Testing of the Survivin Suppressant YM155 in a Large Panel of Drug-Resistant Neuroblastoma Cell Lines
Abstract
1. Introduction
2. Results
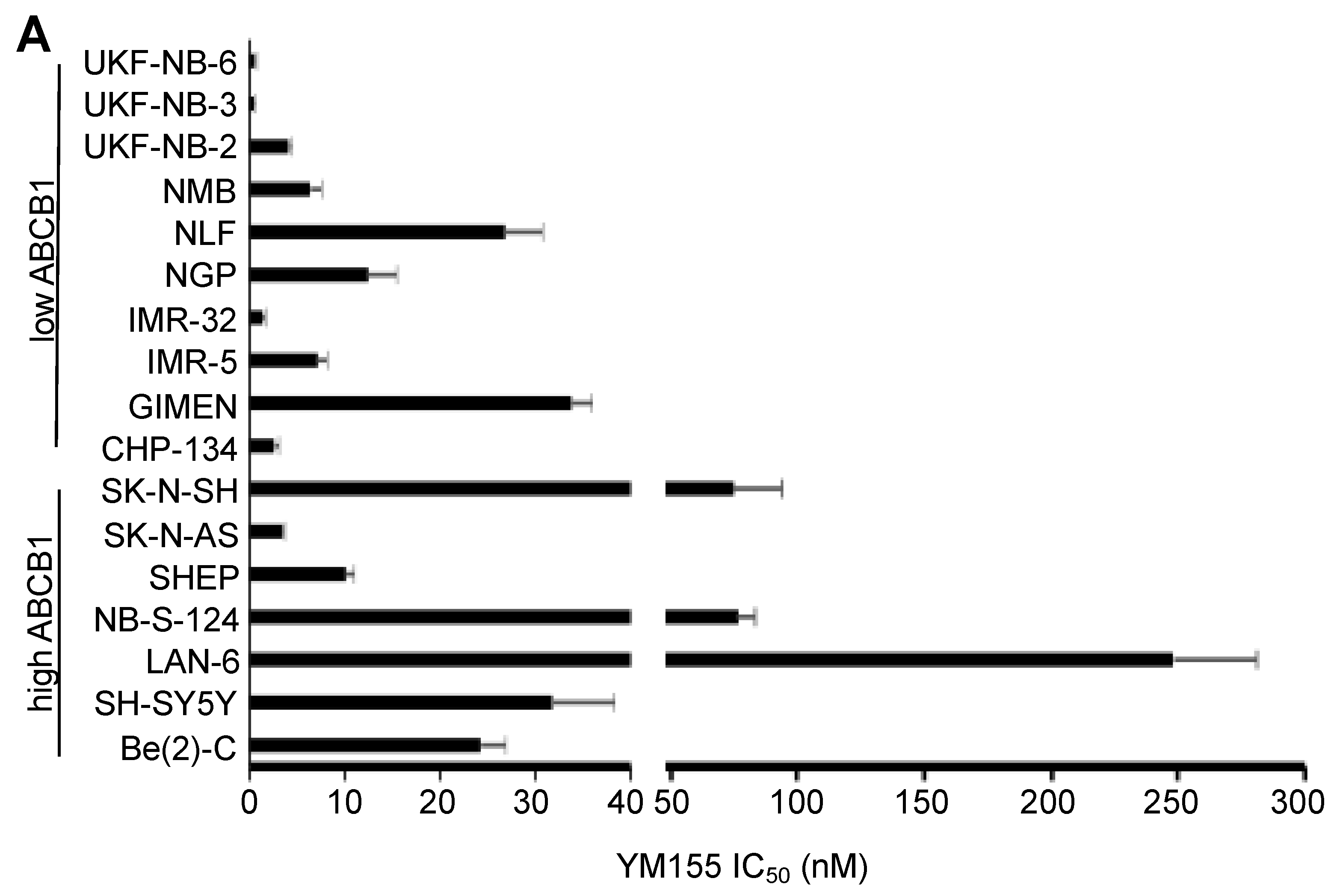
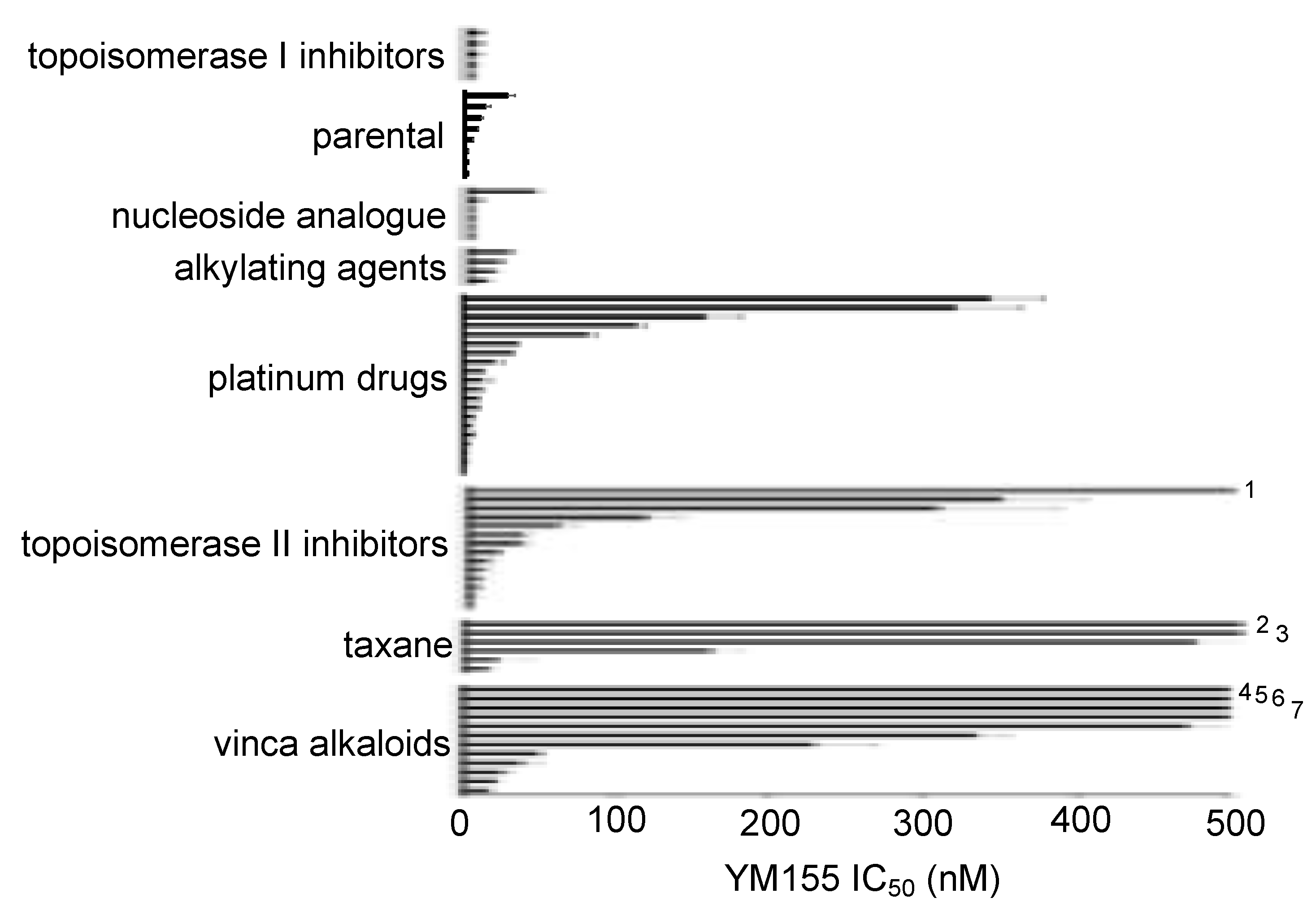
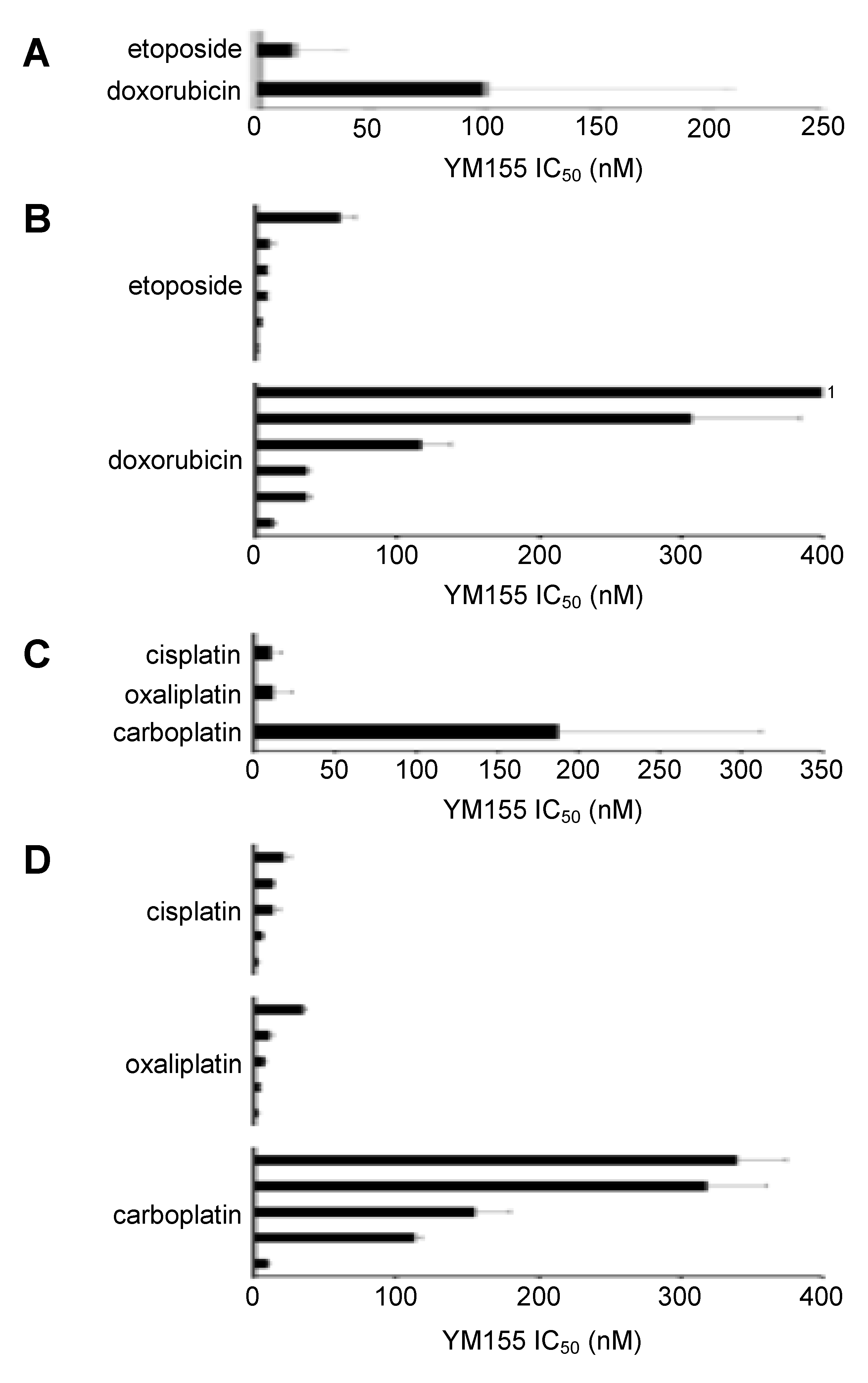
2.1. Effects of YM155 on the Viability of Parental Neuroblastoma Cell Lines
2.2. ABCB1-Expressing Neuroblastoma Cell Lines Display Low Sensitivity to YM155
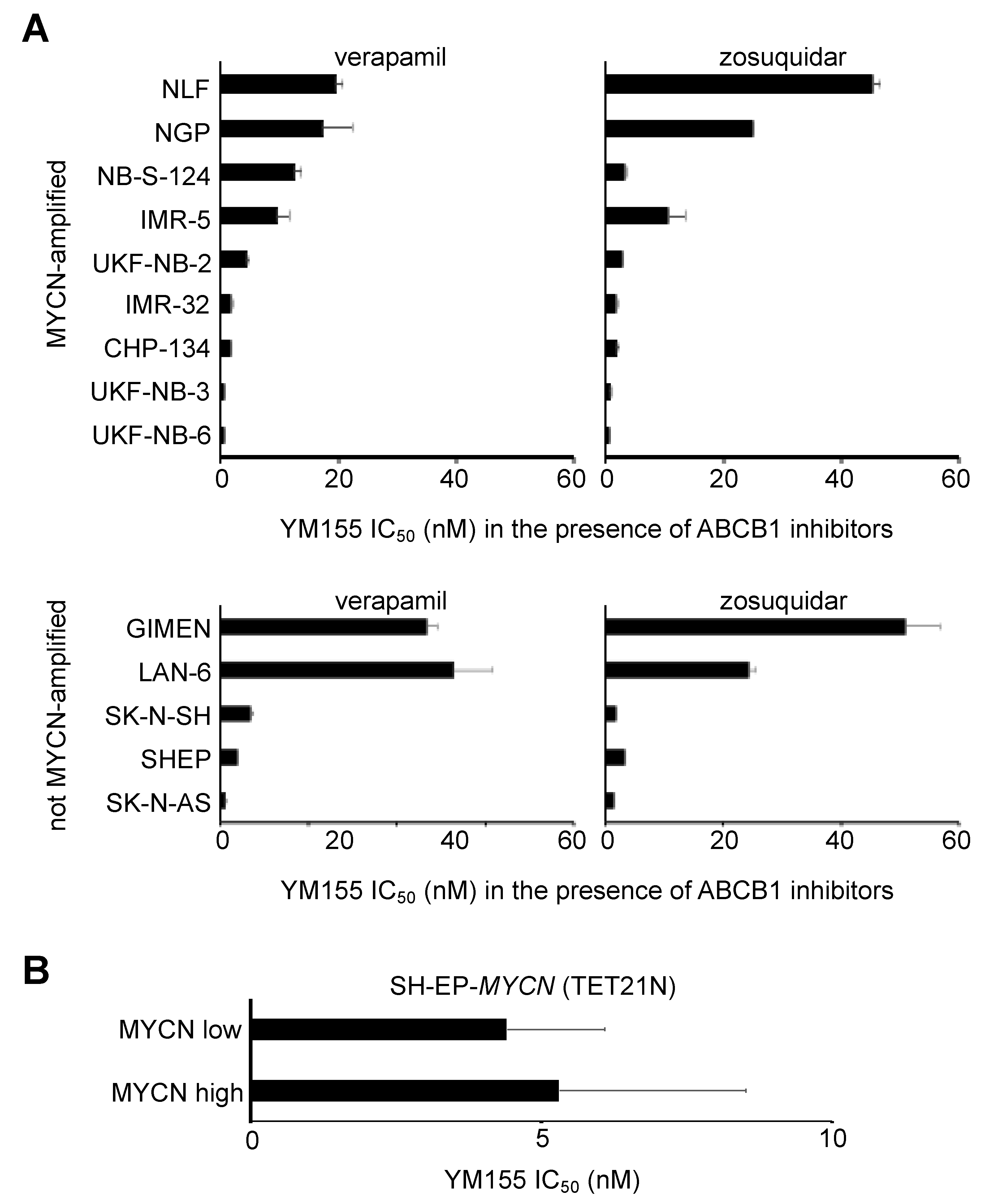
2.3. MYCN Status Does Not Influence Neuroblastoma Cell Sensitivity to YM155
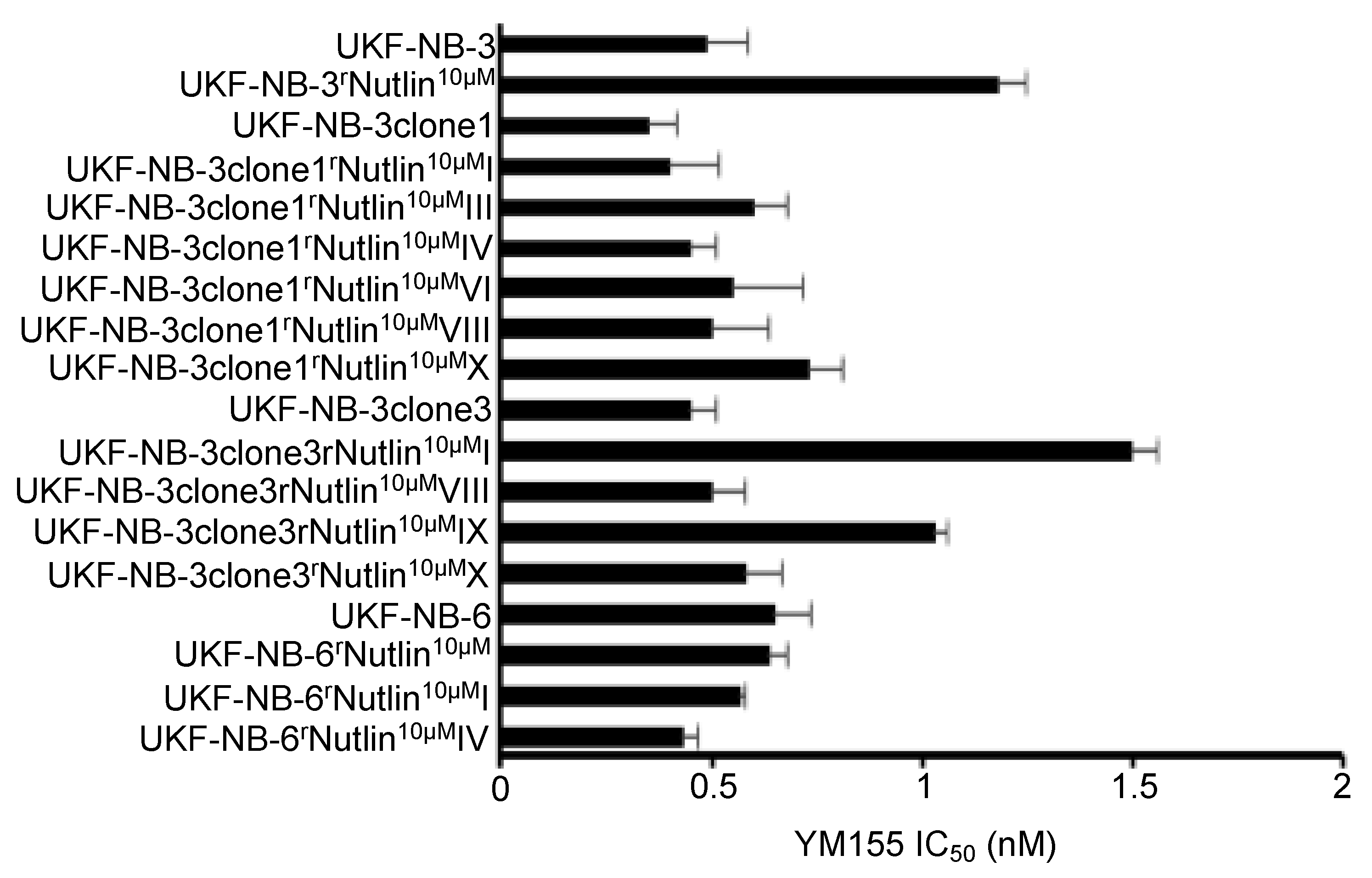
2.4. TP53 Status Does Not Predict Neuroblastoma Cell Sensitivity to YM155
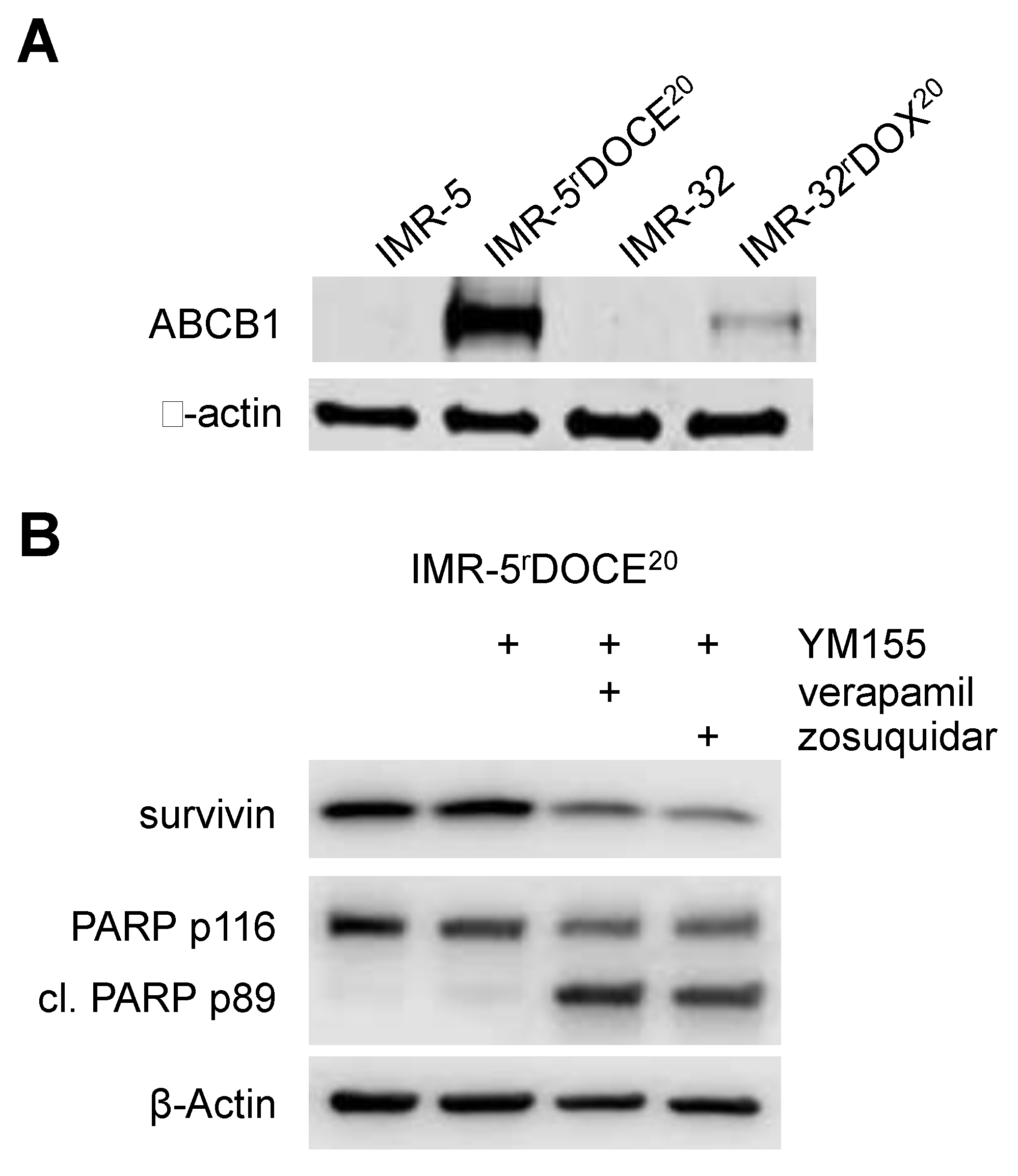
2.5. Effects of YM155 on the Viability of Neuroblastoma Cell Lines with Acquired Drug Resistance
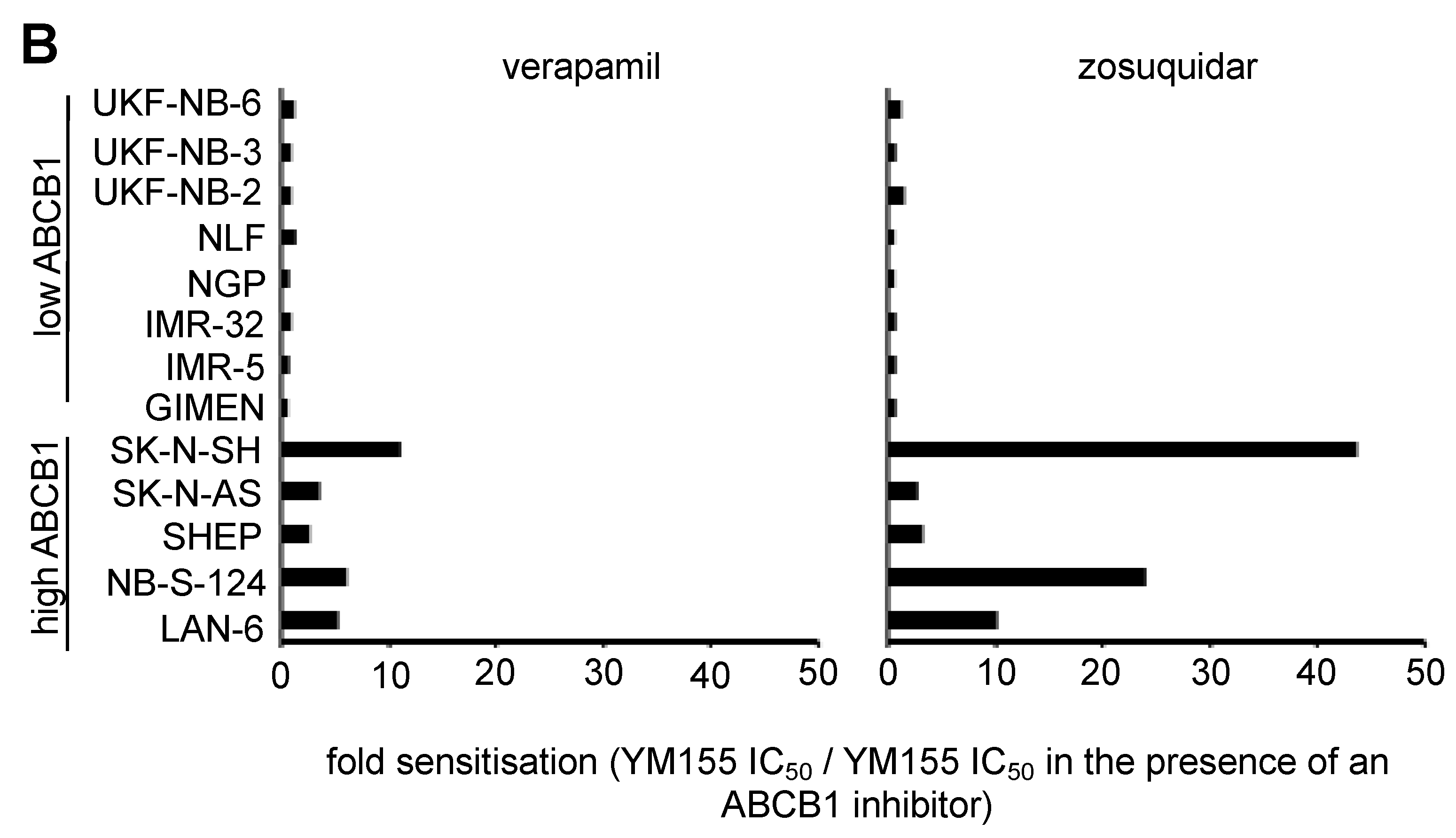
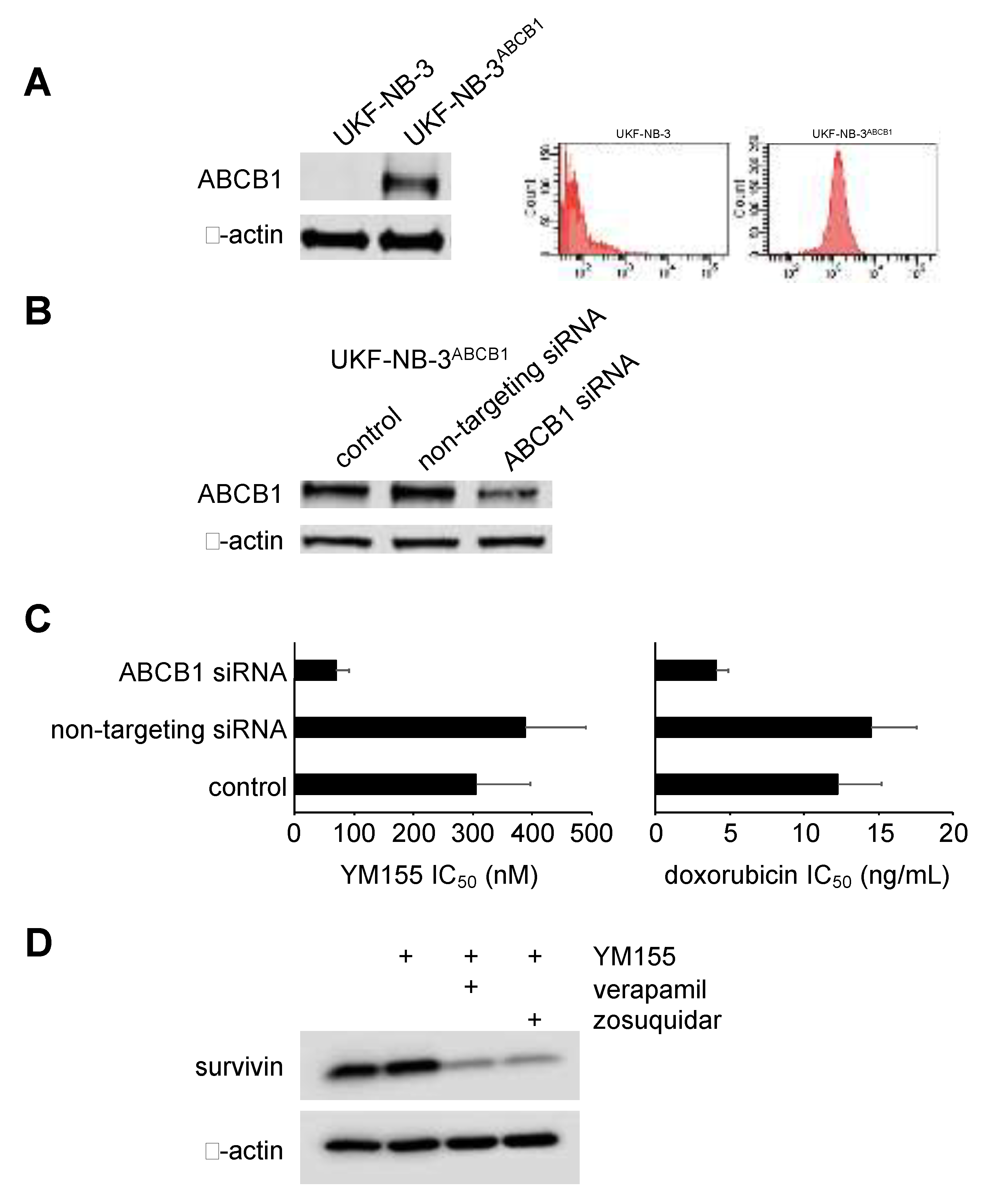
2.6. Role of ABCB1 in the YM155 response of Drug-Adapted Neuroblastoma Cells
2.7. ABCC1 Mediates Resistance to YM155
2.8. Cross Resistance to YM155 is Caused by Multiple Resistance Mechanisms in Drug-Adapted Neuroblastoma Cells
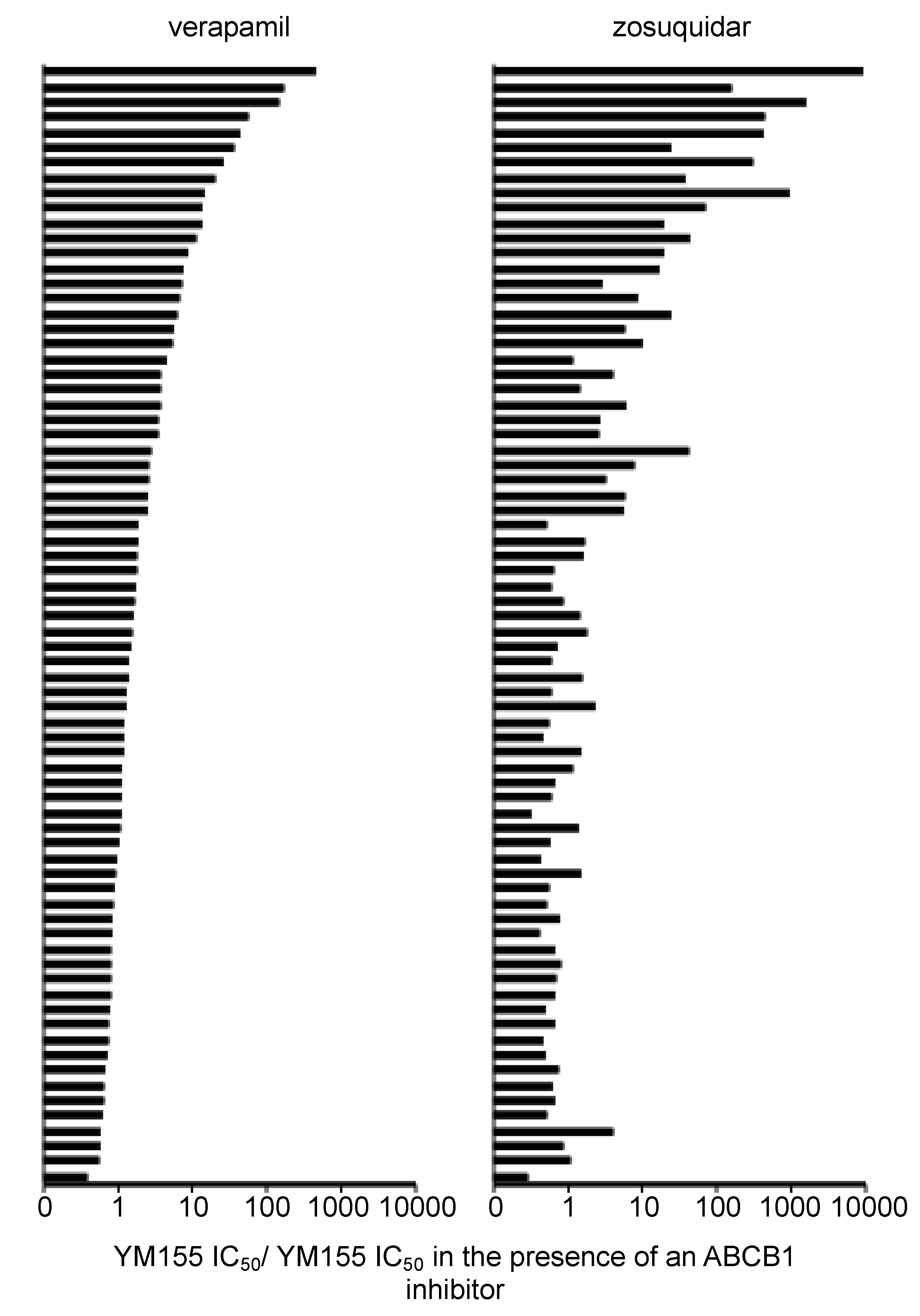
2.9. Discrepancies in the Effects of the ABCB1 Inhibitors Verapamil and Zosuquidar on Neuroblastoma Cell Sensitivity to YM155
3. Discussion
4. Materials and Methods
4.1. Drugs
4.2. Cells
4.3. Viability Assay
4.4. Western Blot
4.5. RNA Interference (RNAi)
4.6. TP53 Sequencing
4.7. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rödel, F.; Sprenger, T.; Kaina, B.; Liersch, T.; Rödel, C.; Fulda, S.; Hehlgans, S. Survivin as a Prognostic/Predictive Marker and Molecular Target in Cancer Therapy. Curr. Med. Chem. 2012, 19, 3679–3688. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Aljahdali, I.; Ling, X. Cancer therapeutics using survivin BIRC5 as a target: What can we do after over two decades of study? J. Exp. Clin. Cancer Res. 2019, 38, 368. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, S.P.; Altieri, D.C. Survivin at a glance. J. Cell. Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, K.C.; Hogarty, M.D. Targeting programmed cell death pathways with experimental therapeutics: Opportunities in high-risk neuroblastoma. Cancer Lett. 2005, 228, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Lamers, F.; van der Ploeg, I.; Schild, L.; Ebus, M.E.; Koster, J.; Hansen, B.R.; Koch, T.; Versteeg, R.; Caron, H.N.; Molenaar, J.J. Knockdown of survivin (BIRC5) causes apoptosis in neuroblastoma via mitotic catastrophe. Endocr. Relat. Cancer 2011, 18, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Lamers, F.; Schild, L.; Koster, J.; Versteeg, R.; Caron, H.N.; Molenaar, J.J. Targeted BIRC5 silencing using YM155 causes cell death in neuroblastoma cells with low ABCB1 expression. Eur. J. Cancer 2012, 48, 763–771. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, L.; Xu, R.; Ju, X.L. Silencing of survivin using YM155 induces apoptosis and chemosensitization in neuroblastomas cells. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 2909–2915. [Google Scholar]
- Rauch, A.; Hennig, D.; Schäfer, C.; Wirth, M.; Marx, C.; Heinzel, T.; Schneider, G.; Krämer, O.H. Survivin and YM155: How faithful is the liaison? Biochim. Biophys. Acta. 2014, 1845, 202–220. [Google Scholar] [CrossRef]
- Hagenbuchner, J.; Kiechl-Kohlendorfer, U.; Obexer, P.; Ausserlechner, M.J. BIRC5/Survivin as a target for glycolysis inhibition in high-stage neuroblastoma. Oncogene 2016, 35, 2052–2061. [Google Scholar] [CrossRef]
