Latest Insights into Marek’s Disease Virus Pathogenesis and Tumorigenesis
Abstract
1. Introduction
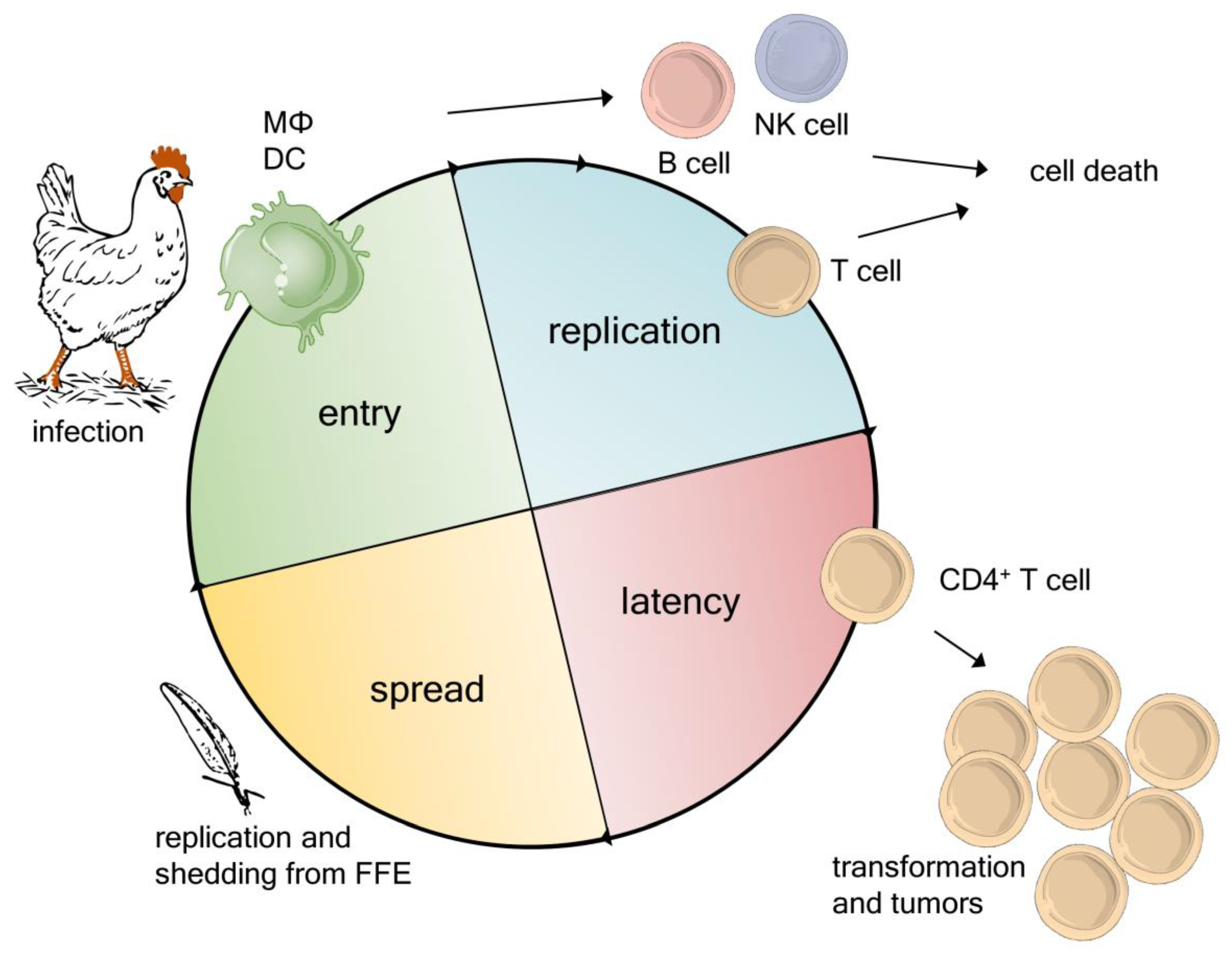
2. Novel Insights into the MDV Life Cycle
3. Virulence Factors in the MDV Genome
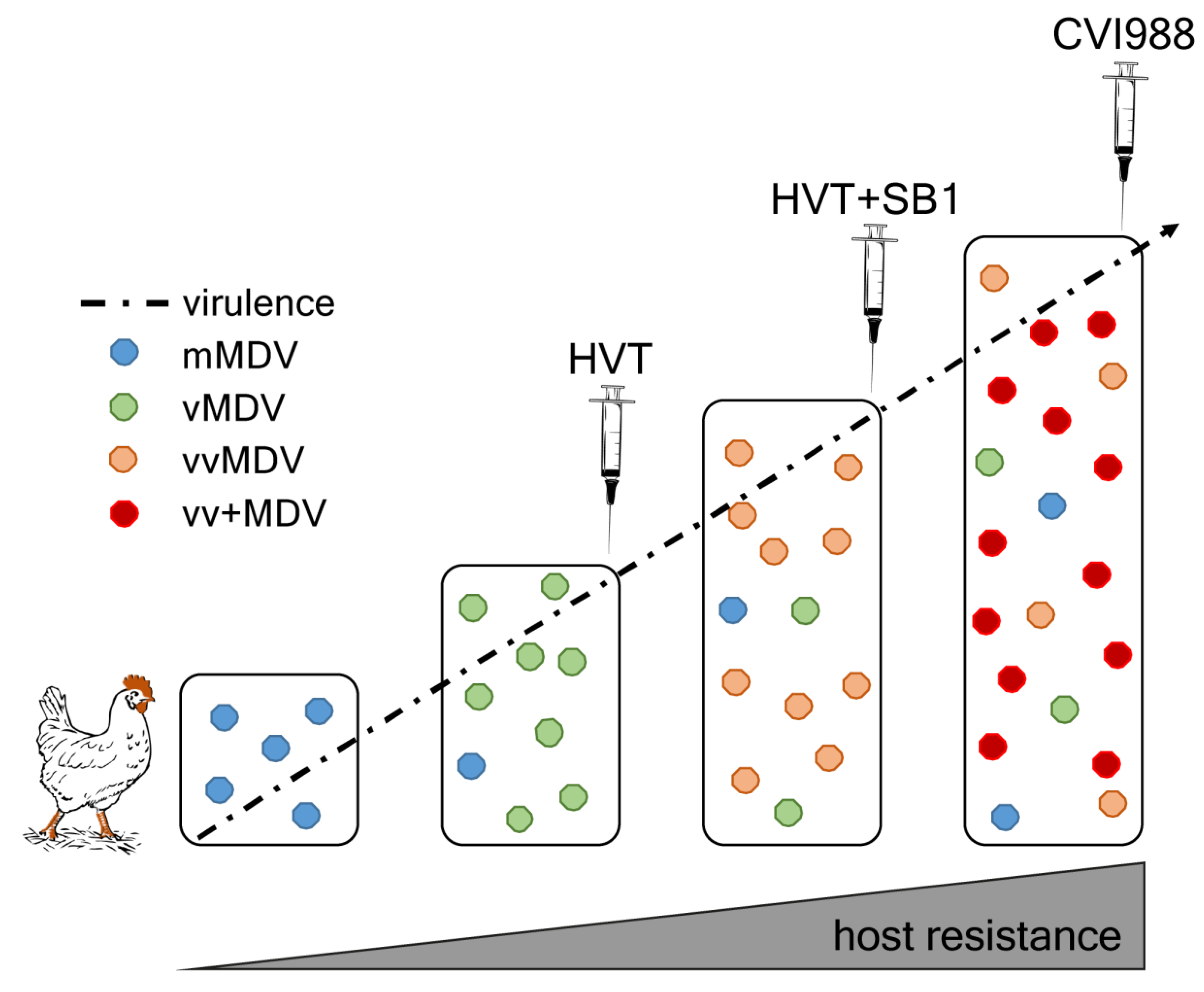
4. MDV Evolution and Increase in Virulence
5. Future Perspectives and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Purchase, H.G. Clinical Disease and Its Economic Impact. In Marek’s Disease; Payne, L.N., Ed.; Springer: Boston, MA, USA, 1985; pp. 17–42. [Google Scholar]
- Davison, T.F.; Nair, V. Marek’s Disease: An Evolving Problem; Elsevier Academic Press: London, UK, 2004. [Google Scholar]
- Davison, A.J. Herpesvirus systematics. Vet. Microbiol. 2010, 143, 52–69. [Google Scholar] [CrossRef] [PubMed]
- Marek, J. Multiple Nervenentzündung (Polyneuritis) bei Hühnern. Dtsch. Tierärztl. Wochenschr. 1907, 15, 417–421. [Google Scholar]
- Osterrieder, N.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Trapp, S. Marek’s disease virus: From miasma to model. Nat. Rev. Microbiol. 2006, 4, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Payne, L.N.; Venugopal, K. Neoplastic diseases: Marek’s disease, avian leukosis and reticuloendotheliosis. Revue. Sci. Tech. 2000, 19, 544–564. [Google Scholar] [CrossRef]
- Rushton, J. The Economics of Animal Health and Production; CABI Publishing: Oxfordshire, UK, 2008. [Google Scholar]
- Dunn, J.R.; Gimeno, I.M. Current status of Marek’s disease in the United States and worldwide based on a questionnaire survey. Avian. Dis. 2013, 57, 483–490. [Google Scholar] [CrossRef]
- Rozins, C.; Day, T.; Greenhalgh, S. Managing Marek’s disease in the egg industry. Epidemics 2019. [Google Scholar] [CrossRef]
- Gimeno, I.M.; Schat, K.A. Virus-Induced Immunosuppression in Chickens. Avian. Dis. 2018, 62, 272–285. [Google Scholar] [CrossRef]
- Morrow, C.; Fehler, F. Marek’s Disease: A worldwide problem. Marek’s Disease 2004. [Google Scholar]
- Gimeno, I.M. Marek’s disease vaccines: A solution for today but a worry for tomorrow? Vaccine 2008, 26, C31–C41. [Google Scholar] [CrossRef]
- Schat, K.A.; Nair, V. Neoplastic Diseases. In Diseases of Poultry; Swayne, D.E., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 513–673. [Google Scholar]
- Davison, F.; Nair, V. Use of Marek’s disease vaccines: Could they be driving the virus to increasing virulence? Expert Rev. Vaccines 2005, 4, 77–88. [Google Scholar] [CrossRef]
- Baigent, S.J.; Smith, L.P.; Nair, V.K.; Currie, R.J. Vaccinal control of Marek’s disease: Current challenges, and future strategies to maximize protection. Vet. Immunol. Immunopathol. 2006, 112, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Zou, H.; Shi, H.; Shao, H.; Ye, J.; Miao, J.; Wu, G.; Qin, A. Outbreak of Marek’s disease in a vaccinated broiler breeding flock during its peak egg-laying period in China. BMC Vet. Res. 2015, 11, 157. [Google Scholar] [CrossRef] [PubMed]
- OIE-Listed Diseases, Infections and Infestations in Force in 2020. Available online: https://www.oie.int/animal-health-in-the-world/oie-listed-diseases-2020/ (accessed on 19 February 2020).
