BNIP3L-Dependent Mitophagy Promotes HBx-Induced Cancer Stemness of Hepatocellular Carcinoma Cells via Glycolysis Metabolism Reprogramming
Abstract
1. Introduction
2. Results
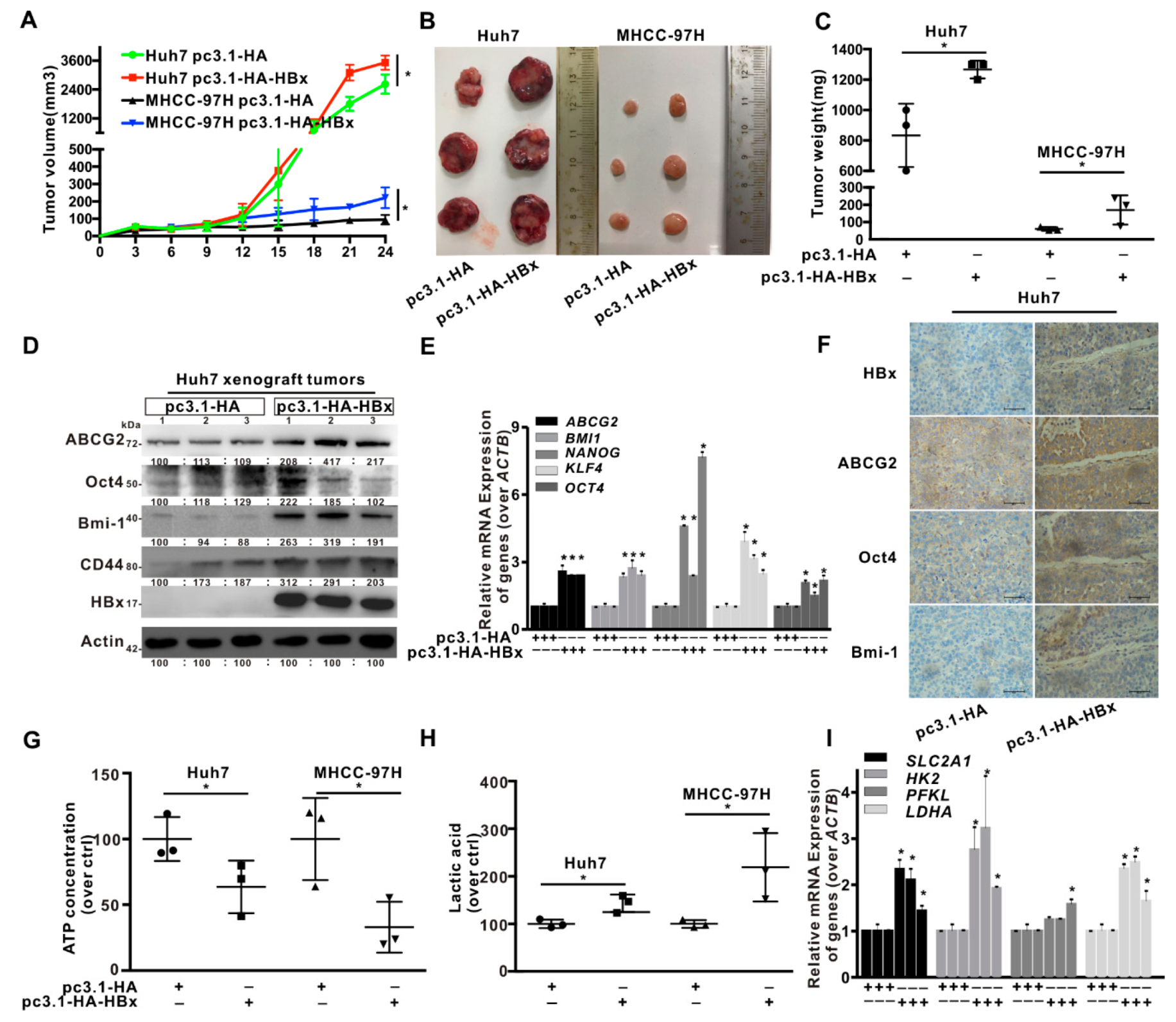
2.1. HBx Promoted HCC Cells Xenograft Tumors Growth via Upregulated Glycolytic Metabolism In Vivo
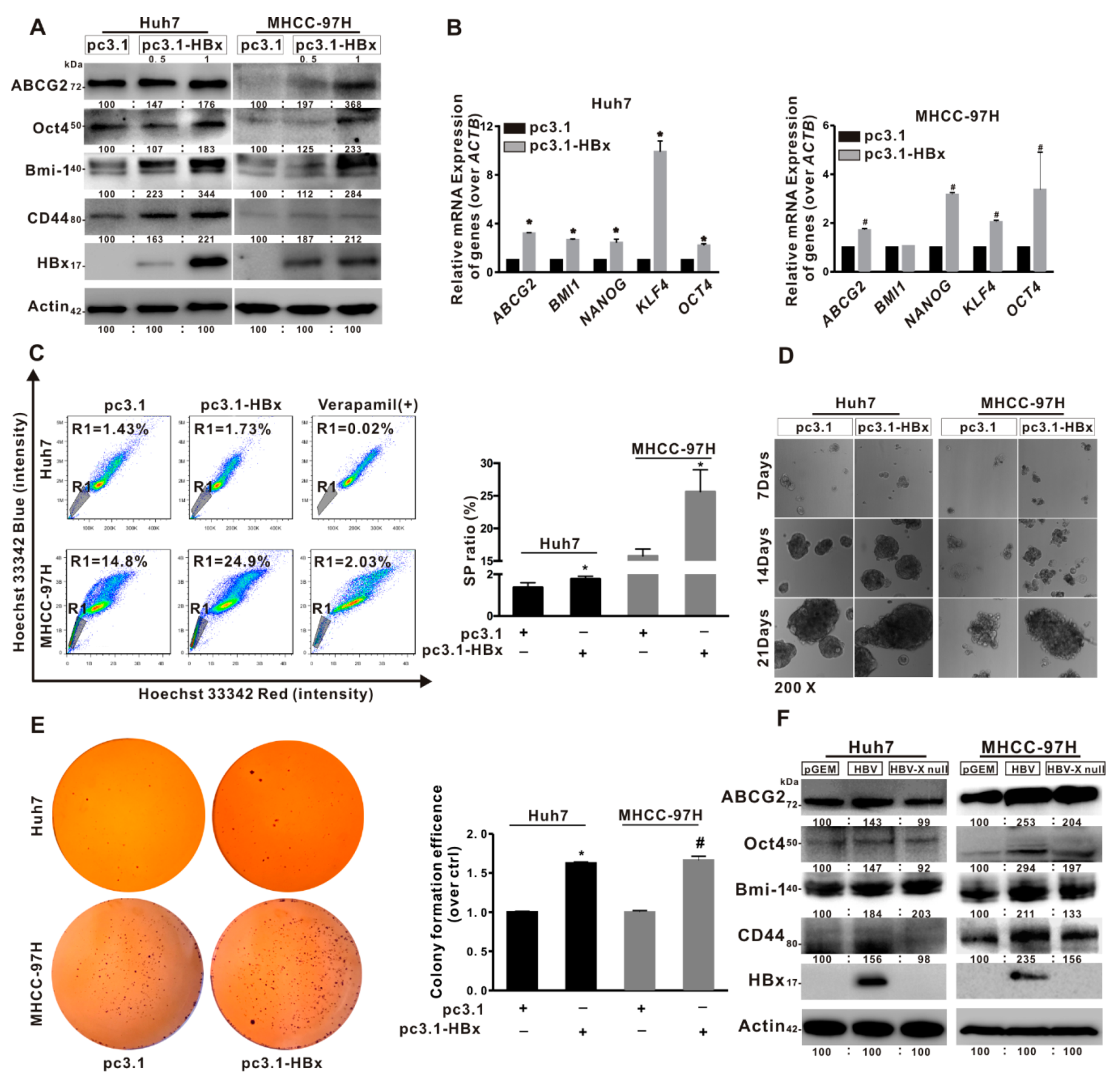
2.2. HBx Promoted Cancer Stemness Phenotype of HCC Cells
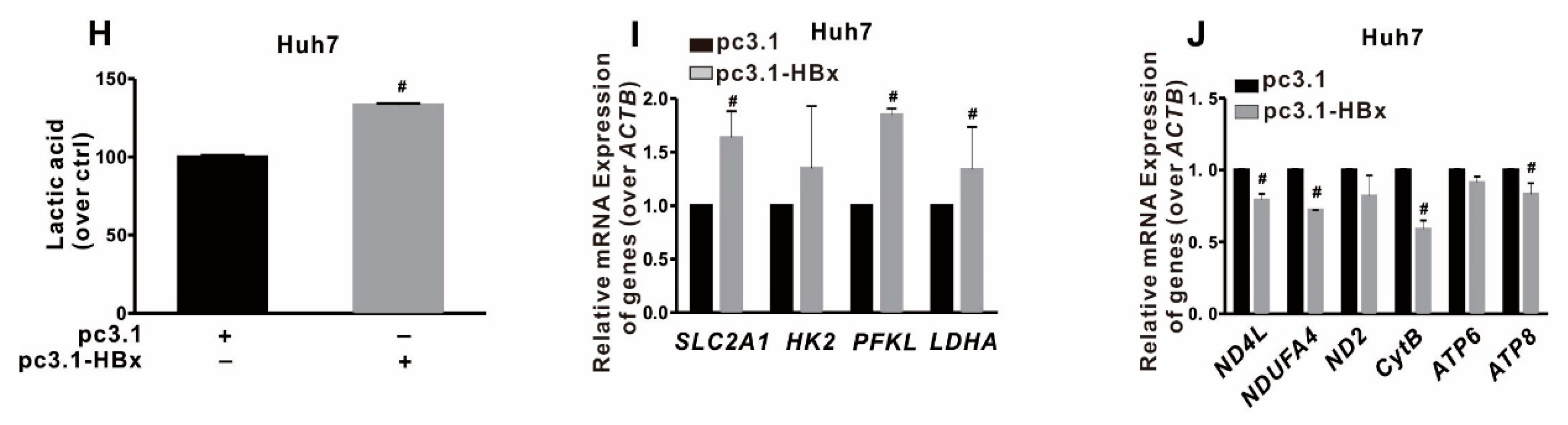
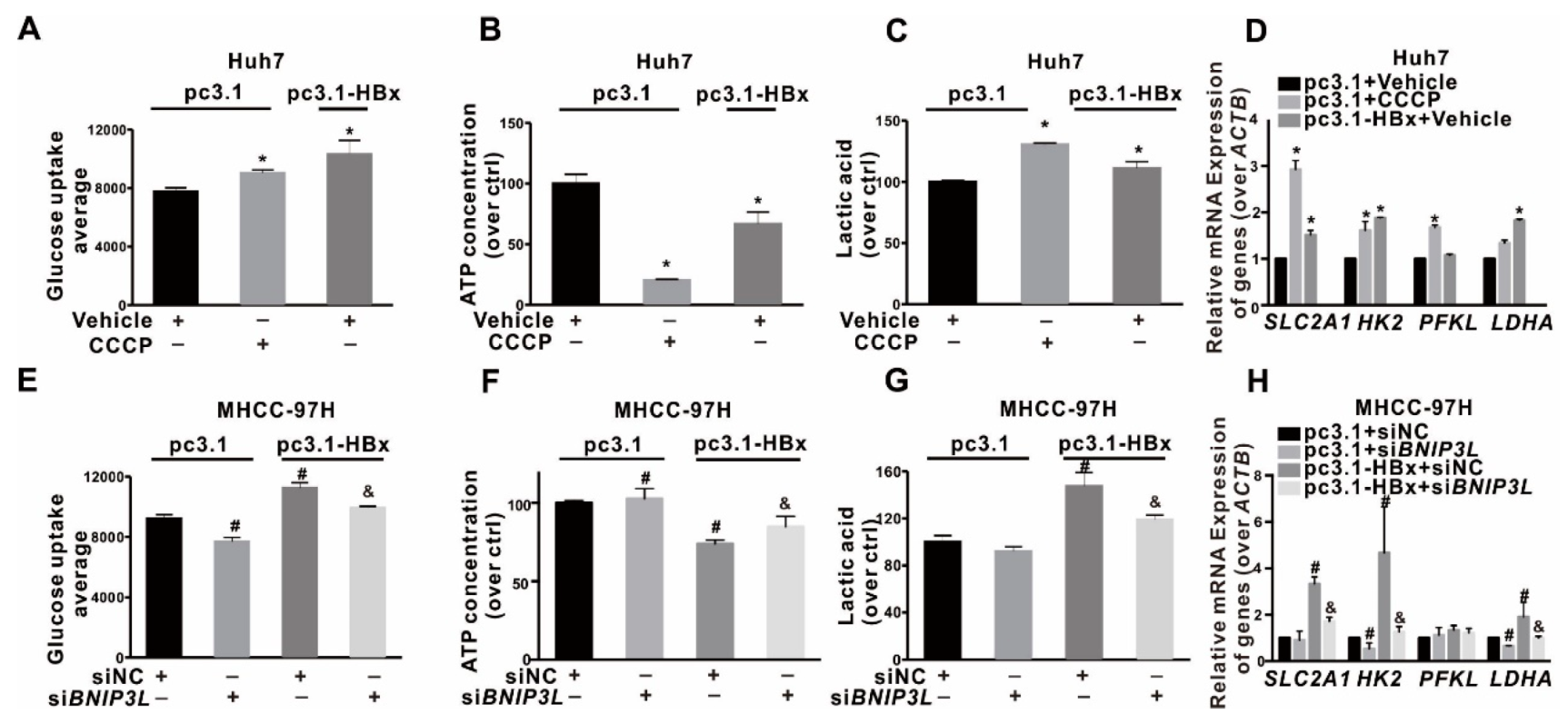
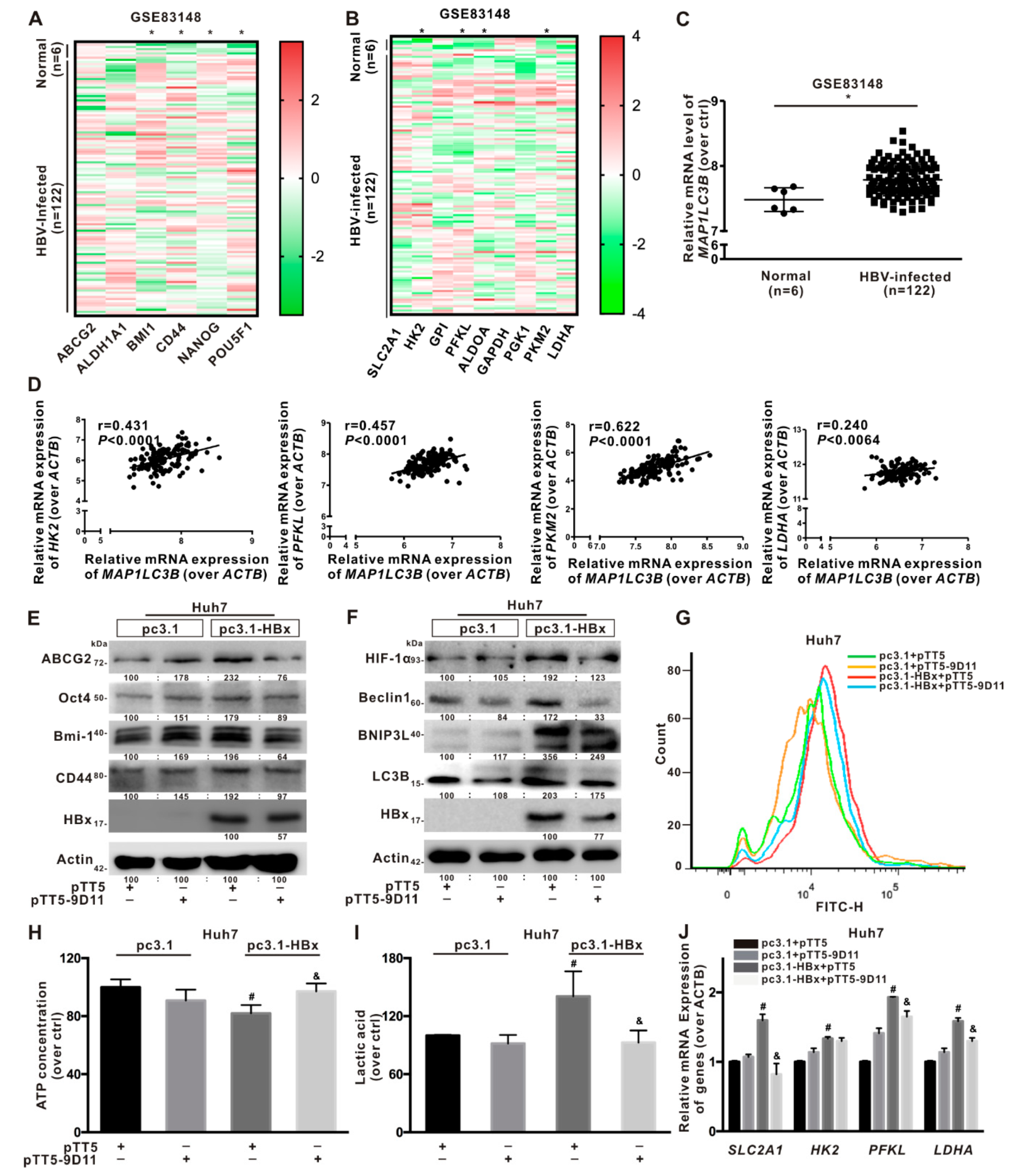
2.3. Glycolytic Metabolism Was Reprogrammed in the HBx-Expressing HCC Cells and LCSCs
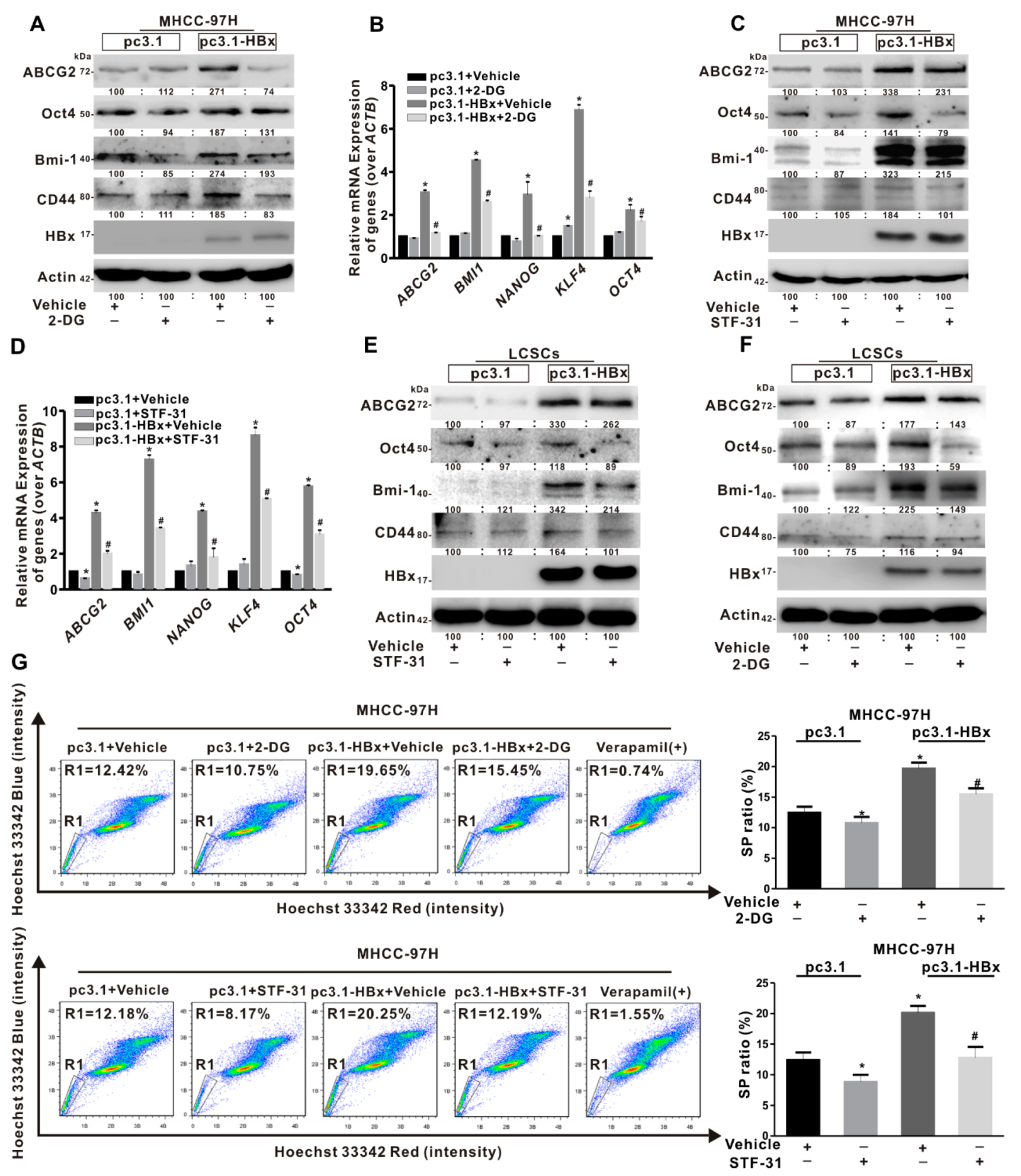
2.4. Glycolysis Metabolism Reprogramming Regulated Cancer Stemness of HCC Cells and LCSCs
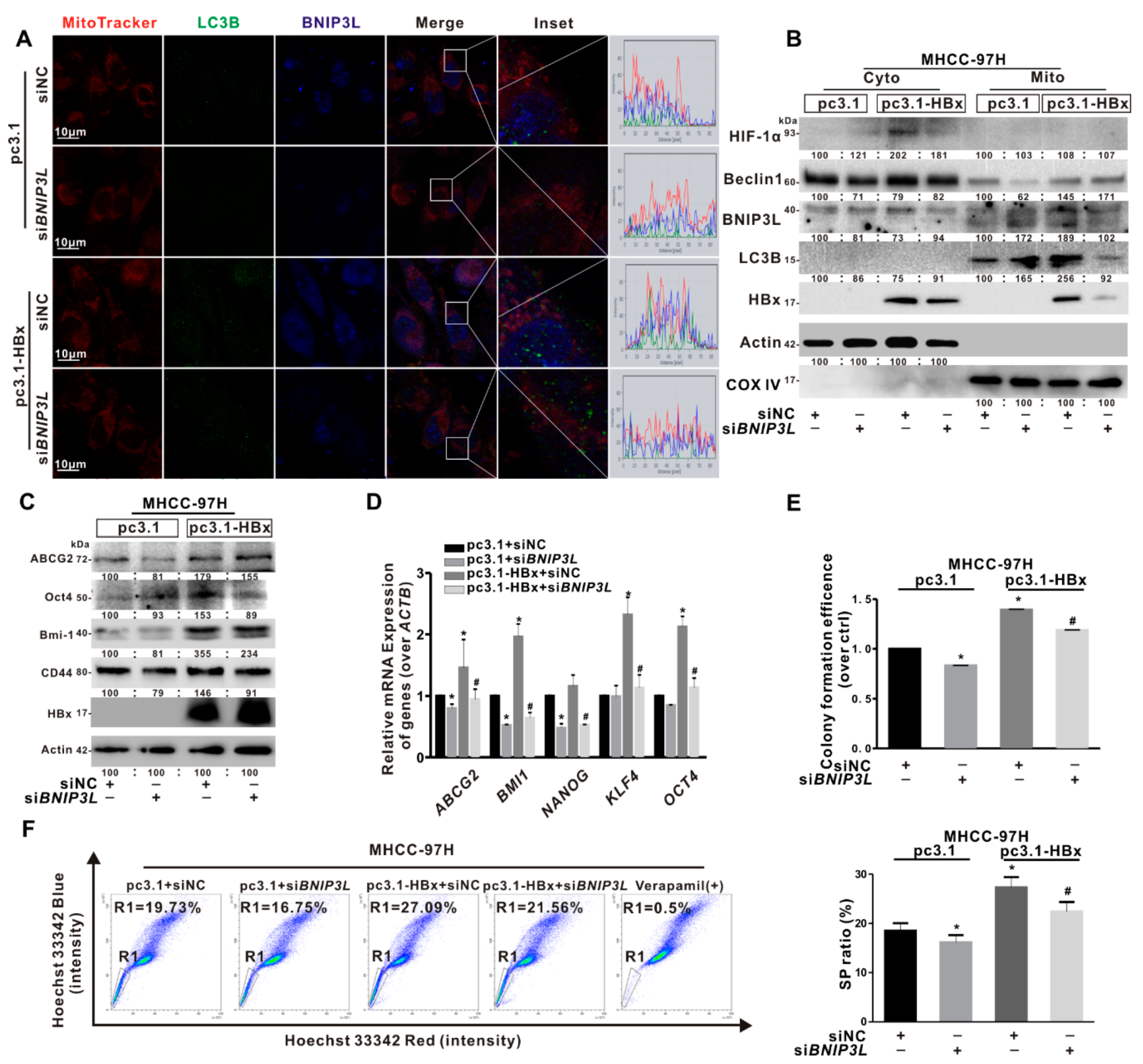
2.5. BNIP3L-Dependent Mitophagy Was Induced in HBx-Expressing HCC Cells and LCSCs
2.6. BNIP3L-Dependent Mitophagy Regulated Cancer Stemness of the HBx-Expressing HCC Cells
2.7. BNIP3L-Dependent Mitophagy Induced by HBx-Expressing Reprogrammed the Glycolytic Metabolism
2.8. Anti-HBx Targeting Intervention to Intracellular HBx Inhibited the Hepatocarcinogenesis Associated with BNIP3L-Dependent Mitophagy
3. Discussion
4. Materials and Methods
4.1. Gene Expression Omnibus (GEO) Data Analysis
4.2. Xenograft Tumor Study
4.3. Cell Culture
4.4. Sphere-Formation Assay
4.5. Cell Transfection
4.6. Sorting and Analysis of SP Cells and Main Population (MP) Cells
4.7. Anchorage-Independent Growth Assay
4.8. Colony Formation Assay
4.9. Glucose Transport Assay
4.10. ATP Content and Lactic Acid Secretion Assay
4.11. Transmission Electron Microscope (TEM) Assay
4.12. Immunofluorescence (IF) Assay and Analysis
4.13. Immunohistochemistry (IHC) Assay
4.14. Isolation of Mitochondrial-Cytosolic and Nuclear-Cytosolic Fractions
4.15. Western Blots (WB)
4.16. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2-DG | 2-deoxy-D-glucose | AFP | alpha fetoprotein |
| 2-NBDG | 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-2-deoxyglucose | ||
| ABCG2 | ATP-binding cassette sub-family G member 2 | ||
| AKT | protein kinase B | apoB | apolipoprotein B |
| ATP | adenosine triphosphate | bHLH | basic helix–loop–helix |
| Bmi-1 | B lymphoma Mo-MLV insertion region 1 homolog | ||
| BNIP3L | B-cell lymphoma 2/adenovirus E1B interacting 19 kDa-interacting protein 3-like protein | ||
| CCCP | carbonyl cyanide m-chlorophenylhydrazone | CSCs | cancer stem cells |