- Voges, Y.; Michaelis, M.; Rothweiler, F.; Schaller, T.; Schneider, C.; Politt, K.; Mernberger, M.; Nist, A.; Stiewe, T.; Wass, M.N.; et al. Effects of YM155 on survivin levels and viability in neuroblastoma cells with acquired drug resistance. Cell Death Dis. 2016, 7, e2410. [Google Scholar] [CrossRef]
- Morgenstern, D.A.; Baruchel, S.; Irwin, M.S. Current and future strategies for relapsed neuroblastoma: Challenges on the road to precision therapy. J. Pediatr. Hematol. Oncol. 2013, 35, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Bagatell, R.; London, W.B.; Maris, J.M.; Cohn, S.L.; Mattay, K.K.; Hogarty, M.; COG Neuroblastoma Committee. Children’s Oncology Group’s 2013 blueprint for research: Neuroblastoma. Pediatr. Blood Cancer 2013, 60, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, T.; Kita, A.; Yamanaka, K.; Mori, M.; Amino, N.; Takeuchi, M.; Tominaga, F.; Hatakeyama, S.; Kinoyama, I.; Matsuhisa, A.; et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007, 67, 8011–8014. [Google Scholar] [CrossRef] [PubMed]
- Glaros, T.G.; Stockwin, L.H.; Mullendore, M.E.; Smith, B.; Morrison, B.L.; Newton, D.L. The "survivin suppressants" NSC 80467 and YM155 induce a DNA damage response. Cancer Chemother. Pharmacol. 2012, 70, 207–212. [Google Scholar] [CrossRef]
- Chang, B.H.; Johnson, K.; LaTocha, D.; Rowley, J.S.; Bryant, J.; Burke, R.; Smith, R.L.; Loriaux, M.; Müschen, M.; Mullighan, C.; et al. YM155 potently kills acute lymphoblastic leukemia cells through activation of the DNA damage pathway. J. Hematol. Oncol. 2015, 8, 39. [Google Scholar] [CrossRef]
- Tang, H.; Shao, H.; Yu, C.; Hou, J. Mcl-1 downregulation by YM155 contributes to its synergistic anti-tumor activities with ABT-263. Biochem. Pharmacol. 2011, 82, 1066–1072. [Google Scholar] [CrossRef]
