Crosstalk between HER2 and PD-1/PD-L1 in Breast Cancer: From Clinical Applications to Mathematical Models
Abstract
1. Introduction
2. Current HER2+-Targeted Therapeutic Agents and Drug Resistance
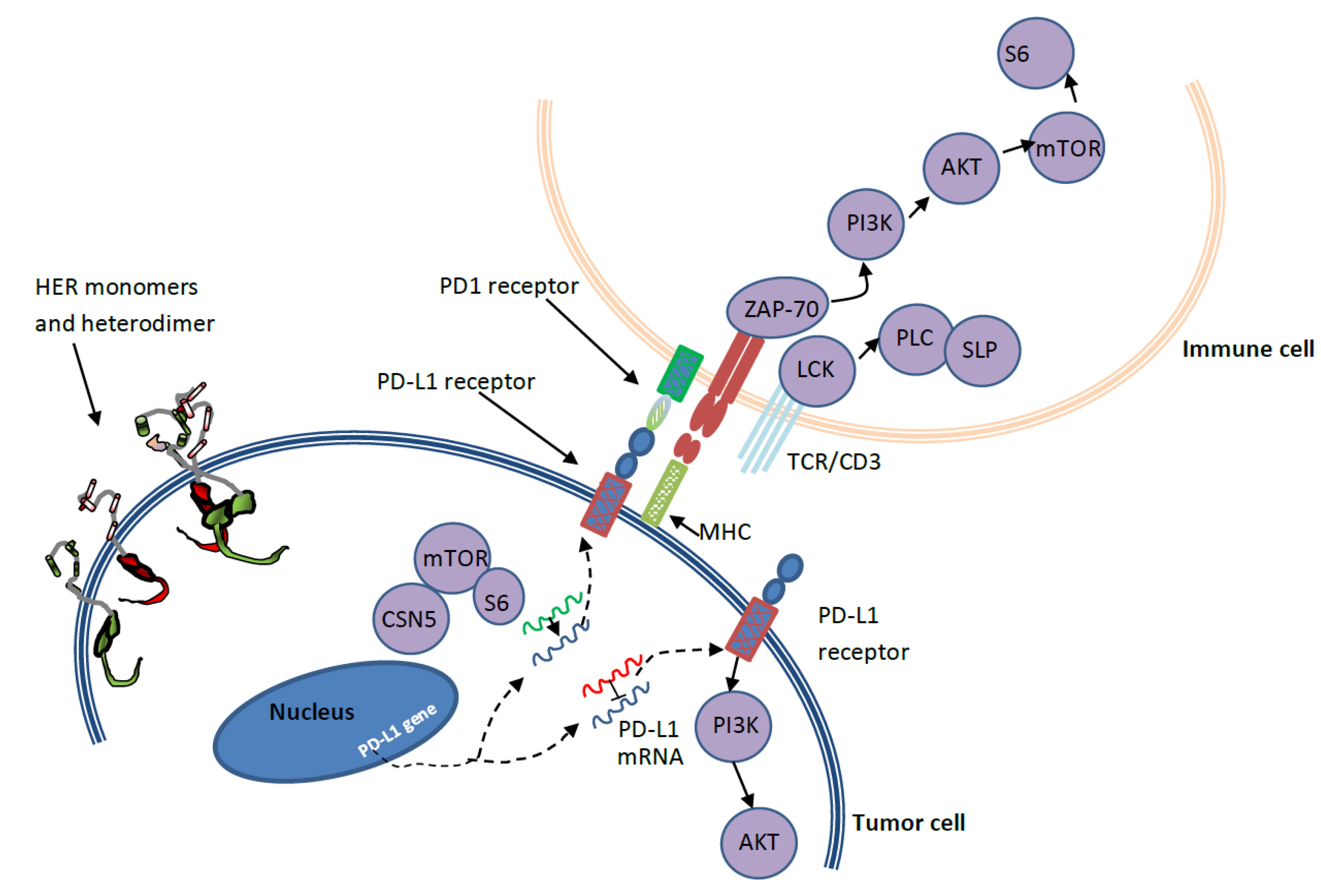
3. PD-1/PD-L1 and HER2 Crosstalk in Breast Cancer
4. Mathematical Models Used for Breast Cancer Management
- Out of the 5 FDA-approved anti-HER2 drugs, only the dynamics of trastuzumab and T-DM1 have been studied using a mathematical model to a certain extent. The dynamics of other drugs have yet to be explored on a quantitative basis. Such drug-specific models can be used for treatment planning and dose optimization [30,34,35,64,160,187].
- Developing mathematical models in terms of the biomarkers related to disease prognosis (e.g., PDL1 expression + high Tregs + less TILs = poor prognosis), and treatment response (e.g., presence of TILs favors response to trastuzumab) can help to identify patient cohorts that will benefit from a certain therapy [146].
- Mathematical models can be used to quantify drug dynamics of potential new drugs and different combinations and to explore possible additive or synergistic drug interaction when used in combinations [52].
5. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Wei, H.C. Mathematical Modeling of Tumor Growth: The MCF-7 Breast Cancer Cell Line. Math. Biosci. Eng. 2019, 16, 6512–6535. [Google Scholar] [PubMed]
- Schnitt, S.J. Classification and Prognosis of Invasive Breast Cancer: From Morphology to Molecular Taxonomy. Mod. Pathol. 2010, 23, S60. [Google Scholar] [PubMed]
- Emens, L.A.; Braiteh, F.S.; Cassier, P.; Delord, J.; Eder, J.P.; Fasso, M.; Xiao, Y.; Wang, Y.; Molinero, L.; Chen, D.S. Abstract 2859: Inhibition of Pd-L1 by mpdl3280a Leads to Clinical Activity in Patients with Metastatic Triple-Negative Breast Cancer (TNBC). Inhibition of PD-L1 by MPDL3280A leads to clinical activity in patients with metastatic triple-negative breast cancer (TNBC). Cancer Res. 2015, 75, 2859. [Google Scholar]
- Smith, S.E.; Mellor, P.; Ward, A.K.; Kendall, S.; McDonald, M.; Vizeacoumar, F.S.; Vizeacoumar, F.J.; Napper, S.; Anderson, D.H. Molecular Characterization of Breast Cancer Cell Lines through Multiple Omic Approaches. Breast Cancer Res. 2017, 19, 65. [Google Scholar] [PubMed]
- Gatalica, Z.; Vranic, S.; Krušlin, B.; Poorman, K.; Stafford, P.; Kacerovska, D.; Senarathne, W.; Florento, E.; Contreras, E.; Leary, A. Comparison of the Biomarkers for Targeted Therapies in Primary Extra-mammary and Mammary Paget’s Disease. Cancer Med. 2020, 9, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Daemen, A.; Manning, G. HER2 is Not a Cancer Subtype but rather a Pan-Cancer Event and is Highly Enriched in AR-Driven Breast Tumors. Breast Cancer Res. 2018, 20, 8. [Google Scholar]
- Vranic, S.; Feldman, R.; Gatalica, Z. Apocrine Carcinoma of the Breast: A Brief Update on the Molecular Features and Targetable Biomarkers. Bosn. J. Basic Med. Sci. 2017, 17, 9–11. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. Arch. Pathol. Lab. Med. 2018, 142, 1364–1382. [Google Scholar] [CrossRef]
- Ortiz, A.G.; Muñoz, A.S.; Parrado, M.R.C.; Pérez, M.Á.; Entrena, N.R.; Dominguez, A.R.; Conejo, E.A. Deciphering HER2 Breast Cancer Disease: Biological and Clinical Implications. Front. Oncol. 2019, 9, 1124. [Google Scholar] [CrossRef]
- He, L.; Du, Z.; Xiong, X.; Ma, H.; Zhu, Z.; Gao, H.; Cao, J.; Li, T.; Li, H.; Yang, K. Targeting Androgen Receptor in Treating HER2 Positive Breast Cancer. Sci. Rep. 2017, 7, 14584. [Google Scholar]
- Rexer, B.N.; Arteaga, C.L. Intrinsic and Acquired Resistance to HER2-Targeted Therapies in HER2 Gene-Amplified Breast Cancer: Mechanisms and Clinical Implications. Crit. Rev. Oncog. 2012, 17. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Bang, Y. HER2-Targeted Therapies—a Role Beyond Breast Cancer. Nat. Rev. Clin. Oncol. 2020, 17, 33–48. [Google Scholar] [PubMed]
- Nixon, N.; Hannouf, M.; Verma, S. A Review of the Value of Human Epidermal Growth Factor Receptor 2 (HER2)-Targeted Therapies in Breast Cancer. Eur. J. Cancer 2018, 89, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.G.; O’Neill, S.; Salimu, J.; Breslin, S.; Clayton, A.; Crown, J.; O’Driscoll, L. Resistance to HER2-Targeted Anti-Cancer Drugs is Associated with Immune Evasion in Cancer Cells and their Derived Extracellular Vesicles. Oncoimmunology 2017, 6, e1362530. [Google Scholar] [CrossRef] [PubMed]
- Vernieri, C.; Milano, M.; Brambilla, M.; Mennitto, A.; Maggi, C.; Cona, M.S.; Prisciandaro, M.; Fabbroni, C.; Celio, L.; Mariani, G. Resistance Mechanisms to Anti-HER2 Therapies in HER2-Positive Breast Cancer: Current Knowledge, New Research Directions and Therapeutic Perspectives. Crit. Rev. Oncol. 2019. [Google Scholar] [CrossRef]
- Ayoub, N.M.; Al-Shami, K.M.; Yaghan, R.J. Immunotherapy for HER2-Positive Breast Cancer: Recent Advances and Combination Therapeutic Approaches. Breast Cancer Targets Ther. 2019, 11, 53. [Google Scholar] [CrossRef]
- Doi, T.; Iwata, H.; Tsurutani, J.; Takahashi, S.; Park, H.; Redfern, C.H.; Shitara, K.; Shimizu, C.; Taniguchi, H.; Iwasa, T. Single Agent Activity of DS-8201a, a HER2-Targeting Antibody-Drug Conjugate, in Heavily Pretreated HER2 Expressing Solid Tumors. J. Clin. Oncol. 2017, 35, 108. [Google Scholar] [CrossRef]
- Puglisi, F.; Fontanella, C.; Amoroso, V.; Bianchi, G.V.; Bisagni, G.; Falci, C.; Fontana, A.; Generali, D.; Gianni, L.; Grassadonia, A. Current Challenges in HER2-Positive Breast Cancer. Crit. Rev. Oncol. 2016, 98, 211–221. [Google Scholar] [CrossRef]
- Luen, S.J.; Savas, P.; Fox, S.B.; Salgado, R.; Loi, S. Tumour-Infiltrating Lymphocytes and the Emerging Role of Immunotherapy in Breast Cancer. Pathology 2017, 49, 141–155. [Google Scholar] [CrossRef]
- Yu, W.; Sun, G.; Li, J.; Xu, J.; Wang, X. Mechanisms and Therapeutic Potentials of Cancer Immunotherapy in Combination with Radiotherapy and/or Chemotherapy. Cancer Lett. 2019, 452, 66–70. [Google Scholar] [CrossRef]
- Galluzzi, L.; Buque, A.; Kepp, O.; Zitvogel, L.; Kroemer, G. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell 2015, 28, 690–714. [Google Scholar] [CrossRef]
- Muenst, S.; Schaerli, A.; Gao, F.; Däster, S.; Trella, E.; Droeser, R.; Muraro, M.; Zajac, P.; Zanetti, R.; Gillanders, W. Expression of Programmed Death Ligand 1 (PD-L1) is Associated with Poor Prognosis in Human Breast Cancer. Breast Cancer Res. Treat. 2014, 146, 15–24. [Google Scholar] [CrossRef]