- Read, A.F.; Baigent, S.J.; Powers, C.; Kgosana, L.B.; Blackwell, L.; Smith, L.P.; Kennedy, D.A.; Walkden-Brown, S.W.; Nair, V.K. Imperfect Vaccination Can Enhance the Transmission of Highly Virulent Pathogens. PLoS Biol. 2015, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.A.; Read, A.F. Why the evolution of vaccine resistance is less of a concern than the evolution of drug resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 12878–12886. [Google Scholar] [CrossRef]
- Weiss, R.A. The oncologist’s debt to the chicken. Avian. Pathol. 1998, 27, S8–S15. [Google Scholar] [CrossRef]
- Calnek, B.W. Pathogenesis of Marek’s disease virus infection. Curr. Top. Microbiol. Immunol. 2001, 255, 25–55. [Google Scholar]
- Baaten, B.J.; Staines, K.A.; Smith, L.P.; Skinner, H.; Davison, T.F.; Butter, C. Early replication in pulmonary B cells after infection with Marek’s disease herpesvirus by the respiratory route. Viral. Immunol. 2009, 22, 431–444. [Google Scholar] [CrossRef]
- Chakraborty, P.; Vervelde, L.; Dalziel, R.G.; Wasson, P.S.; Nair, V.; Dutia, B.M.; Kaiser, P. Marek’s disease virus infection of phagocytes: A de novo in vitro infection model. J. Gen. Virol. 2017, 98, 1080–1088. [Google Scholar] [CrossRef]
- Barrow, A.D.; Burgess, S.C.; Baigent, S.J.; Howes, K.; Nair, V.K. Infection of macrophages by a lymphotropic herpesvirus: A new tropism for Marek’s disease virus. J. Gen. Virol. 2003, 84, 2635–2645. [Google Scholar] [CrossRef]
- Parcells, M.S.; Lin, S.F.; Dienglewicz, R.L.; Majerciak, V.; Robinson, D.R.; Chen, H.C.; Wu, Z.; Dubyak, G.R.; Brunovskis, P.; Hunt, H.D.; et al. Marek’s disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): Characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol. 2001, 75, 5159–5173. [Google Scholar] [CrossRef]
- Kaiser, P.; Hughes, S.; Bumstead, N. The chicken 9E3/CEF4 CXC chemokine is the avian orthologue of IL8 and maps to chicken Chromosome 4 syntenic with genes flanking the mammalian chemokine cluster. Immunogenetics 1999, 49, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.T.; Selvaraj, R.K.; Kamil, J.P.; Osterrieder, N.; Kaufer, B.B. Marek’s disease viral interleukin-8 promotes lymphoma formation through targeted recruitment of B cells and CD4+ CD25+ T cells. J. Virol. 2012, 86, 8536–8545. [Google Scholar] [CrossRef] [PubMed]
- Haertle, S.; Alzuheir, I.; Busalt, F.; Waters, V.; Kaiser, P.; Kaufer, B.B. Identification of the Receptor and Cellular Ortholog of the Marek’s Disease Virus (MDV) CXC Chemokine. Front Microbiol. 2017, 8, 2543. [Google Scholar] [CrossRef] [PubMed]
- Baigent, S.J.; Ross, L.J.; Davison, T.F. A flow cytometric method for identifying Marek’s disease virus pp38 expression in lymphocyte subpopulations. Avian. Pathol. 1996, 25, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Baigent, S.J.; Ross, L.J.; Davison, T.F. Differential susceptibility to Marek’s disease is associated with differences in number, but not phenotype or location, of pp38+ lymphocytes. J. Gen. Virol. 1998, 79, 2795–2802. [Google Scholar] [CrossRef]