- Wagner, V.; Hose, D.; Seckinger, A.; Weiz, L.; Meißner, T.; Rème, T.; Breitkreutz, I.; Podar, K.; Ho, A.D.; Goldschmidt, H.; et al. Preclinical efficacy of sepantronium bromide (YM155) in multiple myeloma is conferred by down regulation of Mcl-1. Oncotarget 2014, 5, 10237–10250. [Google Scholar] [CrossRef]
- de Necochea-Campion, R.; Diaz Osterman, C.J.; Hsu, H.W.; Fan, J.; Mirshahidi, S.; Wall, N.R.; Chen, C.S. AML sensitivity to YM155 is modulated through AKT and Mcl-1. Cancer Lett. 2015, 366, 44–51. [Google Scholar] [CrossRef]
- Iwai, M.; Minematsu, T.; Li, Q.; Iwatsubo, T.; Usui, T. Utility of P-glycoprotein and organic cation transporter 1 double-transfected LLC-PK1 cells for studying the interaction of YM155 monobromide, novel small-molecule survivin suppressant, with P-glycoprotein. Drug Metab. Dispos. 2011, 39, 2314–2320. [Google Scholar] [CrossRef]
- Winter, G.E.; Radic, B.; Mayor-Ruiz, C.; Blomen, V.A.; Trefzer, C.; Kandasamy, R.K.; Huber, K.V.M.; Gridling, M.; Chen, D.; Klampfl, T.; et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat. Chem. Biol. 2014, 10, 768–773. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Mita, A.; Lewis, L.D.; Garrett, C.R.; Till, E.; Daud, A.I.; Patnaik, A.; Papadopoulos, K.; Takimoto, C.; Bartels, P.; et al. Phase I and pharmacokinetic study of YM155, a small-molecule inhibitor of survivin. J. Clin. Oncol. 2008, 26, 5198–5203. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Okamoto, I.; Miyazaki, M.; Morinaga, R.; Tsuya, A.; Hasegawa, Y.; Terashima, M.; Ueda, S.; Fukuoka, M.; Ariyoshi, Y.; et al. Phase I study of YM155, a novel survivin suppressant, in patients with advanced solid tumors. Clin. Cancer Res. 2009, 15, 3872–3880. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, Y.; Kaibara, A.; Takada, A.; Nishimura, T.; Katashima, M.; Sawamoto, T. Population pharmacokinetic modeling of sepantronium bromide (YM155), a small molecule survivin suppressant, in patients with non-small cell lung cancer, hormone refractory prostate cancer, or unresectable stage III or IV melanoma. Invest. New Drugs 2013, 31, 443–451. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, C.A.; Callaghan, R. How can we best use structural information on P-glycoprotein to design inhibitors? Pharmacol. Ther. 2007, 113, 429–441. [Google Scholar] [CrossRef]
- Lutz, W.; Stöhr, M.; Schürmann, J.; Wenzel, A.; Löhr, A.; Schwab, M. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene 1996, 13, 803–812. [Google Scholar]
- Michaelis, M.; Rothweiler, F.; Barth, S.; Cinatl, J.; van Rikxoort, M.; Löschmann, N.; Voges, Y.; Breitling, R.; von Deimling, A.; Rödel, F.; et al. Adaptation of cancer cells from different entities to the MDM2 inhibitor nutlin-3 results in the emergence of p53-mutated multi-drug-resistant cancer cells. Cell Death Dis. 2011, 2, e243. [Google Scholar] [CrossRef]
- Michaelis, M.; Rothweiler, F.; Agha, B.; Barth, S.; Voges, Y.; Löschmann, N.; von Deimling, A.; Breitling, R.; Doerr, H.W.; Rödel, F.; et al. Human neuroblastoma cells with acquired resistance to the p53 activator RITA retain functional p53 and sensitivity to other p53 activating agents. Cell Death Dis. 2012, 3, e294. [Google Scholar] [CrossRef]
- Löschmann, N.; Michaelis, M.; Rothweiler, F.; Zehner, R.; Cinatl, J.; Voges, Y.; Sharifi, M.; Riecken, K.; Meyer, J.; von Deimling, A.; et al. Testing of SNS-032 in a panel of human neuroblastoma cell lines with acquired resistance to a broad range of drugs. Transl. Oncol. 2013, 6, 685–696. [Google Scholar] [CrossRef]