- McLemore, L.E.; Janakiram, M.; Albanese, J.; Shapiro, N.; Lo, Y.; Zang, X.; Fineberg, S. An Immunoscore using PD-L1, CD68, and Tumor-Infiltrating Lymphocytes (TILs) to Predict Response to Neoadjuvant Chemotherapy in Invasive Breast Cancer. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Joneja, U.; Vranic, S.; Swensen, J.; Feldman, R.; Chen, W.; Kimbrough, J.; Xiao, N.; Reddy, S.; Palazzo, J.; Gatalica, Z. Comprehensive Profiling of Metaplastic Breast Carcinomas Reveals Frequent Overexpression of Programmed Death-Ligand 1. J. Clin. Pathol. 2017, 70, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, F.; Finetti, P.; Birnbaum, D.; Mamessier, E. The PD1/PDL1 Axis, a Promising Therapeutic Target in Aggressive Breast Cancers. Oncoimmunology 2016, 5, e1085148. [Google Scholar] [CrossRef] [PubMed]
- McKenna, M.T.; Weis, J.A.; Barnes, S.L.; Tyson, D.R.; Miga, M.I.; Quaranta, V.; Yankeelov, T.E. A Predictive Mathematical Modeling Approach for the Study of Doxorubicin Treatment in Triple Negative Breast Cancer. Sci. Rep. 2017, 7, 5725. [Google Scholar] [CrossRef]
- Quaranta, V.; Weaver, A.M.; Cummings, P.T.; Anderson, A.R. Mathematical Modeling of Cancer: The Future of Prognosis and Treatment. Clin. Chim. Acta 2005, 357, 173–179. [Google Scholar] [CrossRef]
- Enderling, H.; Chaplain, M.A.; Anderson, A.R.; Vaidya, J.S. A Mathematical Model of Breast Cancer Development, Local Treatment and Recurrence. J. Theor. Biol. 2007, 246, 245–259. [Google Scholar] [CrossRef]
- Benzekry, S.; Lamont, C.; Beheshti, A.; Tracz, A.; Ebos, J.M.; Hlatky, L.; Hahnfeldt, P. Classical Mathematical Models for Description and Prediction of Experimental Tumor Growth. PLoS Comput. Biol. 2014, 10, e1003800. [Google Scholar] [CrossRef]
- Mkango, S.B.; Shaban, N.; Mureithi, E.; Ngoma, T. Dynamics of Breast Cancer Under Different Rates of Chemoradiotherapy. Comput. Math. Methods Med. 2019, 2019. [Google Scholar] [CrossRef]
- Moore, H. How to Mathematically Optimize Drug Regimens using Optimal Control. J. Pharmacokinet. Pharmacodyn. 2018, 45, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Deisboeck, T.S. Mathematical Modeling in Cancer Drug Discovery. Drug Discov. Today 2014, 19, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Dogra, P.; Butner, J.D.; Chuang, Y.; Caserta, S.; Goel, S.; Brinker, C.J.; Cristini, V.; Wang, Z. Mathematical Modeling in Cancer Nanomedicine: A Review. Biomed. Microdevices 2019, 21, 40. [Google Scholar] [CrossRef]
- Dey, S.K.; Dey, S.C. Mathematical Modeling of Breast Cancer Treatment. In Applied Mathematics; Springer: Berlin, Germany, 2015; pp. 149–160. [Google Scholar]
- Yazdjerdi, P.; Meskin, N.; Al-Naemi, M.; Al Moustafa, A.; Kovács, L. Reinforcement Learning-Based Control of Tumor Growth Under Anti-Angiogenic Therapy. Comput. Methods Programs Biomed. 2019, 173, 15–26. [Google Scholar] [CrossRef]
- Oke, S.; Matadi, M.; Xulu, S. Optimal Control Analysis of a Mathematical Model for Breast Cancer. Math. Comput. Appl. 2018, 23, 21. [Google Scholar]
- Brocato, T.A.; Brown-Glaberman, U.; Wang, Z.; Selwyn, R.G.; Wilson, C.M.; Wyckoff, E.F.; Lomo, L.C.; Saline, J.L.; Hooda-Nehra, A.; Pasqualini, R.; et al. Predicting Breast Cancer Response to Neoadjuvant Chemotherapy Based on Tumor Vascular Features in Needle Biopsies. JCI Insight 2019, 5. [Google Scholar] [CrossRef]
- Liu, L.; Lam, C.K.; Alderson, R.; Long, V.; Yang, Y.; Burns, R.; Widjaja, L.; Li, J.; Wolf, C.; Ciccarone, V. Selection of a Bispecific Trivalent HER2 X CD137 TRIDENT Format Providing Optimal Tumor-Anchored Immune Co-Stimulation. Cancer Res. 2019, 79. [Google Scholar] [CrossRef]
- Centanni, M.; Moes, D.J.A.; Trocóniz, I.F.; Ciccolini, J.; van Hasselt, J.C. Clinical Pharmacokinetics and Pharmacodynamics of Immune Checkpoint Inhibitors. Clin. Pharmacokinet. 2019, 58, 835–857. [Google Scholar] [CrossRef]
- Chase, J.G.; Preiser, J.; Dickson, J.L.; Pironet, A.; Chiew, Y.S.; Pretty, C.G.; Shaw, G.M.; Benyo, B.; Moeller, K.; Safaei, S. Next-Generation, Personalised, Model-Based Critical Care Medicine: A State-of-the Art Review of in Silico Virtual Patient Models, Methods, and Cohorts, and how to Validation Them. Biomed. Eng. Online 2018, 17, 24. [Google Scholar] [CrossRef]
- Okines, A.F.; Cunningham, D. Trastuzumab: A Novel Standard Option for Patients with HER-2-Positive Advanced Gastric or Gastro-Oesophageal Junction Cancer. Ther. Adv. Gastroenterol. 2012, 5, 301–318. [Google Scholar] [CrossRef]
- Rinnerthaler, G.; Gampenrieder, S.P.; Greil, R. HER2 Directed Antibody-Drug-Conjugates Beyond T-DM1 in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 1115. [Google Scholar] [CrossRef] [PubMed]
- Pegram, M.D.; Konecny, G.E.; O’Callaghan, C.; Beryt, M.; Pietras, R.; Slamon, D.J. Rational Combinations of Trastuzumab with Chemotherapeutic Drugs used in the Treatment of Breast Cancer. J. Natl. Cancer Inst. 2004, 96, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Parakh, S.; Gan, H.K.; Parslow, A.C.; Burvenich, I.J.; Burgess, A.W.; Scott, A.M. Evolution of Anti-HER2 Therapies for Cancer Treatment. Cancer Treat. Rev. 2017, 59, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, C.L.; Sliwkowski, M.X.; Osborne, C.K.; Perez, E.A.; Puglisi, F.; Gianni, L. Treatment of HER2-Positive Breast Cancer: Current Status and Future Perspectives. Nat. Rev. Clin. Oncol. 2012, 9, 16. [Google Scholar] [CrossRef]
- Muller, P.; Kreuzaler, M.; Khan, T.; Thommen, D.S.; Martin, K.; Glatz, K.; Savic, S.; Harbeck, N.; Nitz, U.; Gluz, O.; et al. Trastuzumab Emtansine (T-DM1) Renders HER2+ Breast Cancer Highly Susceptible to CTLA-4/PD-1 Blockade. Sci. Transl. Med. 2015, 7, 315ra188. [Google Scholar] [CrossRef]
- Escrivá-de-Romaní, S.; Arumí, M.; Bellet, M.; Saura, C. HER2-Positive Breast Cancer: Current and New Therapeutic Strategies. Breast 2018, 39, 80–88. [Google Scholar] [CrossRef]
- Bedard, P.L.; Li, S.; Wisinski, K.B.; Yang, E.S.; Limaye, S.A.; Mitchell, E.P.; Zwiebel, J.A.; Moscow, J.; Gray, R.J.; McShane, L.M. NCI Molecular Analysis for Therapy Choice (NCI-MATCH EAY131) Arm B: Phase II Study of Afatinib in Patients (Pts) with HER2 (ERBB2) Activating Mutations. Cancer Res. 2016, 69, S137. [Google Scholar]