- Berthault, C.; Larcher, T.; Härtle, S.; Vautherot, J.F.; Trapp-Fragnet, L.; Denesvre, C. Atrophy of primary lymphoid organs induced by Marek’s disease virus during early infection is associated with increased apoptosis, inhibition of cell proliferation and a severe B-lymphopenia. Vet. Res. 2018, 49, 31. [Google Scholar] [CrossRef]
- Bertzbach, L.D.; Laparidou, M.; Härtle, S.; Etches, R.J.; Kaspers, B.; Schusser, B.; Kaufer, B.B. Unraveling the role of B cells in the pathogenesis of an oncogenic avian herpesvirus. Proc. Natl. Acad. Sci. USA 2018, 115, 11603–11607. [Google Scholar] [CrossRef]
- Schermuly, J.; Greco, A.; Härtle, S.; Osterrieder, N.; Kaufer, B.B.; Kaspers, B. In vitro model for lytic replication, latency, and transformation of an oncogenic alphaherpesvirus. Proc. Natl. Acad. Sci. USA 2015, 112, 7279–7284. [Google Scholar] [CrossRef]
- Bertzbach, L.D.; van Haarlem, D.A.; Härtle, S.; Kaufer, B.B.; Jansen, C.A. Marek’s Disease Virus Infection of Natural Killer Cells. Microorganisms 2019, 7, 12. [Google Scholar] [CrossRef]
- Xing, Z.; Schat, K.A. Inhibitory effects of nitric oxide and gamma interferon on in vitro and in vivo replication of Marek’s disease virus. J. Virol. 2000, 74, 3605–3612. [Google Scholar] [CrossRef]
- Bertzbach, L.D.; Harlin, O.; Härtle, S.; Fehler, F.; Vychodil, T.; Kaufer, B.B.; Kaspers, B. IFNα and IFNγ Impede Marek’s Disease Progression. Viruses 2019, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Lion, A.; Esnault, E.; Kut, E.; Guillory, V.; Trapp-Fragnet, L.; Soubies, S.M.; Chanteloup, N.; Niepceron, A.; Guabiraba, R.; Marc, D.; et al. Chicken endothelial cells are highly responsive to viral innate immune stimuli and are susceptible to infections with various avian pathogens. Avian. Pathol. 2019, 48, 121–134. [Google Scholar] [CrossRef]
- Morgan, R.W.; Xie, Q.; Cantello, J.L.; Miles, A.M.; Bernberg, E.L.; Kent, J.; Anderson, A. Marek’s disease virus latency. Curr. Top. Microbiol. Immunol. 2001, 255, 223–243. [Google Scholar]
- Nair, V. Latency and tumorigenesis in Marek’s disease. Avian. Dis. 2013, 57, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, W.N.; Smith, L.P.; Baigent, S.J.; Beal, R.K.; Nair, V.; Smith, A.L. Clonal structure of rapid-onset MDV-driven CD4+ lymphomas and responding CD8+ T cells. PLoS Pathog. 2011, 7, e1001337. [Google Scholar] [CrossRef] [PubMed]
- Delecluse, H.J.; Hammerschmidt, W. Status of Marek’s disease virus in established lymphoma cell lines: Herpesvirus integration is common. J. Virol. 1993. (0022-538X (Print)). [Google Scholar] [CrossRef]
- Kaufer, B.B.; Jarosinski, K.W.; Osterrieder, N. Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation. J. Exp. Med. 2011, 208, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Osterrieder, N.; Wallaschek, N.; Kaufer, B.B. Herpesvirus Genome Integration into Telomeric Repeats of Host Cell Chromosomes. Annu. Rev. Virol. 2014, 1, 215–235. [Google Scholar] [CrossRef]
- Pauker, V.I.; Bertzbach, L.D.; Hohmann, A.; Kheimar, A.; Teifke, J.P.; Mettenleiter, T.C.; Karger, A.; Kaufer, B.B. Imaging Mass Spectrometry and Proteome Analysis of Marek’s Disease Virus-Induced Tumors. mSphere 2019, 4, 1. [Google Scholar] [CrossRef]