- Löschmann, N.; Michaelis, M.; Rothweiler, F.; Voges, Y.; Balónová, B.; Blight, B.A.; Cinatl, J., Jr. ABCB1 as predominant resistance mechanism in cells with acquired SNS-032 resistance. Oncotarget 2016, 7, 58051–58064. [Google Scholar] [CrossRef][Green Version]
- Abe, T.; Koike, K.; Ohga, T.; Kubo, T.; Wada, M.; Kohno, K.; Mori, T.; Hidaka, K.; Kuwano, M. Chemosensitisation of spontaneous multidrug resistance by a 1,4-dihydropyridine analogue and verapamil in human glioma cell lines overexpressing MRP or MDR1. Br. J. Cancer 1995, 72, 418–423. [Google Scholar] [CrossRef]
- Dantzig, A.H.; Shepard, R.L.; Law, K.L.; Tabas, L.; Pratt, S.; Gillespie, J.S.; Binkley, S.N.; Kuhfeld, M.T.; Starling, J.J.; Wrighton, S.A. Selectivity of the multidrug resistance modulator, LY335979, for P-glycoprotein and effect on cytochrome P-450 activities. J. Pharmacol. Exp. Ther. 1999, 290, 854–862. [Google Scholar] [PubMed]
- Yu, D.M.; Huynh, T.; Truong, A.M.; Haber, M.; Norris, M.D. ABC transporters and neuroblastoma. Adv. Cancer Res. 2015, 125, 139–170. [Google Scholar] [CrossRef] [PubMed]
- Pituch-Noworolska, A.; Zaremba, M.; Wieczorek, A. Expression of proteins associated with therapy resistance in rhabdomyosarcoma and neuroblastoma tumour cells. Pol. J. Pathol. 2009, 60, 168–173. [Google Scholar] [PubMed]
- Oue, T.; Yoneda, A.; Uehara, S.; Yamanaka, H.; Fukuzawa, M. Increased expression of multidrug resistance-associated genes after chemotherapy in pediatric solid malignancies. J. Pediatr. Surg. 2009, 44, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, M.; Rothweiler, F.; Löschmann, N.; Sharifi, M.; Ghafourian, T.; Cinatl, J., Jr. Enzastaurin inhibits ABCB1-mediated drug efflux independently of effects on protein kinase C signalling and the cellular p53 status. Oncotarget 2015, 6, 17605–17620. [Google Scholar] [CrossRef] [PubMed]