| EBV | Epstein-Barr virus | EBNA-2 | EBV nuclear antigen 2 |
| EpCAM | epithelial cell adhesion molecule | FCM | flow cytometry |
| FUNDC1 | FUN14 domain containing 1 | G6PD | glucose-6-phosphate dehydrogenase |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | GEO | gene expression omnibus |
| GLUT1 | glucose transporter 1 | HBV | hepatitis B virus |
| HBx | hepatitis B virus x protein | HCC | hepatocellular carcinoma |
| HIF-1α | hypoxia-inducible factor-1α | HKs | hexokinases |
| IF | immunofluorescence | IHC | immunohistochemistry |
| iPSCs | induced pluripotent stem cells | LC3B | microtubule associated protein 1 light chain 3 beta |
| LCSCs | liver cancer stem cells | LDHA | lactate dehydrogenase A |
| LMP1 | latent membrane protein 1 | MAP1LC3B | encoding gene for LC3B |
| mcAb | monoclonal antibody | MEFs | mouse embryonic fibroblasts |
| MP | main population | MQC | mitochondrial quality control |
| Oct4 | octamer-binding transcription factor 4 | OXPHOS | oxidative phosphorylation |
| PBS | phosphate buffered saline | PFKL | phosphofructokinase, liver type |
| PI3K | phosphatidylinositide 3-kinases | PINK1 | PTEN-induced kinase 1 |
| qRT-PCR | quantitative real-time polymerase chain reaction | ||
| SD | standard deviation | siRNA | small interfering RNA |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis | ||
| SP | side population | SREBP-1a | sterol regulatory element binding protein-1a |
| TEM | transmission electron microscope | WB | western blotting |
References
- Koike, K. Hepatitis B virus X gene is implicated in liver carcinogenesis. Cancer Lett. 2009, 286, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Gong, P. Hepatitis B virus X protein in the proliferation of hepatocellular carcinoma cells. Front. Biosci. 2013, 18, 1256. [Google Scholar] [CrossRef] [PubMed]
- Arzumanyan, A.; Friedman, T.; Ng, I.O.-L.; Clayton, M.M.; Lian, Z.; Feitelson, M.A. Does the hepatitis B antigen HBx promote the appearance of liver cancer stem cells? Cancer Res. 2011, 71, 3701–3708. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Li, W.; Lu, Y.; Dong, X.; Lin, B.; Chen, Y.; Zhang, X.; Guo, J.; Li, M.-S. HBx drives alpha fetoprotein expression to promote initiation of liver cancer stem cells through activating PI3K/AKT signal pathway. Int. J. Cancer 2017, 140, 1346–1355. [Google Scholar] [CrossRef]