- Baigent, S.J.; Kgosana, L.B.; Gamawa, A.A.; Smith, L.P.; Read, A.F.; Nair, V.K. Relationship between levels of very virulent MDV in poultry dust and in feather tips from vaccinated chickens. Avian. Dis. 2013, 57, 440–447. [Google Scholar] [CrossRef]
- Baigent, S.J.; Smith, L.P.; Currie, R.J.; Nair, V.K. Replication kinetics of Marek’s disease vaccine virus in feathers and lymphoid tissues using PCR and virus isolation. J. Gen. Virol. 2005, 86, 2989–2998. [Google Scholar] [CrossRef]
- Couteaudier, M.; Denesvre, C. Marek’s disease virus and skin interactions. Vet. Res. 2014, 45, 36. [Google Scholar] [CrossRef] [PubMed]
- Carrozza, J.H.; Fredrickson, T.N.; Prince, R.P.; Luginbuhl, R.E. Role of Desquamated Epithelial Cells in Transmission of Marek’s Disease. Avian Dis. 1973, 17, 4. [Google Scholar] [CrossRef]
- Witter, R.L.; Burgoyne, G.H.; Burmester, B.R. Survival of Marek’s Disease Agent in Litter and Droppings. Avian Dis. 1968, 12, 3. [Google Scholar] [CrossRef]
- Osterrieder, N.; Vautherot, J.F. The genome content of Marek’s disease-like viruses. Curr. Top. Microbiol. 2004, 17–31. [Google Scholar]
- Bertzbach, L.D.; Pfaff, F.; Pauker, V.I.; Kheimar, A.M.; Höper, D.; Härtle, S.; Karger, A.; Kaufer, B.B. The Transcriptional Landscape of Marek’s Disease Virus in Primary Chicken B Cells Reveals Novel Splice Variants and Genes. Viruses 2019, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Bertzbach, L.D.; Kheimar, A.; Ali, F.A.Z.; Kaufer, B.B. Viral Factors Involved in Marek’s Disease Virus (MDV) Pathogenesis. Curr. Clin. Microbiol. Rep. 2018, 5, 238–244. [Google Scholar] [CrossRef]
- Chbab, N.; Egerer, A.; Veiga, I.; Jarosinski, K.W.; Osterrieder, N. Viral control of vTR expression is critical for efficient formation and dissemination of lymphoma induced by Marek’s disease virus (MDV). Vet. Res. 2010, 41, 56. [Google Scholar] [CrossRef]
- Fragnet, L.; Blasco, M.A.; Klapper, W.; Rasschaert, D. The RNA subunit of telomerase is encoded by Marek’s disease virus. J. Virol. 2003, 77, 5985–5996. [Google Scholar] [CrossRef]
- Kaufer, B.B.; Arndt, S.; Trapp, S.; Osterrieder, N.; Jarosinski, K.W. Herpesvirus telomerase RNA (vTR) with a mutated template sequence abrogates herpesvirus-induced lymphomagenesis. PLoS Pathog. 2011, 7, e1002333. [Google Scholar] [CrossRef]
- Kaufer, B.B.; Trapp, S.; Jarosinski, K.W.; Osterrieder, N. Herpesvirus telomerase RNA(vTR)-dependent lymphoma formation does not require interaction of vTR with telomerase reverse transcriptase (TERT). PLoS Pathog. 2010, 6, e1001073. [Google Scholar] [CrossRef] [PubMed]
- Kheimar, A.; Previdelli, R.L.; Wight, D.J.; Kaufer, B.B. Telomeres and Telomerase: Role in Marek’s Disease Virus Pathogenesis, Integration and Tumorigenesis. Viruses 2017, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Trapp, S.; Parcells, M.S.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Kumar, P.M.; Nair, V.K.; Osterrieder, N. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 2006, 203, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Nair, V. Role of virus-encoded microRNAs in Avian viral diseases. Viruses 2014, 6, 1379–1394. [Google Scholar] [CrossRef]
