [42] |
| HOTAIR | [21] | CCAT1 | [6] | MALAT1 | [94] |
| HOTAIRM1 | [42] | CCAT2 | [6] | MIR22HG | [6] |
| LINC01210 | [27] | EIBC | [6] | MNX1-AS1 | [6] |
| LOC100190986 | [42] | FLJ33360 | [66] | NEAT1 | [6] |
| MALAT1 | [21] | HOXA11-AS | [6] | SNHG15 | [67] |
| RUNX1-IT1 | [42] | HOXD-AS1 | [6] | SPRY4-IT1 | [6] |
| TC0101441 | [92] | lncARSR | [6] | UCA1 | [26] |
| ZFAS1 | [101] | lncBRM | [6] | XIST | [42,54] |
| Platinum Salts | Taxanes | ||||||
|---|---|---|---|---|---|---|---|
| + | – | + | – | ||||
| LncRNA | Ref. | LncRNA | Ref. | LncRNA | Ref. | LncRNA | Ref. |
| ANRIL | [18] | BC200 | [21] | CTD-2589M5.4* | [102] | KB-1471A8.2 | [103] |
| CCAT1 | [104] | GAS5 | [46] | FER1L4 | [105] | SNHG5 | [62] |
| CCAT2 | [106] | Linc00312 | [6] | NEAT1 | [105] | XIST | [26] |
| CTD-2589M5.4* | [102] | LINC00515 | [21] | PVT1 | [105] | ||
| DNM3OS | [107] | LINC01125 | [73] | UCA1 | [105,108] | ||
| ENST00000457645 | [6] | linc-TNFRSF19-1 | [21] | ||||
| FER1L4 | [105] | MEG3 | [42] | ||||
| H19 | [6,109] | RP11-135L22.1 | [48] | ||||
| HOTAIR | [105,110] | XIST | [111] | ||||
| LINC00152 | [112] | ||||||
| Linc00161 | [113] | ||||||
| LINC00961 | [21] | ||||||
| linc-CARS2-2 | [21] | ||||||
| linc-RECK-3 | [21] | ||||||
| LUCAT1 | [21] | ||||||
| MALAT1 | [107] | ||||||
| NCK1-AS1 | [114] | ||||||
| NEAT1 | [105] | ||||||
| PVT1 | [107] | ||||||
| SNHG15 | [67] | ||||||
| UCA1 | [105] | ||||||
| ZFAS1 | [6] | ||||||
| lncRNA | miRNA | mRNA | Ref. |
|---|---|---|---|
| ADAMTS9-AS2 | miR-182-5p | FOXF2 | [19] |
| ANRIL | let-7a | HMGA2 | [18] |
| BLACAT1 | miR-519d-3p | RPS15A | [142] |
| Casc15 | miR-221 | ARID1A | [12] |
| CCAT1 | miR-454 | Survivin | [104] |
| miR-1290 | - | [6] | |
| miR-130b | STAT3, ZEB1 | ||
| miR-152 | ADAM17, WNT1 | ||
| CCAT2 | miR-424 | - | [6] |
| DANCR | miR-145 | VEGF | [133] |
| DARS-AS1 | miR-532-3p | - | [143] |
| DLEU1 | miR-490-3p | - | [32] |
| DLX6-AS1 | miR-613 | - | [144] |
| EPB41L4A-AS2 | miR-103a | RUNX1T1 | [23] |
| EWSAT1 | miR-330-5p | Pdia3 | [6] |
| FEZF1-AS1 | miR-130a-5p | SOX4 | [20] |
| FLJ33360 | miR-30b-3p | - | [66] |
| FLVCRA1-AS1 | miR-513 | YAP1 | [41] |
| GAS5 | miR-196a-5p | HOXA5 | [22] |
| miR-21 | SPRY2 | [6] | |
| H19 | let-7 | HMGA2, c-MYC, IGF2BP | [31] |
| miR-324-5p | PKM2 | [132] | |
| miR-370-3p | TGF-B | [26] | |
| HAS2-AS1 | miR-466 | RUNX2 | [79] |
| HNF1A-AS1 | miR-214 | SEMA4D, PlexinB1, Tiam1, Rac/1/2/3 | [134] |
| HOST2 | let-7b | HMGA2, c-Myc, Dicer, Imp3 | [31] |
| HOTAIR | miR-1 | MAPK1 | [145] |
| miR-200c | - | [146] | |
| mir-214(-3p) | MAPK1, (PIK3R3) | [145,147] | |
| miR-217 | PIK3R3 | [147] | |
| miR-330-5p | MAPK1 | [145] | |
| miR-373 | Rab22a | [6] | |
| HOXD-AS1 | miR-133a-3p | - | [6] |
| miR-186-5p | PI3KR3 | [26] | |
| HOXD-AS1 | miR-608 | FZD4 | [26] |
| KCNQ1OT1 | miR-142-5p | CAPN10 | [93] |
| miR-212-3p | LCN2 | [35] | |
| LINC00152 | miR-125b | MCL-1 | [6] |
| Linc00161 | miR-194 | MAPK1 | [113] |
| LINC00319 | miR-423-5p | NACC1 | [97] |
| LINC00339 | miR-148a-3p | ROCK1 | [55] |
| LINC00504 | miR-1244 | - | [70] |
| LINC01088 | miR-24-1-5p | PAK4 | [58] |
| LINC01125 | miR-1972 | - | [73] |
| Linc-ROR | miR-145 | FLNB | [59] |
| LNC00908 | miR-495-5p | ANXA3 | [80] |
| LNC01133 | miR-126 | - | [82] |
| lncARSR | miR-200c | ZEB1, ZEB2 | [6] |
| lncBRM | miR-204 | - | [6] |