- Muramatsu, S.; Tanaka, S.; Mogushi, K.; Adikrisna, R.; Aihara, A.; Ban, D.; Ochiai, T.; Irie, T.; Kudo, A.; Nakamura, N.; et al. Visualization of stem cell features in human hepatocellular carcinoma revealsin vivosignificance of tumor-host interaction and clinical course. Hepatology 2013, 58, 218–228. [Google Scholar] [CrossRef]
- Chang, T.-S.; Chen, C.-L.; Wu, Y.-C.; Liu, J.-J.; Kuo, Y.C.; Lee, K.-F.; Lin, S.-Y.; Lin, S.-E.; Tung, S.-Y.; Kuo, L.-M.; et al. Inflammation Promotes Expression of Stemness-Related Properties in HBV-Related Hepatocellular Carcinoma. PLoS ONE 2016, 11, e0149897. [Google Scholar] [CrossRef]
- Chang, T.-S.; Wu, Y.; Chi, C.-C.; Su, W.-C.; Chang, P.-J.; Lee, K.-F.; Tung, T.-H.; Wang, J.; Liu, J.; Tung, S.-Y.; et al. Activation of IL6/IGFIR Confers Poor Prognosis of HBV-Related Hepatocellular Carcinoma through Induction of OCT4/NANOG Expression. Clin. Cancer Res. 2015, 21, 201–210. [Google Scholar] [CrossRef]
- Lee, Y.I.; Hwang, J.M.; Im, J.H.; Lee, Y.I.; Kim, N.-S.; Kim, D.G.; Yu, D.-Y.; Moon, H.B.; Park, S.K. Human Hepatitis B Virus-X Protein Alters Mitochondrial Function and Physiology in Human Liver Cells. J. Boil. Chem. 2004, 279, 15460–15471. [Google Scholar] [CrossRef]
- Wu, H.; Chen, Q. Hypoxia Activation of Mitophagy and Its Role in Disease Pathogenesis. Antioxidants Redox Signal. 2015, 22, 1032–1046. [Google Scholar] [CrossRef]
- Chu, C.; Ji, J.; Dagda, R.; Jiang, J.F.; Tyurina, Y.Y.; Kapralov, O.; Tyurin, V.; Yanamala, N.; Shrivastava, I.H.; Mohammadyani, D.; et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013, 15, 1197–1205. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, H.-N.; Wang, K.; Zhang, L.; Huang, Z.; Liu, J.; Zhang, Z.; Luo, M.; Lei, Y.; Peng, Y.; et al. Ketoconazole exacerbates mitophagy to induce apoptosis by downregulating cyclooxygenase-2 in hepatocellular carcinoma. J. Hepatol. 2019, 70, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Khan, M.; Quan, J.; Till, A.; Subramani, S.; Siddiqui, A. Hepatitis B Virus Disrupts Mitochondrial Dynamics: Induces Fission and Mitophagy to Attenuate Apoptosis. PLOS Pathog. 2013, 9, e1003722. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.-C.; Chen, S.-L.; Lin, S.-L.; Tsai, C.-Y.; Chuang, W.-Y.; Lin, Y.-H.; Huang, Y.-H.; Tsai, M.-M.; Yeh, C.-T.; Lin, K.-H. Thyroid hormone protects hepatocytes from HBx-induced carcinogenesis by enhancing mitochondrial turnover. Oncogene 2017, 36, 5274–5284. [Google Scholar] [CrossRef] [PubMed]