- Zhuang, G.; Sun, A.; Teng, M.; Luo, J. A Tiny RNA that Packs a Big Punch: The Critical Role of a Viral miR-155 Ortholog in Lymphomagenesis in Marek’s Disease. Front Microbiol. 2017, 8, 1169. [Google Scholar] [CrossRef]
- Kheimar, A.; Trimpert, J.; Groenke, N.; Kaufer, B.B. Overexpression of cellular telomerase RNA enhances virus-induced cancer formation. Oncogene 2018, 38, 1778–1786. [Google Scholar] [CrossRef]
- Kheimar, A.; Kaufer, B.B. Epstein-Barr virus-encoded RNAs (EBERs) complement the loss of Herpesvirus telomerase RNA (vTR) in virus-induced tumor formation. Sci. Rep. 2018, 8, 209. [Google Scholar] [CrossRef]
- Bondada, M.S.; Yao, Y.; Nair, V. Multifunctional miR-155 Pathway in Avian Oncogenic Virus-Induced Neoplastic Diseases. Noncoding RNA 2019, 5, 24. [Google Scholar] [CrossRef]
- Yu, Z.H.; Teng, M.; Sun, A.J.; Yu, L.L.; Hu, B.; Qu, L.H.; Ding, K.; Cheng, X.C.; Liu, J.X.; Cui, Z.Z.; et al. Virus-encoded miR-155 ortholog is an important potential regulator but not essential for the development of lymphomas induced by very virulent Marek’s disease virus. Virology 2014, 448, 55–64. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, H.; Yao, Y.; Smith, L.P.; Kgosana, L.; Green, J.; Petherbridge, L.; Baigent, S.J.; Nair, V. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog. 2011, 7, e1001305. [Google Scholar] [CrossRef]
- Xu, S.; Xue, C.; Li, J.; Bi, Y.; Cao, Y. Marek’s disease virus type 1 microRNA miR-M3 suppresses cisplatin-induced apoptosis by targeting Smad2 of the transforming growth factor beta signal pathway. J. Virol. 2011, 85, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Strassheim, S.; Stik, G.; Rasschaert, D.; Laurent, S. mdv1-miR-M7-5p, located in the newly identified first intron of the latency-associated transcript of Marek’s disease virus, targets the immediate-early genes ICP4 and ICP27. J. Gen. Virol. 2012, 93, 1731–1742. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.; Anderson, A.; Bernberg, E.; Kamboj, S.; Huang, E.; Lagasse, G.; Isaacs, G.; Parcells, M.; Meyers, B.C.; et al. Sequence conservation and differential expression of Marek’s disease virus microRNAs. J. Virol. 2008, 82, 12213–12220. [Google Scholar] [CrossRef] [PubMed]
- Parnas, O.; Corcoran, D.L.; Cullen, B.R. Analysis of the mRNA targetome of microRNAs expressed by Marek’s disease virus. Microbiology 2014, 5, e01060–13. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Croce, C.M.; Michaille, J.J. miR-155: on the crosstalk between inflammation and cancer. Int. Rev. Immunol. 2009, 28, 264–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, N.; Luo, J.; Teng, M.; Moffat, K.; Shen, Z.; Watson, M.; Nair, V.; Yao, Y. Marek’s disease virus-encoded miR-155 ortholog critical for the induction of lymphomas is not essential for the proliferation of transformed cell lines. J. Virol. 2019, 93, e00713-19. [Google Scholar] [CrossRef]
- Sadigh, Y.; Tahiri-Alaoui, A.; Spatz, S.; Nair, V.; Ribeca, P. Pervasive differential splicing in Marek’s Disease Virus can discriminate CVI-988 vaccine strain from RB-1B virulent strain in chicken embryonic fibroblasts. bioRxiv 2019. [Google Scholar] [CrossRef]