| Lnc-OC1 | miR-34a | - | [6] |
| miR-34c | - | [6] | |
| LOXL1-AS1 | miR-18b-5p | VMA21 | [89] |
| LUCAT1 | miR-612 | HOXA13 | [30] |
| miR-199a-5p | - | [148] | |
| MAGI1-IT1 | miR-200a | ZEB1, ZEB2 | [34] |
| MAGI2-AS3 | miR-15b miR-374a miR-374b | HOXA5, MTSS1, PTEN, RECK | [149] |
| MALAT1 | miR-143-3p | CMPK | [94] |
| miR-200c | - | [6] | |
| miR-211 | PHF19 | [6] | |
| miR-503-5p | pJak2, pSTAT3 | [150] | |
| miR-506 | iASPP | [6] | |
| MCM3AP-AS1 | miR-28-5p | - | [36] |
| MIAT | miR-150-5p | - | [151] |
| MIF-AS1 | miR-315p | PLCB1 | [98] |
| MEG3 | miR-214 | - | [107] |
| miR-219a-5p | EGFR | [38] | |
| miR-421 | PDGFRA, NOTCH1, HES1, RBPJ | [123] | |
| MLK7-AS1 | miR-375 | YAP1 | [40] |
| NCK1-AS1 | miR-137 | NCK1 | [114] |
| NEAT1 | miR-124 | - | [21] |
| miR-194 | ZEB1 | [105] | |
| miR-382-3p | ROCK1 | [6] | |
| miR-506 | RAD51 | [105] | |
| NORAD | miR-155-5p | - | [115] |
| miR-199a-3p | - | [152] | |
| miR-608 | STAT3 | [116] | |
| PCA3 | miR-106b | RhoC, Bcl/xL, P70S6K, MMP2 | [6] |
| PTAF | miR-25 | SNAI2 | [6] |
| PTAL/AC004988.1 | miR-101 | FN1 | [153] |
| PTAR | miR-101-3p | ZEB1 | [154] |
| PVT1 | miR-133a | - | [6] |
| miR-140 | [74] | ||
| RHPN1-AS1 | miR-596-3p | LETM1 | [77] |
| SNHG12 | miR-129 | SOX4 | [47] |
| SNHG14 | miR-125a-5p | DHX33 | [86] |
| miR-219a-5p | - | [84] | |
| SNHG3 | miR-186a-5p miR-590-3p | - | [88] |
| SNHG5 | miR-23a | - | [62] |
| TDRG1 | miR-93 | RhoC, P70S6K, Bcl-xL, MMP2 | [6] |
| TINCR | miR-335 | FGF2 | [51] |
| TTN-AS1 | miR-139-5p | ROCK2 | [65] |
| miR-15b-5p | FBXW7 | [124] | |
| TUG1 | miR-29b-3p | MDM2 | [155] |
| UCA1 | miR-129 | ABCB1 | [105] |
| miR-485-5p | MMP14 | [21] | |
| miR-654-5p | SIK2 | [108] | |
| WDFY3-AS2 | miR-18a | RORA | [156] |
| XIST | miR-150-5p | PDCD4 | [99] |
| miR-214-3p | PTEN | [105] | |
| ZFAS1 | miR-150-5p | Sp1 | [6] |
| Name | Description | Link | Ref. |
|---|---|---|---|
| GEPIA 2 | Gene Expression Profiling Interactive Analysis | http://gepia2.cancer-pku.cn/#index | [159] |
| CCLE | Cancer Cell Line Encyclopedia | https://portals.broadinstitute.org/ccle | [160] |
| TANRIC | The Atlas of non-coding RNA in Cancer | https://ibl.mdanderson.org/tanric/_design/basic/main.html | [161] |
| LNCipedia | Database for lncRNA | https://lncipedia.org/ | [162] |
| NONCODE | Database dedicated to ncRNA, especially lncRNA | http://www.noncode.org/ | [163] |
| AnnoLnc | Database | http://annolnc.cbi.pku.edu.cn/index.jsp | [164] |
| lncRNAdb | Database | http://lncrnadb.org/ | [165] |
| lncRBase | Database | http://bicresources.jcbose.ac.in/zhumur/lncrbase/ | [166] |
| RNAInter | RNA interactome database | http://www.rna-society.org/rnainter | [167] |
| StarBase-ENCORI | The Encyclopedia of RNA Interactomes | http://starbase.sysu.edu.cn | [168] |
| miRTarBase | microRNA-target interactions database | http://mirtarbase.mbc.nctu.edu.tw/php/index.php | [169] |
| ORTI | Open-access Repository of Transcriptional Interaction | http://orti.sydney.edu.au/index.html | [170] |
| DiseaseMeth | Human disease methylation database | http://bio-bigdata.hrbmu.edu.cn/diseasemeth/ | [171] |
| Lnc2Meth | Relationships between lncRNAs and DNA methylation | http://bio-bigdata.hrbmu.edu.cn/Lnc2Meth/index.jsp | [172] |
| LncRNA2Target | Database of experiments focused on lncRNA in human and mouse | http://123.59.132.21/lncrna2target/ | [173] |