- Kim, H.; Yoon, Y.; Kim, J.; Han, S.; Hur, H.; Park, J.; Lee, J.; Oh, D.; Im, S.; Bang, Y. Lapatinib, a Dual EGFR and HER2 Tyrosine Kinase Inhibitor, Downregulates Thymidylate Synthase by Inhibiting the Nuclear Translocation of EGFR and HER2. PLoS ONE 2009, 4, e5933. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Xie, J.D.; Chen, X.G.; Sim, H.M.; Zhang, X.; Liang, Y.J.; Singh, S.; Talele, T.T.; Sun, Y.; Ambudkar, S.V.; et al. Neratinib Reverses ATP-Binding Cassette B1-Mediated Chemotherapeutic Drug Resistance in vitro, in vivo, and ex vivo. Mol. Pharmacol. 2012, 82, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Madell, R. Her2-Positive Breast Cancer Survival Rates and Other Statistics; Healthline: San Francisco, CA, USA, 2019. [Google Scholar]
- Eladdadi, A.; Isaacson, D. A Mathematical Model for the Effects of HER2 Over-Expression on Cell Cycle Progression in Breast Cancer. Bull. Math. Biol. 2011, 73, 2865–2887. [Google Scholar] [CrossRef]
- Timms, J.F.; White, S.L.; O’Hare, M.J.; Waterfield, M.D. Effects of ErbB-2 Overexpression on Mitogenic Signalling and Cell Cycle Progression in Human Breast Luminal Epithelial Cells. Oncogene 2002, 21, 6573–6586. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, F.; Gavaghan, D.; Osborne, J.; Barrett, I.; You, T.; Ghadially, H.; Sainson, R.; Wilkinson, R.; Byrne, H. A Mathematical Model of Antibody-Dependent Cellular Cytotoxicity (ADCC). J. Theor. Biol. 2018, 436, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Jarrett, A.M.; Shah, A.; Bloom, M.J.; McKenna, M.T.; Hormuth, D.A.; Yankeelov, T.E.; Sorace, A.G. Experimentally-Driven Mathematical Modeling to Improve Combination Targeted and Cytotoxic Therapy for HER2 Breast Cancer. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ogitani, Y.; Hagihara, K.; Oitate, M.; Naito, H.; Agatsuma, T. Bystander Killing Effect of DS-8201a, a Novel Anti-human Epidermal Growth Factor Receptor 2 Antibody–drug Conjugate, in Tumors with Human Epidermal Growth Factor Receptor 2 Heterogeneity. Cancer Sci. 2016, 107, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Goetsch, L.; Dumontet, C.; Corvaïa, N. Strategies and Challenges for the Next Generation of Antibody–drug Conjugates. Nat. Rev. Drug Discov. 2017, 16, 315. [Google Scholar] [CrossRef]
- Li, J.Y.; Perry, S.R.; Muniz-Medina, V.; Wang, X.; Wetzel, L.K.; Rebelatto, M.C.; Hinrichs, M.J.M.; Bezabeh, B.Z.; Fleming, R.L.; Dimasi, N. A Biparatopic HER2-Targeting Antibody-Drug Conjugate Induces Tumor Regression in Primary Models Refractory to Or Ineligible for HER2-Targeted Therapy. Cancer Cell 2016, 29, 117–129. [Google Scholar] [CrossRef]
- van der Lee, M.M.; Groothuis, P.G.; Ubink, R.; van der Vleuten, M.A.; van Achterberg, T.A.; Loosveld, E.M.; Damming, D.; Jacobs, D.C.; Rouwette, M.; Egging, D.F.; et al. The Preclinical Profile of the Duocarmycin-Based HER2-Targeting ADC SYD985 Predicts for Clinical Benefit in Low HER2-Expressing Breast Cancers. Mol. Cancer. Ther. 2015, 14, 692–703. [Google Scholar] [CrossRef]
- Yao, X.; Jiang, J.; Wang, X.; Huang, C.; Li, D.; Xie, K.; Xu, Q.; Li, H.; Li, Z.; Lou, L. A Novel Humanized Anti-HER2 Antibody Conjugated with MMAE Exerts Potent Anti-Tumor Activity. Breast Cancer Res. Treat. 2015, 153, 123–133. [Google Scholar] [CrossRef]
- Faria, M.; Peay, M.; Lam, B.; Ma, E.; Yuan, M.; Waldron, M.; Mylott, W.R.; Liang, M.; Rosenbaum, A.I. Multiplex LC-MS/MS Assays for Clinical Bioanalysis of MEDI4276, an Antibody-Drug Conjugate of Tubulysin Analogue Attached Via Cleavable Linker to a Biparatopic Humanized Antibody Against HER-2. Antibodies 2019, 8, 11. [Google Scholar] [CrossRef]
- Oganesyan, V.; Peng, L.; Bee, J.S.; Li, J.; Perry, S.R.; Comer, F.; Xu, L.; Cook, K.; Senthil, K.; Clarke, L.; et al. Structural Insights into the Mechanism of Action of a Biparatopic Anti-HER2 Antibody. J. Biol. Chem. 2018, 293, 8439–8448. [Google Scholar] [CrossRef]
- Tamura, K.; Tsurutani, J.; Takahashi, S.; Iwata, H.; Krop, I.E.; Redfern, C.; Sagara, Y.; Doi, T.; Park, H.; Murthy, R.K. Trastuzumab Deruxtecan (DS-8201a) in Patients with Advanced HER2-Positive Breast Cancer Previously Treated with Trastuzumab Emtansine: A Dose-Expansion, Phase 1 Study. Lancet Oncol. 2019, 20, 816–826. [Google Scholar] [CrossRef]
- Hurvitz, S.; Galsky, M.; Shahidi, J.; Zhang, G.; Raza, S.; Necchi, A. 370TiP A Phase Ib, Multicenter, Open-Label Study of the Antibody-Drug Conjugate Trastuzumab Deruxtecan (DS-8201a) Combination with Nivolumab for Advanced HER2-Expressing Breast or Urothelial Cancer. Ann. Oncol. 2018, 29, mdy272.358. [Google Scholar] [CrossRef]
- Modi, S.; Saura, C.; Yamashita, T.; Park, Y.H.; Kim, S.; Tamura, K.; Andre, F.; Iwata, H.; Ito, Y.; Tsurutani, J. Trastuzumab Deruxtecan in Previously Treated HER2-Positive Breast Cancer. N. Engl. J. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Iwata, T.N.; Sugihara, K.; Wada, T.; Agatsuma, T. [Fam-] Trastuzumab Deruxtecan (DS-8201a)-Induced Antitumor Immunity is Facilitated by the Anti-CTLA-4 Antibody in a Mouse Model. PLoS ONE 2019, 14, e0222280. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.; Zhao, R.; Haeno, H.; Vivanco, I.; Michor, F. Mathematical Modeling Identifies Optimum Lapatinib Dosing Schedules for the Treatment of Glioblastoma Patients. PLoS Comput. Biol. 2018, 14, e1005924. [Google Scholar] [CrossRef] [PubMed]
- Cadoo, K.A.; Gajria, D.; Suh, E.; Patil, S.; Theodoulou, M.; Norton, L.; Hudis, C.A.; Traina, T.A. Decreased Gastrointestinal Toxicity Associated with a Novel Capecitabine Schedule (7 Days on and 7 Days Off): A Systematic Review. NPJ Breast Cancer 2016, 2, 1–5. [Google Scholar] [CrossRef]
- Gajria, D.; Gonzalez, J.; Feigin, K.; Patil, S.; Chen, C.; Theodoulou, M.; Drullinsky, P.; D’Andrea, G.; Lake, D.; Norton, L. Phase II Trial of a Novel Capecitabine Dosing Schedule in Combination with Lapatinib for the Treatment of Patients with HER2-Positive Metastatic Breast Cancer. Breast Cancer Res. Treat. 2012, 131, 111–116. [Google Scholar] [CrossRef]
- Fehling-Kaschek, M.; Peckys, D.B.; Kaschek, D.; Timmer, J.; de Jonge, N. Mathematical Modeling of Drug-Induced Receptor Internalization in the HER2-Positive SKBR3 Breast Cancer Cell-Line. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef]