- Gennart, I.; Coupeau, D.; Pejakovic, S.; Laurent, S.; Rasschaert, D.; Muylkens, B. Marek’s disease: Genetic regulation of gallid herpesvirus 2 infection and latency. Vet. J. (Lond., Engl. 1997) 2015, 205, 339–348. [Google Scholar] [CrossRef]
- Dunn, J.R.; Black Pyrkosz, A.; Steep, A.; Cheng, H.H. Identification of Marek’s disease virus genes associated with virulence of US strains. J. Gen. Virol. 2019, 100, 1132–1139. [Google Scholar] [CrossRef]
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Pathogenesis of Virus Infections. Fenner White’s Med Virol. 2017, 77–104. [Google Scholar]
- Witter, R.L. Avian tumor viruses: Persistent and evolving pathogens. Acta. Vet. Hung. 1997, 45, 251–266. [Google Scholar]
- Witter, R.L. Evolution of virulence of Marek’s disease virus: Evidence for a novel pathotype. Curr. Res. Marek’s Dis. 1996, 86–91. [Google Scholar]
- Witter, R.L. The changing landscape of Marek’s disease. Avian Pathol. 1998, 27, S46–S53. [Google Scholar] [CrossRef]
- Witter, R.L. Increased virulence of Marek’s disease virus field isolates. Avian Dis. 1997, 41, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Witter, R.L.; Gimeno, I.M.; Reed, W.M.; Bacon, L.D. An acute form of transient paralysis induced by highly virulent strains of Marek’s disease virus. Avian Dis. 1999, 43, 704–720. [Google Scholar] [CrossRef]
- Gimeno, I.M.; Witter, R.L.; Reed, W.M. Four distinct neurologic syndromes in Marek’s disease: Effect of viral strain and pathotype. Avian Dis. 1999, 43, 721–737. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, I.M.; Witter, R.L.; Hunt, H.D.; Lee, L.F.; Reddy, S.M.; Neumann, U. Marek’s disease virus infection in the brain: Virus replication, cellular infiltration, and major histocompatibility complex antigen expression. Vet. Pathol. 2001, 38, 491–503. [Google Scholar] [CrossRef]
- Witter, R.L.; Gimeno, I.M. Susceptibility of adult chickens, with and without prior vaccination, to challenge with Marek’s disease virus. Avian Dis. 2006, 50, 354–365. [Google Scholar] [CrossRef]
- Trimpert, J.; Groenke, N.; Jenckel, M.; He, S.; Kunec, D.; Szpara, M.L.; Spatz, S.J.; Osterrieder, N.; McMahon, D.P. A phylogenomic analysis of Marek’s disease virus reveals independent paths to virulence in Eurasia and North America. Evol. Appl. 2017, 10, 1091–1101. [Google Scholar] [CrossRef]
- Schat, K.A. History of the First-Generation Marek’s Disease Vaccines: The Science and Little-Known Facts. Avian Dis. 2016, 60, 715–724. [Google Scholar] [CrossRef]
- Reddy, S.M.; Izumiya, Y.; Lupiani, B. Marek’s disease vaccines: Current status, and strategies for improvement and development of vector vaccines. Vet. Microbiol. 2017, 206, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.I.; Cheng, H.H.; Chase-Topping, M.; Mays, J.K.; Anacleto, O.; Dunn, J.R.; Doeschl-Wilson, A. Pathogen transmission from vaccinated hosts can cause dose-dependent reduction in virulence. PLoS Biol. 2020, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.M.; Baigent, S.J.; Petherbridge, L.J.; Smith, L.P.; Nair, V.K. Comparative efficacy of BAC-derived recombinant SB-1 vaccine and the parent wild type strain in preventing replication, shedding and disease induced by virulent Marek’s disease virus. Res. Vet. Sci. 2010, 89, 140–145. [Google Scholar] [CrossRef]