- Moon, E.; Jeong, C.-H.; Jeong, J.; Kim, K.R.; Yu, D.-Y.; Murakami, S.; Kim, C.W.; Kim, K. Hepatitis B virus X protein induces angiogenesis by stabilizing hypoxia-inducible factor-1α. FASEB J. 2003, 18, 1–16. [Google Scholar] [CrossRef]
- Novak, I.; Kirkin, V.; McEwan, D.G.; Zhang, J.; Wild, P.; Rozenknop, A.; Rogov, V.; Löhr, F.; Popovic, D.; Occhipinti, A.; et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2009, 11, 45–51. [Google Scholar] [CrossRef]
- Sowter, H.M.; Ratcliffe, P.J.; Watson, P.; Greenberg, A.H.; Harris, A.L. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001, 61, 6669–6673. [Google Scholar]
- Sandoval, H.; Thiagarajan, P.; Dasgupta, S.K.; Schumacher, A.; Prchal, J.T.; Chen, M.; Wang, J. Essential role for Nix in autophagic maturation of erythroid cells. Nature 2008, 454, 232–235. [Google Scholar] [CrossRef]
- Xiang, G.; Yang, L.; Long, Q.; Chen, K.; Tang, H.; Wu, Y.; Liu, Z.; Zhou, Y.; Qi, J.; Zheng, L.; et al. BNIP3L-dependent mitophagy accounts for mitochondrial clearance during 3 factors-induced somatic cell reprogramming. Autophagy 2017, 13, 1543–1555. [Google Scholar] [CrossRef]
- Yan, C.; Luo, L.; Guo, C.-Y.; Goto, S.; Urata, Y.; Shao, J.-H.; Li, T.-S. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett. 2017, 388, 34–42. [Google Scholar] [CrossRef]
- Drake, L.E.; Springer, M.Z.; Poole, L.P.; Kim, C.; MacLeod, K. Expanding perspectives on the significance of mitophagy in cancer. Semin. Cancer Boil. 2017, 47, 110–124. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Pelicano, H.; Martin, D.S.; Xu, R.-H.; Huang, P. Glycolysis inhibition for anticancer treatment. Oncogene 2006, 25, 4633–4646. [Google Scholar] [CrossRef] [PubMed]
- Shang, R.-Z.; Qu, S.-B.; Wang, D.-S. Reprogramming of glucose metabolism in hepatocellular carcinoma: Progress and prospects. World J. Gastroenterol. 2016, 22, 9933–9943. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Fang, M.; He, Z.; Cui, D.; Jia, S.; Lin, Q.X.X.; Xu, X.; Zhou, T.; Liu, W. Hepatitis B virus stimulates G6PD expression through HBx-mediated Nrf2 activation. Cell Death Dis. 2015, 6, e1980. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.K.; Chung, T.-W.; Lee, J.-Y.; Lee, Y.-C.; Morton, R.E.; Kim, C.-H. The Hepatitis B Virus X Protein Inhibits Secretion of Apolipoprotein B by Enhancing the Expression of N-Acetylglucosaminyltransferase III. J. Boil. Chem. 2004, 279, 28106–28112. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Wu, Q.; Lu, X.; Zhou, Y.; Fernández-Alvarez, A.; Ye, L.; Zhang, X.; Han, J.; Casado, M.; Liu, Q. SREBP-1a activation by HBx and the effect on hepatitis B virus enhancer II/core promoter. Biochem. Biophys. Res. Commun. 2013, 432, 643–649. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yue, D.; Zhang, Y.; Cheng, L.; Ma, J.; Xi, Y.; Yang, L.; Su, C.; Shao, B.; Huang, A.; Xiang, R.; et al. Hepatitis B virus X protein (HBx)-induced abnormalities of nucleic acid metabolism revealed by 1H-NMR-based metabonomics. Sci. Rep. 2016, 6, 24430. [Google Scholar] [CrossRef]
- Nayagam, A.S.; Thursz, M.; Sicuri, E.; Conteh, L.; Wiktor, S.; Low-Beer, D.; Hallett, T. Requirements for global elimination of hepatitis B: A modelling study. Lancet Infect. Dis. 2016, 16, 1399–1408. [Google Scholar] [CrossRef]
- Zhang, J.-F.; Xiong, H.-L.; Cao, J.-L.; Wang, S.-J.; Guo, X.-R.; Lin, B.-Y.; Zhang, Y.; Zhao, J.-H.; Wang, Y.-B.; Zhang, T.-Y.; et al. A cell-penetrating whole molecule antibody targeting intracellular HBx suppresses hepatitis B virus via TRIM21-dependent pathway. Theranostics 2018, 8, 549–562. [Google Scholar] [CrossRef]
- Liu, K.; Lee, J.; Kim, J.Y.; Wang, L.; Tian, Y.; Chan, S.T.; Cho, C.; Machida, K.; Chen, D.; Ou, J.-H.J. Mitophagy Controls the Activities of Tumor Suppressor p53 to Regulate Hepatic Cancer Stem Cells. Mol. Cell 2017, 68, 281–292.e5. [Google Scholar] [CrossRef]
- Zhou, C.; Zhong, W.; Zhou, J.; Sheng, F.; Fang, Z.; Wei, Y.; Chen, Y.; Deng, X.; Lou, Z.; Lin, J. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy 2012, 8, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Lu, H.; Xu, J.; Yu, H.; Wang, X.; Zhang, X. Protective roles of autophagy in retinal pigment epithelium under high glucose condition via regulating PINK1/Parkin pathway and BNIP3L. Boil. Res. 2018, 51, 22. [Google Scholar] [CrossRef] [PubMed]
- Ailles, L.E.; Weissman, I.L. Cancer stem cells in solid tumors. Curr. Opin. Biotechnol. 2007, 18, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Wang, X.W. Cancer stem cells in the development of liver cancer. J. Clin. Investig. 2013, 123, 1911–1918. [Google Scholar] [CrossRef]
- Wang, C.; Wang, M.-D.; Cheng, P.; Huang, H.; Dong, W.; Zhang, W.-W.; Li, P.-P.; Lin, C.; Pan, Z.-Y.; Wu, M.-C.; et al. Hepatitis B virus X protein promotes the stem-like properties of OV6+ cancer cells in hepatocellular carcinoma. Cell Death Dis. 2017, 8, e2560. [Google Scholar] [CrossRef]