- Jarrett, A.M.; Bloom, M.J.; Godfrey, W.; Syed, A.K.; Ekrut, D.A.; Ehrlich, L.I.; Yankeelov, T.E.; Sorace, A.G. Mathematical Modelling of Trastuzumab-Induced Immune Response in an in vivo Murine Model of HER2 Breast Cancer. Math. Med. Biol. A J. IMA 2019, 36, 381–410. [Google Scholar] [CrossRef]
- Nahta, R.; Yu, D.; Hung, M.; Hortobagyi, G.N.; Esteva, F.J. Mechanisms of Disease: Understanding Resistance to HER2-Targeted Therapy in Human Breast Cancer. Nat. Rev. Clin. Oncol. 2006, 3, 269. [Google Scholar] [CrossRef]
- Crosby, E.J.; Lei, G.; Wei, J.; Yang, X.Y.; Wang, T.; Liu, C.; Lyerly, H.K.; Hartman, Z.C. Abstract a22: Augmentation of a Novel Adenoviral Vaccine Strategy by Checkpoint Inhibitors. Abstract A22: Augmentation of a novel adenoviral vaccine strategy by checkpoint inhibitors. Cancer Immunol. Res. 2018, 6, A22. [Google Scholar]
- Ishizuka, J.J.; Manguso, R.T.; Cheruiyot, C.K.; Bi, K.; Panda, A.; Iracheta-Vellve, A.; Miller, B.C.; Du, P.P.; Yates, K.B.; Dubrot, J. Loss of ADAR1 in Tumours Overcomes Resistance to Immune Checkpoint Blockade. Nature 2019, 565, 43. [Google Scholar] [CrossRef]
- Colomer, R.; Montero, S.; Lluch, A.; Ojeda, B.; Barnadas, A.; Casado, A.; Massuti, B.; Cortes-Funes, H.; Lloveras, B. Circulating HER2 Extracellular Domain and Resistance to Chemotherapy in Advanced Breast Cancer. Clin. Cancer Res. 2000, 6, 2356–2362. [Google Scholar] [PubMed]
- Molina, M.A.; Saez, R.; Ramsey, E.E.; Garcia-Barchino, M.J.; Rojo, F.; Evans, A.J.; Albanell, J.; Keenan, E.J.; Lluch, A.; Garcia-Conde, J.; et al. NH(2)-Terminal Truncated HER-2 Protein but Not Full-Length Receptor is Associated with Nodal Metastasis in Human Breast Cancer. Clin. Cancer Res. 2002, 8, 347–353. [Google Scholar]
- Lipton, A.; Ali, S.M.; Leitzel, K.; Demers, L.; Chinchilli, V.; Engle, L.; Harvey, H.A.; Brady, C.; Nalin, C.M.; Dugan, M.; et al. Elevated Serum Her-2/Neu Level Predicts Decreased Response to Hormone Therapy in Metastatic Breast Cancer. J. Clin. Oncol. 2002, 20, 1467–1472. [Google Scholar] [CrossRef]
- Jordan, N.V.; Bardia, A.; Wittner, B.S.; Benes, C.; Ligorio, M.; Zheng, Y.; Yu, M.; Sundaresan, T.K.; Licausi, J.A.; Desai, R. HER2 Expression Identifies Dynamic Functional States within Circulating Breast Cancer Cells. Nature 2016, 537, 102. [Google Scholar] [CrossRef] [PubMed]
- Alaoui-Jamali, M.A.; Paterson, J.; Al Moustafa, A.; Yen, L. The Role of ErbB-2 Tyrosine Kinase Receptor in Cellular Intrinsic Chemoresistance: Mechanisms and Implications. Biochem. Cell Biol. 1997, 75, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y.; Lan, K.; Zhou, X.; Tan, M.; Esteva, F.J.; Sahin, A.A.; Klos, K.S.; Li, P.; Monia, B.P.; Nguyen, N.T. PTEN Activation Contributes to Tumor Inhibition by Trastuzumab, and Loss of PTEN Predicts Trastuzumab Resistance in Patients. Cancer Cell 2004, 6, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, C.; Schiff, R. HER2: Biology, Detection, and Clinical Implications. Arch. Pathol. Lab. Med. 2011, 135, 55–62. [Google Scholar] [PubMed]
- Tural, D.; Akar, E.; Mutlu, H.; Kilickap, S. P95 HER2 Fragments and Breast Cancer Outcome. Expert Rev. Anticancer Ther. 2014, 14, 1089–1096. [Google Scholar] [CrossRef]
- Loibl, S.; Gianni, L. HER2-Positive Breast Cancer. Lancet 2017, 389, 2415–2429. [Google Scholar] [CrossRef]
- Loi, S.; Giobbie-Hurder, A.; Gombos, A.; Bachelot, T.; Hui, R.; Curigliano, G.; Campone, M.; Biganzoli, L.; Bonnefoi, H.; Jerusalem, G. Pembrolizumab Plus Trastuzumab in Trastuzumab-Resistant, Advanced, HER2-Positive Breast Cancer (PANACEA): A Single-Arm, Multicentre, Phase 1b–2 Trial. Lancet Oncol. 2019, 20, 371–382. [Google Scholar] [CrossRef]
- Sabbaghi, M.; Gil-Gomez, G.; Guardia, C.; Servitja, S.; Arpi, O.; Garcia-Alonso, S.; Menendez, S.; Arumi-Uria, M.; Serrano, L.; Salido, M.; et al. Defective Cyclin B1 Induction in Trastuzumab-Emtansine (T-DM1) Acquired Resistance in HER2-Positive Breast Cancer. Clin. Cancer Res. 2017, 23, 7006–7019. [Google Scholar] [CrossRef]
- Akhand, S.S.; Purdy, S.C.; Liu, Z.; Anderson, J.; Willey, C.; Wendt, M. Fibroblast Growth Factor Receptor Facilitates Recurrence of Minimal Residual Disease Following Trastuzumab Emtansine Therapy. BioRxiv 2019. [Google Scholar] [CrossRef]
- Elster, N.; Cremona, M.; Morgan, C.; Toomey, S.; Carr, A.; O’Grady, A.; Hennessy, B.T.; Eustace, A.J. A Preclinical Evaluation of the PI3K Alpha/Delta Dominant Inhibitor BAY 80-6946 in HER2-Positive Breast Cancer Models with Acquired Resistance to the HER2-Targeted Therapies Trastuzumab and Lapatinib. Breast Cancer Res. Treat. 2015, 149, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Goyal, S.; Kumari, A.; Singh, A.; Jamal, S.; Grover, A. Structural Investigations on Mechanism of Lapatinib Resistance Caused by HER-2 Mutants. PLoS ONE 2018, 13, e0190942. [Google Scholar] [CrossRef] [PubMed]
- Tse, S.; Liang, Y.; Leung, K.; Lee, K.; Mok, T.S. A Memetic Algorithm for Multiple-Drug Cancer Chemotherapy Schedule Optimization. IEEE Trans. Syst. Man Cybern. Part B 2007, 37, 84–91. [Google Scholar] [CrossRef]
- Rhodes, A.; Hillen, T. Mathematical Modeling of the Role of Survivin on Dedifferentiation and Radioresistance in Cancer. Bull. Math. Biol. 2016, 78, 1162–1188. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, G.; Hirata, Y.; Goldenberg, S.L.; Bruchovsky, N.; Aihara, K. Mathematical Modelling of Prostate Cancer Growth and its Application to Hormone Therapy. Philos. Trans. R. Soc. A 2010, 368, 5029–5044. [Google Scholar] [CrossRef] [PubMed]
- Brocato, T.; Dogra, P.; Koay, E.J.; Day, A.; Chuang, Y.; Wang, Z.; Cristini, V. Understanding Drug Resistance in Breast Cancer with Mathematical Oncology. Curr. Breast Cancer Rep. 2014, 6, 110–120. [Google Scholar] [CrossRef]
- Ideta, A.M.; Tanaka, G.; Takeuchi, T.; Aihara, K. A Mathematical Model of Intermittent Androgen Suppression for Prostate Cancer. J. Nonlinear Sci. 2008, 18, 593. [Google Scholar] [CrossRef]
- Zhang, Q.; Vignali, D.A. Co-Stimulatory and Co-Inhibitory Pathways in Autoimmunity. Immunity 2016, 44, 1034–1051. [Google Scholar] [CrossRef]
- Zerdes, I.; Matikas, A.; Bergh, J.; Rassidakis, G.Z.; Foukakis, T. Genetic, Transcriptional and Post-Translational Regulation of the Programmed Death Protein Ligand 1 in Cancer: Biology and Clinical Correlations. Oncogene 2018, 37, 4639. [Google Scholar] [CrossRef]
- Clinical Trials; U.S. National library of medicine: Bethesda, MD, USA, 2019. Available online: https://clinicaltrials.gov (accessed on 24 December 2019).