- Atkins, K.E.; Read, A.F.; Savill, N.J.; Renz, K.G.; Walkden-Brown, S.W.; Woolhouse, M.E. Modelling Marek’s disease virus (MDV) infection: parameter estimates for mortality rate and infectiousness. BMC Vet. Res. 2011, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.A.; Dunn, P.A.; Read, A.F. Modeling Marek’s disease virus transmission: A framework for evaluating the impact of farming practices and evolution. Epidemics 2018, 23, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Nair, V. Evolution of Marek’s disease—A paradigm for incessant race between the pathogen and the host. Vet. J. (Lond., Engl. 1997) 2005, 170, 175–183. [Google Scholar] [CrossRef]
- Rozins, C.; Day, T. The industrialization of farming may be driving virulence evolution. Evol. Appl. 2017, 10, 189–198. [Google Scholar] [CrossRef]
- Atkins, K.E.; Read, A.F.; Savill, N.J.; Renz, K.G.; Islam, A.F.; Walkden-Brown, S.W.; Woolhouse, M.E. Vaccination and reduced cohort duration can drive virulence evolution: Marek’s disease virus and industrialized agriculture. Evolution 2013, 67, 851–860. [Google Scholar] [CrossRef]
- Shamblin, C.E.; Greene, N.; Arumugaswami, V.; Dienglewicz, R.L.; Parcells, M.S. Comparative analysis of Marek’s disease virus (MDV) glycoprotein-, lytic antigen pp38- and transformation antigen Meq-encoding genes: association of meq mutations with MDVs of high virulence. Vet. Microbiol. 2004, 102, 147–167. [Google Scholar] [CrossRef]
- Firth, C.; Kitchen, A.; Shapiro, B.; Suchard, M.A.; Holmes, E.C.; Rambaut, A. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol. Biol. Evol. 2010, 27, 2038–2051. [Google Scholar] [CrossRef]
- Trimpert, J.; Groenke, N.; Kunec, D.; Eschke, K.; He, S.; McMahon, D.P.; Osterrieder, N. A proofreading-impaired herpesvirus generates populations with quasispecies-like structure. Nat. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Padhi, A.; Parcells, M.S. Positive Selection Drives Rapid Evolution of the meq Oncogene of Marek’s Disease Virus. PLoS ONE 2016, 11, e0162180. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.; Shalem-Meilin, E.; Heller, E.D. Marek’s disease vaccines cause temporary U-lymphocyte dysfunction and reduced resistance to infection in chicks. Avian Pathol. 1992, 21, 621–631. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kleven, S.H.; Eidson, C.S.; Anderson, D.P.; Fletcher, O.J. Decrease of antibody response to Mycoplasma synoviae in chickens infected with Marek’s disease herpesvirus. Am. J. Vet. Res. 1972, 33, 2037–2042. [Google Scholar] [PubMed]
- Spatz, S.J.; Petherbridge, L.; Zhao, Y.; Nair, V. Comparative full-length sequence analysis of oncogenic and vaccine (Rispens) strains of Marek’s disease virus. J. Gen. Virol. 2007, 88, 1080–1096. [Google Scholar] [CrossRef]