- Engelman, J.A.; Zejnullahu, K.; Mitsudomi, T.; Song, Y.; Hyland, C.; Park, J.O.; Lindeman, N.; Gale, C.M.; Zhao, X.; Christensen, J.; et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007, 316, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, R.; Shi, H.; Wang, Q.; Kong, X.; Koya, R.C.; Lee, H.; Chen, Z.; Lee, M.K.; Attar, N.; Sazegar, H.; et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Persaud, Y.; Janakiraman, M.; Kong, X.; Ng, C.; Moriceau, G.; Shi, H.; Atefi, M.; Titz, B.; Gabay, M.T.; et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011, 480, 387–390. [Google Scholar] [CrossRef]
- Domingo-Domenech, J.; Vidal, S.J.; Rodriguez-Bravo, V.; Castillo-Martin, M.; Quinn, S.A.; Rodriguez-Barrueco, R.; Bonal, D.M.; Charytonowicz, E.; Gladoun, N.; de la Iglesia-Vicente, J.; et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell 2012, 22, 373–388. [Google Scholar] [CrossRef]
- Joseph, J.D.; Lu, N.; Qian, J.; Sensintaffar, J.; Shao, G.; Brigham, D.; Moon, M.; Maneval, E.C.; Chen, I.; Darimont, B.; et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov. 2013, 3, 1020–1029. [Google Scholar] [CrossRef]
- Korpal, M.; Korn, J.M.; Gao, X.; Rakiec, D.P.; Ruddy, D.A.; Doshi, S.; Yuan, J.; Kovats, S.G.; Kim, S.; Cooke, V.G.; et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 2013, 3, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Crystal, A.S.; Shaw, A.T.; Sequist, L.V.; Friboulet, L.; Niederst, M.J.; Lockerman, E.L.; Frias, R.L.; Gainor, J.F.; Amzallag, A.; Greninger, P.; et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014, 346, 1480–1486. [Google Scholar] [CrossRef] [PubMed]