- FDA Approvals-Cancer Currents Blog; FDA: Rockville, MD, USA, 2019.
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.; Hwu, W.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K. Safety and Activity of anti–PD-L1 Antibody in Patients with Advanced Cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Dirix, L.Y.; Takacs, I.; Jerusalem, G.; Nikolinakos, P.; Arkenau, H.; Forero-Torres, A.; Boccia, R.; Lippman, M.E.; Somer, R.; Smakal, M. Avelumab, an Anti-PD-L1 Antibody, in Patients with Locally Advanced or Metastatic Breast Cancer: A Phase 1b JAVELIN Solid Tumor Study. Breast Cancer Res. Treat. 2018, 167, 671–686. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Zeng, Y.D.; Qin, G.; Xu, F.; Lu, J.B.; Fang, W.F.; Xue, C.; Zhan, J.H.; Zhang, X.K.; Zheng, Q.F.; et al. High PD-L1 Expression was Associated with Poor Prognosis in 870 Chinese Patients with Breast Cancer. Oncotarget 2015, 6, 33972–33981. [Google Scholar] [CrossRef] [PubMed]
- Gatalica, Z.; Snyder, C.; Maney, T.; Ghazalpour, A.; Holterman, D.A.; Xiao, N.; Overberg, P.; Rose, I.; Basu, G.D.; Vranic, S.; et al. Programmed Cell Death 1 (PD-1) and its Ligand (PD-L1) in Common Cancers and their Correlation with Molecular Cancer Type. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2965–2970. [Google Scholar] [CrossRef]
- Tsang, J.Y.; Au, W.; Lo, K.; Ni, Y.; Hlaing, T.; Hu, J.; Chan, S.; Chan, K.; Cheung, S.; Gary, M.T. PD-L1 Expression and Tumor Infiltrating PD-1 Lymphocytes Associated with Outcome in HER2 Breast Cancer Patients. Breast Cancer Res. Treat. 2017, 162, 19–30. [Google Scholar] [CrossRef]
- Jiang, C.; Cao, S.; Li, N.; Jiang, L.; Sun, T. PD-1 and PD-L1 Correlated Gene Expression Profiles and their Association with Clinical Outcomes of Breast Cancer. Cancer Cell Int. 2019, 19, 233. [Google Scholar] [CrossRef]
- Beckers, R.K.; Selinger, C.I.; Vilain, R.; Madore, J.; Wilmott, J.S.; Harvey, K.; Holliday, A.; Cooper, C.L.; Robbins, E.; Gillett, D. Programmed Death Ligand 1 Expression in Triple-negative Breast Cancer is Associated with Tumour-infiltrating Lymphocytes and Improved Outcome. Histopathology 2016, 69, 25–34. [Google Scholar] [CrossRef]
- Dill, E.A.; Gru, A.A.; Atkins, K.A.; Friedman, L.A.; Moore, M.E.; Bullock, T.N.; Cross, J.V.; Dillon, P.M.; Mills, A.M. PD-L1 Expression and Intratumoral Heterogeneity Across Breast Cancer Subtypes and Stages. Am. J. Surg. Pathol. 2017, 41, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Lee, S.J.; Kim, Y.K.; Park, W.Y.; Kim, J.Y.; Lee, C.H.; Gong, G.; Huh, G.Y.; Choi, K.U. Programmed Death-Ligand 1 (PD-L1) Expression in Tumour Cell and Tumour Infiltrating Lymphocytes of HER2-Positive Breast Cancer and its Prognostic Value. Sci. Rep. 2017, 7, 11671. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Dong, P.; Ren, M.; Song, Y.; Qian, X.; Yang, Y.; Li, S.; Zhang, X.; Liu, F. PD-L1 Expression is Associated with Tumor FOXP3(+) Regulatory T-Cell Infiltration of Breast Cancer and Poor Prognosis of Patient. J. Cancer 2016, 7, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Ubago, J.M.; Blanco, L.Z.; Shen, T.; Siziopikou, K.P. The PD-1/PD-L1 Axis in HER2 Ductal Carcinoma in Situ (DCIS) of the Breast. Am. J. Clin. Pathol. 2019, 152, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Liu, Z.; Yu, Q.; Wang, X.; Bian, M.; Yu, Z.; Yu, J. Expression of PD-1/PD-L1 in Primary Breast Tumours and Metastatic Axillary Lymph Nodes and its Correlation with Clinicopathological Parameters. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Okabe, M.; Toh, U.; Iwakuma, N.; Saku, S.; Akashi, M.; Kimitsuki, Y.; Seki, N.; Kawahara, A.; Ogo, E.; Itoh, K. Predictive Factors of the Tumor Immunological Microenvironment for Long-term Follow-up in Early Stage Breast Cancer. Cancer Sci. 2017, 108, 81–90. [Google Scholar] [CrossRef]
- Mittal, D.; Vijayan, D.; Neijssen, J.; Kreijtz, J.; Habraken, M.M.; Van Eenennaam, H.; Van Elsas, A.; Smyth, M.J. Blockade of ErbB2 and PD-L1 using a Bispecific Antibody to Improve Targeted Anti-ErbB2 Therapy. OncoImmunology 2019, 8, e1648171. [Google Scholar] [CrossRef]
- Bae, S.B.; Cho, H.D.; Oh, M.; Lee, J.; Jang, S.; Hong, S.A.; Cho, J.; Kim, S.Y.; Han, S.W.; Lee, J.E. Expression of Programmed Death Receptor Ligand 1 with High Tumor-Infiltrating Lymphocytes is Associated with Better Prognosis in Breast Cancer. J. Breast Cancer 2016, 19, 242–251. [Google Scholar] [CrossRef]
- Mittendorf, E.A.; Philips, A.V.; Meric-Bernstam, F.; Qiao, N.; Wu, Y.; Harrington, S.; Su, X.; Wang, Y.; Gonzalez-Angulo, A.M.; Akcakanat, A.; et al. PD-L1 Expression in Triple-Negative Breast Cancer. Cancer Immunol. Res. 2014, 2, 361–370. [Google Scholar] [CrossRef]
- Ghebeh, H.; Mohammed, S.; Al-Omair, A.; Qattant, A.; Lehe, C.; Al-Qudaihi, G.; Elkum, N.; Alshabanah, M.; Amer, S.B.; Tulbah, A. The B7-H1 (PD-L1) T Lymphocyte-Inhibitory Molecule is Expressed in Breast Cancer Patients with Infiltrating Ductal Carcinoma: Correlation with Important High-Risk Prognostic Factors. Neoplasia 2006, 8, 190–198. [Google Scholar] [CrossRef]
- Kurozumi, S.; Inoue, K.; Matsumoto, H.; Fujii, T.; Horiguchi, J.; Oyama, T.; Kurosumi, M.; Shirabe, K. Clinicopathological Values of PD-L1 Expression in HER2-Positive Breast Cancer. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Nitta, H.; Wei, L.; Banks, P.M.; Parwani, A.V.; Li, Z. Evaluation of Immune Reaction and PD-L1 Expression using Multiplex Immunohistochemistry in HER2-Positive Breast Cancer: The Association with Response to Anti-HER2 Neoadjuvant Therapy. Clin. Breast Cancer 2018, 18, e237–e244. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Nitta, H.; Wei, L.; Banks, P.M.; Lustberg, M.; Wesolowski, R.; Ramaswamy, B.; Parwani, A.V.; Li, Z. PD-L1 Expression and CD8-positive T Cells are Associated with Favorable Survival in HER2-positive Invasive Breast Cancer. Breast J. 2018, 24, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, R.; Finetti, P.; Mamessier, E.; Adelaide, J.; Chaffanet, M.; Ali, H.R.; Viens, P.; Caldas, C.; Birnbaum, D.; Bertucci, F. Prognostic and Predictive Value of PDL1 Expression in Breast Cancer. Oncotarget 2015, 6, 5449–5464. [Google Scholar] [CrossRef] [PubMed]