- McPherson, M.C.; Cheng, H.H.; Delany, M.E. Marek’s disease herpesvirus vaccines integrate into chicken host chromosomes yet lack a virus-host phenotype associated with oncogenic transformation. Vaccine 2016, 34, 5554–5561. [Google Scholar] [CrossRef]
- Conradie, A.M.; Bertzbach, L.D.; Bhandari, N.; Parcells, M.; Kaufer, B.B. A Common Live-Attenuated Avian Herpesvirus Vaccine Expresses a Very Potent Oncogene. mSphere 2019, 4, 5. [Google Scholar] [CrossRef]
- Deng, X.; Li, X.; Shen, Y.; Qiu, Y.; Shi, Z.; Shao, D.; Jin, Y.; Chen, H.; Ding, C.; Li, L.; et al. The Meq oncoprotein of Marek’s disease virus interacts with p53 and inhibits its transcriptional and apoptotic activities. Virol. J. 2010, 7, 348. [Google Scholar] [CrossRef]
- Li, K.; Liu, Y.; Xu, Z.; Zhang, Y.; Luo, D.; Gao, Y.; Qian, Y.; Bao, C.; Liu, C.; Zhang, Y.; et al. Avian oncogenic herpesvirus antagonizes the cGAS-STING DNA-sensing pathway to mediate immune evasion. PLoS Pathog. 2019, 15, e1007999. [Google Scholar] [CrossRef]
- Bertzbach, L.D.; Conradie, A.M.; Hahn, F.; Wild, M.; Marschall, M.; Kaufer, B.B. Artesunate derivative TF27 inhibits replication and pathogenesis of an oncogenic avian alphaherpesvirus. Antiviral. Res. 2019, 171, 104606. [Google Scholar] [CrossRef]
- Smith, J.; Gheyas, A.; Burt, D.W. Animal genomics and infectious disease resistance in poultry. Revue. Sci. Tech. 2016, 35, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; He, Y.; Ding, Y.; Liu, G.E.; Zhang, H.; Cheng, H.H.; Taylor, R.L., Jr.; Song, J. Genetic assessment of inbred chicken lines indicates genomic signatures of resistance to Marek’s disease. J. Anim. Sci. Biotechnol. 2018, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Heidari, M.; Zhang, L.; Zhang, H. MicroRNA profiling in the bursae of Marek’s disease virus-infected resistant and susceptible chicken lines. Genomics 2020. [Google Scholar] [CrossRef]
- Bai, H.; He, Y.; Ding, Y.; Chang, S.; Zhang, H.; Chen, J.; Song, J. Parent-of-origin has no detectable effect on survival days of Marek’s disease virus infected White Leghorns. Poult. Sci. 2019, 98, 4498–4503. [Google Scholar] [CrossRef] [PubMed]
- Sid, H.; Schusser, B. Applications of Gene Editing in Chickens: A New Era Is on the Horizon. Front Genet. 2018, 9, 456. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertzbach, L.D.; Conradie, A.M.; You, Y.; Kaufer, B.B. Latest Insights into Marek’s Disease Virus Pathogenesis and Tumorigenesis. Cancers 2020, 12, 647. https://doi.org/10.3390/cancers12030647
Bertzbach LD, Conradie AM, You Y, Kaufer BB. Latest Insights into Marek’s Disease Virus Pathogenesis and Tumorigenesis. Cancers. 2020; 12(3):647. https://doi.org/10.3390/cancers12030647
Chicago/Turabian StyleBertzbach, Luca D., Andelé M. Conradie, Yu You, and Benedikt B. Kaufer. 2020. "Latest Insights into Marek’s Disease Virus Pathogenesis and Tumorigenesis" Cancers 12, no. 3: 647. https://doi.org/10.3390/cancers12030647
APA StyleBertzbach, L. D., Conradie, A. M., You, Y., & Kaufer, B. B. (2020). Latest Insights into Marek’s Disease Virus Pathogenesis and Tumorigenesis. Cancers, 12(3), 647. https://doi.org/10.3390/cancers12030647