- Ching, R.H.H.; Sze, K.M.F.; Lau, E.Y.T.; Chiu, Y.-T.; Lee, J.M.F.; Ng, I.O.-L.; Lee, T.K.W. C-terminal truncated hepatitis B virus X protein regulates tumorigenicity, self-renewal and drug resistance via STAT3/Nanog signaling pathway. Oncotarget 2017, 8, 23507–23516. [Google Scholar] [CrossRef]
- Zhou, T.-J.; Zhang, S.-L.; He, C.-Y.; Zhuang, Q.-Y.; Han, P.-Y.; Jiang, S.-W.; Yao, H.; Huang, Y.-J.; Ling, W.-H.; Lin, Y.-C.; et al. Downregulation of mitochondrial cyclooxygenase-2 inhibits the stemness of nasopharyngeal carcinoma by decreasing the activity of dynamin-related protein 1. Theranostics 2017, 7, 1389–1406. [Google Scholar] [CrossRef]
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Boil. 2011, 12, 9–14. [Google Scholar] [CrossRef]
- McLelland, G.-L.; Soubannier, V.; Chen, C.X.; McBride, H.M.; Fon, A.E. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014, 33, 282–295. [Google Scholar] [CrossRef]
- Lu, H.; Li, G.; Liu, L.; Feng, L.; Wang, X.; Jin, H. Regulation and function of mitophagy in development and cancer. Autophagy 2013, 9, 1720–1736. [Google Scholar] [CrossRef]
- Vazquez-Martin, A.; Haute, C.V.D.; Cufí, S.; Corominas-Faja, B.; Cuyàs, E.; Bonet, E.L.; Rodríguez-Gallego, E.; Fernández-Arroyo, S.; Joven, J.; Baekelandt, V.; et al. Mitophagy-driven mitochondrial rejuvenation regulates stem cell fate. Aging 2016, 8, 1330–1349. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.L.; Terzic, A. Energy metabolism in the acquisition and maintenance of stemness. Semin. Cell Dev. Boil. 2016, 52, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Peiris-Pagès, M.; Martinez-Outschoorn, U.; Pestell, R.G.; Sotgia, F.; Lisanti, M. Cancer stem cell metabolism. Breast Cancer Res. 2016, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Son, M.J.; Kwon, Y.; Son, M.-Y.; Seol, B.; Choi, H.-S.; Ryu, S.-W.; Choi, C.; Cho, Y.S. Mitofusins deficiency elicits mitochondrial metabolic reprogramming to pluripotency. Cell Death Differ. 2015, 22, 1957–1969. [Google Scholar] [CrossRef]
- Peng, F.; Wang, J.-H.; Fan, W.-J.; Meng, Y.-T.; Li, M.-M.; Li, T.-T.; Cui, B.; Wang, H.-F.; Zhao, Y.; An, F.; et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 2017, 37, 1062–1074. [Google Scholar] [CrossRef]
- Zhao, H.; Duan, Q.; Zhang, Z.; Li, H.; Wu, H.; Shen, Q.; Wang, C.; Yin, T. Up-regulation of glycolysis promotes the stemness and EMT phenotypes in gemcitabine-resistant pancreatic cancer cells. J. Cell. Mol. Med. 2017, 21, 2055–2067. [Google Scholar] [CrossRef]