- Kitano, A.; Ono, M.; Yoshida, M.; Noguchi, E.; Shimomura, A.; Shimoi, T.; Kodaira, M.; Yunokawa, M.; Yonemori, K.; Shimizu, C. Tumour-Infiltrating Lymphocytes are Correlated with Higher Expression Levels of PD-1 and PD-L1 in Early Breast Cancer. ESMO Open 2017, 2, e000150. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Campbell, C.; Savas, P.; Nuciforo, P.; Aura, C.; De Azambuja, E.; Eidtmann, H.; Ellis, C.E.; Baselga, J. Tumor-Infiltrating Lymphocytes and Associations with Pathological Complete Response and Event-Free Survival in HER2-Positive Early-Stage Breast Cancer Treated with Lapatinib and Trastuzumab: A Secondary Analysis of the NeoALTTO Trial. JAMA Oncol. 2015, 1, 448–455. [Google Scholar] [CrossRef]
- Krasniqi, E.; Barchiesi, G.; Pizzuti, L.; Mazzotta, M.; Venuti, A.; Maugeri-Saccà, M.; Sanguineti, G.; Massimiani, G.; Sergi, D.; Carpano, S. Immunotherapy in HER2-Positive Breast Cancer: State of the Art and Future Perspectives. J. Hematol. Oncol. 2019, 12, 111. [Google Scholar] [CrossRef]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M. Use of Chemotherapy Plus a Monoclonal Antibody Against HER2 for Metastatic Breast Cancer that Overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef]
- Esteva, F.J.; Valero, V.; Booser, D.; Guerra, L.T.; Murray, J.L.; Pusztai, L.; Cristofanilli, M.; Arun, B.; Esmaeli, B.; Fritsche, H.A. Phase II Study of Weekly Docetaxel and Trastuzumab for Patients with HER-2–overexpressing Metastatic Breast Cancer. J. Clin. Oncol. 2002, 20, 1800–1808. [Google Scholar] [CrossRef]
- Price-Schiavi, S.A.; Jepson, S.; Li, P.; Arango, M.; Rudland, P.S.; Yee, L.; Carraway, K.L. Rat Muc4 (Sialomucin Complex) Reduces Binding of anti-ErbB2 Antibodies to Tumor Cell Surfaces, a Potential Mechanism for Herceptin Resistance. Int. J. Cancer 2002, 99, 783–791. [Google Scholar] [CrossRef]
- Loi, S.; Michiels, S.; Salgado, R.; Sirtaine, N.; Jose, V.; Fumagalli, D.; Kellokumpu-Lehtinen, P.; Bono, P.; Kataja, V.; Desmedt, C. Tumor Infiltrating Lymphocytes are Prognostic in Triple Negative Breast Cancer and Predictive for Trastuzumab Benefit in Early Breast Cancer: Results from the FinHER Trial. Ann. Oncol. 2014, 25, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
- Ingold Heppner, B.; Untch, M.; Denkert, C.; Pfitzner, B.M.; Lederer, B.; Schmitt, W.; Eidtmann, H.; Fasching, P.A.; Tesch, H.; Solbach, C.; et al. Tumor-Infiltrating Lymphocytes: A Predictive and Prognostic Biomarker in Neoadjuvant-Treated HER2-Positive Breast Cancer. Clin. Cancer Res. 2016, 22, 5747–5754. [Google Scholar] [CrossRef] [PubMed]
- Kurozumi, S.; Inoue, K.; Matsumoto, H.; Fujii, T.; Horiguchi, J.; Oyama, T.; Kurosumi, M.; Shirabe, K. Prognostic Utility of Tumor-Infiltrating Lymphocytes in Residual Tumor After Neoadjuvant Chemotherapy with Trastuzumab for HER2-Positive Breast Cancer. Sci. Rep. 2019, 9, 1583. [Google Scholar] [CrossRef] [PubMed]
- Nocera, N.F.; Lee, M.C.; De La Cruz, L.M.; Rosemblit, C.; Czerniecki, B.J. Restoring Lost Anti-HER-2 Th1 Immunity in Breast Cancer: A Crucial Role for Th1 Cytokines in Therapy and Prevention. Front. Pharmacol. 2016, 7, 356. [Google Scholar] [CrossRef] [PubMed]
- Kodumudi, K.N.; Wiener, D.; Basu, A.; Czerniecki, B. Abstract 2545: Antitumor Efficacy of Type i Polarized Dendritic Cells in Combination with Immune Checkpoint Blockade in a Preclinical Model of Breast Cancer. Antitumor efficacy of Type I polarized dendritic cells in combination with immune checkpoint blockade in a preclinical model of breast cancer. Cancer Res. 2018, 78, 2545. [Google Scholar]
- Tang, F.; Zheng, P. Tumor Cells Versus Host Immune Cells: Whose PD-L1 Contributes to PD-1/PD-L1 Blockade Mediated Cancer Immunotherapy? Cell Biosci. 2018, 8, 34. [Google Scholar] [CrossRef]
- Park, S.E.; Park, K.; Lee, E.; Kim, J.; Ahn, J.S.; Im, Y.; Lee, C.; Jung, H.; Cho, S.Y.; Park, W. Clinical Implication of Tumor Mutational Burden in Patients with HER2-Positive Refractory Metastatic. Breast Cancer Oncoimmunol. 2018, 7, e1466768. [Google Scholar] [CrossRef]
- Klempner, S.J.; Fabrizio, D.; Bane, S.; Reinhart, M.; Peoples, T.; Ali, S.M.; Sokol, E.S.; Frampton, G.; Schrock, A.B.; Anhorn, R.; et al. Tumor Mutational Burden as a Predictive Biomarker for Response to Immune Checkpoint Inhibitors: A Review of Current Evidence. Oncologist 2020, 25, 147–159. [Google Scholar] [CrossRef]
- Bernadou, G.; Campone, M.; Merlin, J.; Gouilleux-Gruart, V.; Bachelot, T.; Lokiec, F.; Rezai, K.; Arnedos, M.; Diéras, V.; Jimenez, M. Influence of Tumour Burden on Trastuzumab Pharmacokinetics in HER2 Positive Non-metastatic Breast Cancer. Br. J. Clin. Pharmacol. 2016, 81, 941–948. [Google Scholar] [CrossRef]
- Polk, A.; Svane, I.; Andersson, M.; Nielsen, D. Checkpoint Inhibitors in Breast Cancer–current Status. Cancer Treat. Rev. 2018, 63, 122–134. [Google Scholar] [CrossRef]
- Guzik, K.; Tomala, M.; Muszak, D.; Konieczny, M.; Hec, A.; Błaszkiewicz, U.; Pustuła, M.; Butera, R.; Dömling, A.; Holak, T.A. Development of the Inhibitors that Target the PD-1/PD-L1 Interaction—A Brief Look at Progress on Small Molecules, Peptides and Macrocycles. Molecules 2019, 24, 2071. [Google Scholar] [CrossRef] [PubMed]
- Ugolkov, A.; Gaisina, I.; Zhang, J.; Billadeau, D.D.; White, K.; Kozikowski, A.; Jain, S.; Cristofanilli, M.; Giles, F.; O’Halloran, T. GSK-3 Inhibition Overcomes Chemoresistance in Human Breast Cancer. Cancer Lett. 2016, 380, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, A.; Ahmed, M.; Okoye, I.; Arutyunova, E.; Babu, D.; Turnbull, W.L.; Kundu, J.K.; Shields, J.; Agopsowicz, K.C.; Xu, L. Comprehensive in Vitro Characterization of PD-L1 Small Molecule Inhibitors. Sci. Rep. 2019, 9, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhu, Z.; Lv, H.; Li, F.; Sun, S.; Li, J.; Lee, C. Immune Checkpoint Blockade Mediated by a Small-Molecule Nanoinhibitor Targeting the PD-1/PD-L1 Pathway Synergizes with Photodynamic Therapy to Elicit Antitumor Immunity and Antimetastatic Effects on Breast Cancer. Small 2019, 15, 1903881. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Li, X.; Yao, H.; Lin, K. The Characteristics of PD-L1 Inhibitors, from Peptides to Small Molecules. Molecules 2019, 24, 1940. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Friedman, A. Combination Therapy of Cancer with Cancer Vaccine and Immune Checkpoint Inhibitors: A Mathematical Model. PLoS ONE 2017, 12, e0178479. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.; Esteva, F.; Beresford, M.; Saura, C.; De Laurentiis, M.; Kim, S. Results from KATE2, a Randomized Phase 2 Study of Atezolizumab (Atezo) Trastuzumab Emtansine (T-DM1) vs. Placebo (Pbo) T-DM1 in Previously Treated HER2 Advanced Breast Cancer (BC). In Proceedings of the San Antonio Breast Cancer Symposium, San Antonio, TX, USA, 4–8 December 2018. [Google Scholar]