| CRISPRlnc | Validated CRISPR/Cas9 sgRNAs for lncRNAs from all species | http://www.crisprlnc.org/ | [174] |
| SmProt | Small Proteins (< 100 aa) especially encoded by non-coding RNAs | http://bioinfo.ibp.ac.cn/SmProt/index.htm | [175] |
| LncTarD | Database for functional lncRNA-target regulation in human diseases | http://biocc.hrbmu.edu.cn/LncTarD/ | [176] |
| LncRNADisease | The LncRNA and Disease Database | http://www.rnanut.net/lncrnadisease/ | [177] |
| LncMAP | LncRNA Modulator Atlas in Pan-cancer | http://bio-bigdata.hrbmu.edu.cn/LncMAP/ | [178] |
| Lnc2Cancer | Experimentally supported associations between lncRNA and human cancer. | http://www.bio-bigdata.com/lnc2cancer/ | [179] |
| CRlncRNA | Cancer-related lncRNA Database | http://crlnc.xtbg.ac.cn | [180] |
| EMT-Regulome | Database for EMT-related regulatory interactions, motifs and network | http://www.medsysbio.org/EMTRegulome | [181] |
| RHPCG | Regulation of the Hippo Pathway in Cancer Genome | http://www.medsysbio.org/RHPCG | [182] |
| Name | Description | Link | Ref. |
|---|---|---|---|
| RPISeq | RNA-Protein Interaction Prediction | http://pridb.gdcb.iastate.edu/RPISeq/ | [183] |
| RPI-Pred | RNA-protein interaction prediction server | http://ctsb.is.wfubmc.edu/projects/rpi-pred/ | [184] |
| lncPro | Prediction of lncRNA-protein interactions | http://bioinfo.bjmu.edu.cn/lncpro/ | [185] |
| catRAPID | Algorithm to estimate the binding propensity of protein-RNA pairs | http://s.tartaglialab.com/page/catrapid_group | [186] |
| mirDIP | microRNA Data Integration Portal | http://ophid.utoronto.ca/mirDIP/ | [187] |
| LncRRISearch | lncRNA-RNA interaction prediction | http://rtools.cbrc.jp/LncRRIsearch/ | [188] |
| LongTarget & LongMan | Predict a lncRNA’s DNA binding motifs and binding sites, locally or at genome-wide scale | http://lncrna.smu.edu.cn/ | [189] |
| RNAfold web server | Predicts RNA secondary structures | http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi | [190] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salamini-Montemurri, M.; Lamas-Maceiras, M.; Barreiro-Alonso, A.; Vizoso-Vázquez, Á.; Rodríguez-Belmonte, E.; Quindós-Varela, M.; Cerdán, M.E. The Challenges and Opportunities of LncRNAs in Ovarian Cancer Research and Clinical Use. Cancers 2020, 12, 1020. https://doi.org/10.3390/cancers12041020
Salamini-Montemurri M, Lamas-Maceiras M, Barreiro-Alonso A, Vizoso-Vázquez Á, Rodríguez-Belmonte E, Quindós-Varela M, Cerdán ME. The Challenges and Opportunities of LncRNAs in Ovarian Cancer Research and Clinical Use. Cancers. 2020; 12(4):1020. https://doi.org/10.3390/cancers12041020
Chicago/Turabian StyleSalamini-Montemurri, Martín, Mónica Lamas-Maceiras, Aida Barreiro-Alonso, Ángel Vizoso-Vázquez, Esther Rodríguez-Belmonte, María Quindós-Varela, and María Esperanza Cerdán. 2020. "The Challenges and Opportunities of LncRNAs in Ovarian Cancer Research and Clinical Use" Cancers 12, no. 4: 1020. https://doi.org/10.3390/cancers12041020
APA StyleSalamini-Montemurri, M., Lamas-Maceiras, M., Barreiro-Alonso, A., Vizoso-Vázquez, Á., Rodríguez-Belmonte, E., Quindós-Varela, M., & Cerdán, M. E. (2020). The Challenges and Opportunities of LncRNAs in Ovarian Cancer Research and Clinical Use. Cancers, 12(4), 1020. https://doi.org/10.3390/cancers12041020