- Shen, Y.-A.; Wang, C.-Y.; Hsieh, Y.-T.; Chen, Y.-J.; Wei, Y.-H. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle 2014, 14, 86–98. [Google Scholar] [CrossRef]
- Janiszewska, M.; Suvà, M.L.; Riggi, N.; Houtkooper, R.H.; Auwerx, J.; Clément-Schatlo, V.; Radovanovic, I.; Rheinbay, E.; Provero, P.; Stamenkovic, I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012, 26, 1926–1944. [Google Scholar] [CrossRef]
- Wong, T.L.; Che, N.; Ma, S.K.Y. Reprogramming of central carbon metabolism in cancer stem cells. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1728–1738. [Google Scholar] [CrossRef]
- Mushtaq, M.; Darekar, S.; Kashuba, E. DNA Tumor Viruses and Cell Metabolism. Oxidative Med. Cell. Longev. 2016, 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sung, W.-W.; Chen, P.-R.; Liao, M.-H.; Lee, J.-W. Enhanced aerobic glycolysis of nasopharyngeal carcinoma cells by Epstein-Barr virus latent membrane protein 1. Exp. Cell Res. 2017, 359, 94–100. [Google Scholar] [CrossRef]
- Lai, D.; Tan, C.L.; Gunaratne, J.; Quek, L.S.; Nei, W.L.; Thierry, F.; Bellanger, S. Localization of HPV-18 E2 at Mitochondrial Membranes Induces ROS Release and Modulates Host Cell Metabolism. PLoS ONE 2013, 8, e75625. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Q.; Zhou, T.; He, C.; Zhang, S.; Qiu, Y.; Luo, B.; Zhao, R.; Liu, H.; Lin, Y.-C.; Lin, Z.-N. Protein phosphatase 2A-B55δ enhances chemotherapy sensitivity of human hepatocellular carcinoma under the regulation of microRNA-133b. J. Exp. Clin. Cancer Res. 2016, 35, 67. [Google Scholar] [CrossRef] [PubMed]
- Maier, S.; Staffler, G.; Hartmann, A.; Höck, J.; Henning, K.; Grabusic, K.; Mailhammer, R.; Hoffmann, R.; Wilmanns, M.; Lang, R.; et al. Cellular Target Genes of Epstein-Barr Virus Nuclear Antigen 2. J. Virol. 2006, 80, 9761–9771. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, W.; Ma, Y.; Zhang, J.; Hu, J.; Zhang, M.; Wang, Y.; Li, Y.; Wu, L.; Pan, Y.; Zhang, Y.; et al. Predictive model for inflammation grades of chronic hepatitis B: Large-scale analysis of clinical parameters and gene expressions. Liver Int. 2017, 37, 1632–1641. [Google Scholar] [CrossRef]
- A Liu, Y.; Tong, Z.; Li, T.; Chen, Q.; Zhuo, L.; Li, W.; Wu, R.-C.; Yu, C. Hepatitis B virus X protein stabilizes amplified in breast cancer 1 protein and cooperates with it to promote human hepatocellular carcinoma cell invasiveness. Hepatology 2012, 56, 1015–1024. [Google Scholar] [CrossRef]