- Bang, Y.J.; Giaccone, G.; Im, S.; Oh, D.; Bauer, T.; Nordstrom, J.; Li, H.; Chichili, G.; Moore, P.; Hong, S. First-in-Human Phase 1 Study of Margetuximab (MGAH22), an Fc-Modified Chimeric Monoclonal Antibody, in Patients with HER2-Positive Advanced Solid Tumors. Ann. Oncol. 2017, 28, 855–861. [Google Scholar] [CrossRef]
- Rugo, H.S.; Pegram, M.D.; Gradishar, W.J.; Cortes, J.; Curigliano, G.; Wigginton, J.M.; Lechleider, R.J.; Cardoso, F. SOPHIA: A Phase 3, Randomized Study of Margetuximab (M) Plus Chemotherapy (CTX) vs. Trastuzumab (T) Plus CTX in the Treatment of Patients with HER2 Metastatic Breast Cancer (MBC). J. Clin. Oncol. 2016, 34, TPS630. [Google Scholar] [CrossRef]
- Nordstrom, J.L.; Gorlatov, S.; Zhang, W.; Yang, Y.; Huang, L.; Burke, S.; Li, H.; Ciccarone, V.; Zhang, T.; Stavenhagen, J. Anti-Tumor Activity and Toxicokinetics Analysis of MGAH22, an Anti-HER2 Monoclonal Antibody with Enhanced Fcγ Receptor Binding Properties. Breast Cancer Res. 2011, 13, R123. [Google Scholar] [CrossRef]
- Saunders, K.O. Conceptual Approaches to Modulating Antibody Effector Functions and Circulation Half-Life. Front. Immunol. 2019, 10, 1296. [Google Scholar] [CrossRef]
- Hinner, M.J.; Aiba, R.S.B.; Jaquin, T.J.; Berger, S.; Durr, M.C.; Schlosser, C.; Allersdorfer, A.; Wiedenmann, A.; Matschiner, G.; Schuler, J.; et al. Tumor-Localized Costimulatory T-Cell Engagement by the 4-1BB/HER2 Bispecific Antibody-Anticalin Fusion PRS-343. Clin. Cancer Res. 2019, 25, 5878–5889. [Google Scholar] [CrossRef] [PubMed]
- Ott, P.A.; Bang, Y.; Piha-Paul, S.; Razak, A.R.A.; Bennouna, J.; Soria, J.; Rugo, H.S.; Cohen, R.B.; O’Neil, B.H.; Mehnert, J.M.; et al. T-Cell–Inflamed Gene-Expression Profile, Programmed Death Ligand 1 Expression, and Tumor Mutational Burden Predict Efficacy in Patients Treated with Pembrolizumab Across 20 Cancers: KEYNOTE-028. JCO 2019, 37, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Chaganty, B.K.; Qiu, S.; Gest, A.; Lu, Y.; Ivan, C.; Calin, G.A.; Weiner, L.M.; Fan, Z. Trastuzumab Upregulates PD-L1 as a Potential Mechanism of Trastuzumab Resistance through Engagement of Immune Effector Cells and Stimulation of IFNγ Secretion. Cancer Lett. 2018, 430, 47–56. [Google Scholar] [CrossRef]
- Esteva, F.J.; Hubbard-Lucey, V.M.; Tang, J.; Pusztai, L. Immunotherapy and Targeted Therapy Combinations in Metastatic Breast Cancer. Lancet Oncol. 2019, 20, e175–e186. [Google Scholar] [CrossRef]
- Vranic, S.; Cyprian, F.S.; Gatalica, Z.; Palazzo, J. PD-L1 Status in Breast Cancer: Current View and Perspectives. In Seminars in Cancer Biology; Elsevier Ltd.: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Han, W.; Carpenter, R.L.; Cao, X.; Lo, H. STAT1 Gene Expression is Enhanced by Nuclear EGFR and HER2 Via Cooperation with STAT3. Mol. Carcinog. 2013, 52, 959–969. [Google Scholar] [CrossRef]
- Zerdes, I.; Wallerius, M.; Sifakis, E.G.; Wallmann, T.; Betts, S.; Bartish, M.; Tsesmetzis, N.; Tobin, N.P.; Coucoravas, C.; Bergh, J. STAT3 Activity Promotes Programmed-Death Ligand 1 Expression and Suppresses Immune Responses in Breast Cancer. Cancers 2019, 11, 1479. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Gu, J.; Gao, Y.; Wang, Z.; Zhang, K.; Mu, N.; Huang, T.; Li, W.; Hao, Q. Synergistic Tumoricidal Effect of Combined hPD-L1 Vaccine and HER2 Gene Vaccine. Biochem. Biophys. Res. Commun. 2018, 497, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Byrne, H.M. Dissecting Cancer through Mathematics: From the Cell to the Animal Model. Nat. Rev. Cancer 2010, 10, 221. [Google Scholar] [CrossRef]
- Spang-Thomsen, M.; Rygaard, K.; Hansen, L.; Halvorsen, A.; Vindeløv, L.; Brünner, N. Growth Kinetics of Four Human Breast Carcinomas Grown in Nude Mice. Breast Cancer Res. Treat. 1989, 14, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Afenya, E.K.; Ouifki, R.; Mundle, S.D. Mathematical Modeling of Bone Marrow–peripheral Blood Dynamics in the Disease State Based on Current Emerging Paradigms, Part II. J. Theor. Biol. 2019, 460, 37–55. [Google Scholar] [CrossRef]
- ARMITAGE, P.; DOLL, R. The Age Distribution of Cancer and a Multi-Stage Theory of Carcinogenesis. Br. J. Cancer 1954, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Engelhart, M.; Lebiedz, D.; Sager, S. Optimal Control for Selected Cancer Chemotherapy ODE Models: A View on the Potential of Optimal Schedules and Choice of Objective Function. Math. Biosci. 2011, 229, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Cardilin, T.; Almquist, J.; Jirstrand, M.; Zimmermann, A.; El Bawab, S.; Gabrielsson, J. Model-Based Evaluation of Radiation and Radiosensitizing Agents in Oncology. CPT Pharmacomet. Syst. Pharmacol. 2018, 7, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Barkal, A.A.; Brewer, R.E.; Markovic, M.; Kowarsky, M.; Barkal, S.A.; Zaro, B.W.; Krishnan, V.; Hatakeyama, J.; Dorigo, O.; Barkal, L.J. CD24 Signalling through Macrophage Siglec-10 is a Target for Cancer Immunotherapy. Nature 2019, 572, 392–396. [Google Scholar] [CrossRef]
- Hirata, Y.; Morino, K.; Akakura, K.; Higano, C.S.; Aihara, K. Personalizing Androgen Suppression for Prostate Cancer using Mathematical Modeling. Sci. Rep. 2018, 8, 2673. [Google Scholar] [CrossRef]
- Rieger, H.; Welter, M. Integrative Models of Vascular Remodeling during Tumor Growth. Wiley Interdiscip. Rev. Syst. Biol. Med. 2015, 7, 113–129. [Google Scholar] [CrossRef]
- Robbins, P.F.; Morgan, R.A.; Feldman, S.A.; Yang, J.C.; Sherry, R.M.; Dudley, M.E.; Wunderlich, J.R.; Nahvi, A.V.; Helman, L.J.; Mackall, C.L.; et al. Tumor Regression in Patients with Metastatic Synovial Cell Sarcoma and Melanoma using Genetically Engineered Lymphocytes Reactive with NY-ESO-1. J. Clin. Oncol. 2011, 29, 917–924. [Google Scholar] [CrossRef]