- Cao, L.; Zhou, Y.; Zhai, B.; Liao, J.; Xu, W.; Zhang, R.; Li, J.; Zhang, Y.; Chen, L.; Qian, H.; et al. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol. 2011, 11, 71. [Google Scholar] [CrossRef]
- Collak, F.K.; Demir, U.; Sagir, F. YAP1 Is Involved in Tumorigenic Properties of Prostate Cancer Cells. Pathol. Oncol. Res. 2019, 1–10. [Google Scholar] [CrossRef]
- Liao, K.; Xia, B.; Zhuang, Q.-Y.; Hou, M.-J.; Zhang, Y.-J.; Luo, B.; Qiu, Y.; Gao, Y.-F.; Li, X.-J.; Chen, H.-F.; et al. Parthenolide Inhibits Cancer Stem-Like Side Population of Nasopharyngeal Carcinoma Cells via Suppression of the NF-κB/COX-2 Pathway. Theranostics 2015, 5, 302–321. [Google Scholar] [CrossRef]
- Anandhan, A.; Lei, S.; Levytskyy, R.; Pappa, A.; Panayiotidis, M.I.; Cerny, R.L.; Khalimonchuk, O.; Powers, R.; Franco, R.; Oleh, K. Glucose Metabolism and AMPK Signaling Regulate Dopaminergic Cell Death Induced by Gene (α-Synuclein)-Environment (Paraquat) Interactions. Mol. Neurobiol. 2016, 54, 3825–3842. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-Y.; Wang, W.-H.; Che, L.; Lan, Y.; Zhang, L.-Y.; Zhan, D.-L.; Huang, Z.-Y.; Lin, Z.-N.; Lin, Y.-C. BNIP3L-Dependent Mitophagy Promotes HBx-Induced Cancer Stemness of Hepatocellular Carcinoma Cells via Glycolysis Metabolism Reprogramming. Cancers 2020, 12, 655. https://doi.org/10.3390/cancers12030655
Chen Y-Y, Wang W-H, Che L, Lan Y, Zhang L-Y, Zhan D-L, Huang Z-Y, Lin Z-N, Lin Y-C. BNIP3L-Dependent Mitophagy Promotes HBx-Induced Cancer Stemness of Hepatocellular Carcinoma Cells via Glycolysis Metabolism Reprogramming. Cancers. 2020; 12(3):655. https://doi.org/10.3390/cancers12030655
Chicago/Turabian StyleChen, Yuan-Yuan, Wei-Hua Wang, Lin Che, You Lan, Li-Yin Zhang, Deng-Lin Zhan, Zi-Yan Huang, Zhong-Ning Lin, and Yu-Chun Lin. 2020. "BNIP3L-Dependent Mitophagy Promotes HBx-Induced Cancer Stemness of Hepatocellular Carcinoma Cells via Glycolysis Metabolism Reprogramming" Cancers 12, no. 3: 655. https://doi.org/10.3390/cancers12030655
APA StyleChen, Y.-Y., Wang, W.-H., Che, L., Lan, Y., Zhang, L.-Y., Zhan, D.-L., Huang, Z.-Y., Lin, Z.-N., & Lin, Y.-C. (2020). BNIP3L-Dependent Mitophagy Promotes HBx-Induced Cancer Stemness of Hepatocellular Carcinoma Cells via Glycolysis Metabolism Reprogramming. Cancers, 12(3), 655. https://doi.org/10.3390/cancers12030655