- Piretto, E.; Delitala, M.; Ferraro, M. Combination Therapies and Intra-Tumoral Competition: Insights from Mathematical Modeling. J. Theor. Biol. 2018, 446, 149–159. [Google Scholar] [CrossRef]
- Botesteanu, D.; Lipkowitz, S.; Lee, J.; Levy, D. Mathematical Models of Breast and Ovarian Cancers. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 337–362. [Google Scholar] [CrossRef]
- Barber, F.D. Recent Developments in Oncology Immunotherapy, Adverse Effects Part 2. J. Nurse Pract. 2018, 14, 259–266. [Google Scholar] [CrossRef]
- Rivaz, A.; Azizian, M.; Soltani, M. Various Mathematical Models of Tumor Growth with Reference to Cancer Stem Cells: A Review. Iran. J. Sci. Technol. Trans. A Sci. 2019, 43, 687–700. [Google Scholar] [CrossRef]
- Altrock, P.M.; Liu, L.L.; Michor, F. The Mathematics of Cancer: Integrating Quantitative Models. Nat. Rev. Cancer 2015, 15, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.; Alhussain, H.; Hasan, A.; Badie Ahmed, M.C.; Yalcin, H.; Al Moustafa, A. A Novel in Ovo Model to Study Cancer Metastasis using Chicken Embryos and GFP Expressing Cancer Cells. BJBMS 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, H.C.; Benslimane, F.M.; Kawakami, K. The First International Zebrafish Conference/Workshop in Qatar. Zebrafish 2019, 16, 493–495. [Google Scholar] [CrossRef]
- Lesi, A.A.; Heilmann, S.; White, R.M.; Rumschitzki, D.S. A New Mathematical Model for Tumor Growth, Reduction and Metastasis, Validation with Zebrafish Melanoma and Potential Implications for Dormancy and Recurrence. BioRxiv 2019. [Google Scholar] [CrossRef]
- Cantini, L.; Pistelli, M.; Savini, A.; Bastianelli, L.; Della Mora, A.; Merloni, F.; Burattini, M.; Berardi, R. Long-responders to anti-HER2 Therapies: A Case Report and Review of the Literature. Mol. Clin. Oncol. 2018, 8, 147–152. [Google Scholar] [CrossRef]
- Annan, K.; Nagel, M.; Brock, H.A. A Mathematical Model of Breast Cancer and Mediated Immune System Interactions. J. Math. Syst. Sci. 2012, 2, 430–446. [Google Scholar]
- Birkhead, B.G.; Rankin, E.M.; Gallivan, S.; Dones, L.; Rubens, R.D. A Mathematical Model of the Development of Drug Resistant to Cancer Chemotherapy. Eur. J. Cancer Clin. Oncol. 1987, 23, 1421–1427. [Google Scholar] [CrossRef]
- Wang, H.; Milberg, O.; Bartelink, I.H.; Vicini, P.; Wang, B.; Narwal, R.; Roskos, L.; Santa-Maria, C.A.; Popel, A.S. In Silico Simulation of a Clinical Trial with Anti-CTLA-4 and Anti-PD-L1 Immunotherapies in Metastatic Breast Cancer using a Systems Pharmacology Model. R. Soc. Open Sci. 2019, 6, 190366. [Google Scholar] [CrossRef]
- Solís-Pérez, J.; Gómez-Aguilar, J.; Atangana, A. A Fractional Mathematical Model of Breast Cancer Competition Model. Chaos Solitons Fractals 2019, 127, 38–54. [Google Scholar] [CrossRef]
- Tyuryumina, E.Y.; Neznanov, A.A. Consolidated Mathematical Growth Model of the Primary Tumor and Secondary Distant Metastases of Breast Cancer (CoMPaS). PLoS ONE 2018, 13, e0200148. [Google Scholar] [CrossRef] [PubMed]
- Roe-Dale, R.; Isaacson, D.; Kupferschmid, M. A Mathematical Model of Breast Cancer Treatment with CMF and Doxorubicin. Bull. Math. Biol. 2011, 73, 585–608. [Google Scholar] [CrossRef] [PubMed]
- Bertelsen, V.; Stang, E. The Mysterious Ways of ErbB2/HER2 Trafficking. Membranes 2014, 4, 424–446. [Google Scholar] [CrossRef] [PubMed]
- Badrinath, N.; Yoo, S.Y. Recent Advances in Cancer Stem Cell-Targeted Immunotherapy. Cancers 2019, 11, 310. [Google Scholar] [CrossRef] [PubMed]
- Darvin, P.; Sasidharan Nair, V.; Elkord, E. PD-L1 Expression in Human Breast Cancer Stem Cells is Epigenetically Regulated through Posttranslational Histone Modifications. J. Oncol. 2019. [Google Scholar] [CrossRef]
- Nikolopoulou, E.; Johnson, L.R.; Harris, D.; Nagy, J.D.; Stites, E.C.; Kuang, Y. Tumour-Immune Dynamics with an Immune Checkpoint Inhibitor. Lett. Biomath. 2018, 5, S137–S159. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling Cancer in Microfluidic Human Organs-on-Chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef]
- Sun, W.; Luo, Z.; Lee, J.; Kim, H.; Lee, K.; Tebon, P.; Feng, Y.; Dokmeci, M.R.; Sengupta, S.; Khademhosseini, A. Organ-on-a-Chip for Cancer and Immune Organs Modeling. Adv. Healthc. Mater. 2019, 8, 1801363. [Google Scholar] [CrossRef]
- McAleer, C.W.; Pointon, A.; Long, C.J.; Brighton, R.L.; Wilkin, B.D.; Bridges, L.R.; Sriram, N.N.; Fabre, K.; McDougall, R.; Muse, V.P. On the Potential of in Vitro Organ-Chip Models to Define Temporal Pharmacokinetic-Pharmacodynamic Relationships. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Padmanabhan, R.; Meskin, N.; Haddad, W.M. Reinforcement Learning-Based Control of Drug Dosing for Cancer Chemotherapy Treatment. Math. Biosci. 2017, 293, 11–20. [Google Scholar] [CrossRef]
- Serre, R.; Benzekry, S.; Padovani, L.; Meille, C.; Andre, N.; Ciccolini, J.; Barlesi, F.; Muracciole, X.; Barbolosi, D. Mathematical Modeling of Cancer Immunotherapy and its Synergy with Radiotherapy. Cancer Res. 2016, 76, 4931–4940. [Google Scholar] [CrossRef] [PubMed]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padmanabhan, R.; Kheraldine, H.S.; Meskin, N.; Vranic, S.; Al Moustafa, A.-E. Crosstalk between HER2 and PD-1/PD-L1 in Breast Cancer: From Clinical Applications to Mathematical Models. Cancers 2020, 12, 636. https://doi.org/10.3390/cancers12030636
Padmanabhan R, Kheraldine HS, Meskin N, Vranic S, Al Moustafa A-E. Crosstalk between HER2 and PD-1/PD-L1 in Breast Cancer: From Clinical Applications to Mathematical Models. Cancers. 2020; 12(3):636. https://doi.org/10.3390/cancers12030636
Chicago/Turabian StylePadmanabhan, Regina, Hadeel Shafeeq Kheraldine, Nader Meskin, Semir Vranic, and Ala-Eddin Al Moustafa. 2020. "Crosstalk between HER2 and PD-1/PD-L1 in Breast Cancer: From Clinical Applications to Mathematical Models" Cancers 12, no. 3: 636. https://doi.org/10.3390/cancers12030636
APA StylePadmanabhan, R., Kheraldine, H. S., Meskin, N., Vranic, S., & Al Moustafa, A.-E. (2020). Crosstalk between HER2 and PD-1/PD-L1 in Breast Cancer: From Clinical Applications to Mathematical Models. Cancers, 12(3), 636. https://doi.org/10.3390/cancers12030636