- Niederst, M.J.; Sequist, L.V.; Poirier, J.T.; Mermel, C.H.; Lockerman, E.L.; Garcia, A.R.; Katayama, R.; Costa, C.; Ross, K.N.; Moran, T.; et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat. Commun. 2015, 6, 6377. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Lee, J.S.; Dickson, M.A.; Schwartz, G.K.; Le Cesne, A.; Varga, A.; Bahleda, R.; Wagner, A.J.; Choy, E.; de Jonge, M.J.; et al. TP53 mutations emerge with HDM2 inhibitor SAR405838 treatment in de-differentiated liposarcoma. Nat. Commun. 2016, 7, 12609. [Google Scholar] [CrossRef]
- Schneider, C.; Oellerich, T.; Baldauf, H.M.; Schwarz, S.M.; Thomas, D.; Flick, R.; Bohnenberger, H.; Kaderali, L.; Stegmann, L.; Cremer, A.; et al. SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat. Med. 2017, 23, 250–255. [Google Scholar] [CrossRef]
- Dhooge, C.R.; De Moerloose, B.M.; Benoit, Y.C.; Van Roy, N.; Philippé, J.; Laureys, G.G. Expression of the MDR1 gene product P-glycoprotein in childhood neuroblastoma. Cancer 1997, 80, 1250–1257. [Google Scholar] [CrossRef][Green Version]
- Palmeira, A.; Sousa, E.; Vasconcelos, M.H.; Pinto, M.M. Three decades of P-gp inhibitors: Skimming through several generations and scaffolds. Curr. Med. Chem. 2012, 19, 1946–2025. [Google Scholar] [CrossRef]
- Sissung, T.M.; Baum, C.E.; Kirkland, C.T.; Gao, R.; Gardner, E.R.; Figg, W.D. Pharmacogenetics of membrane transporters: An update on current approaches. Mol. Biotechnol. 2010, 44, 152–167. [Google Scholar] [CrossRef]
- Ieiri, I. Functional significance of genetic polymorphisms in P-glycoprotein (MDR1, ABCB1) and breast cancer resistance protein (BCRP, ABCG2). Drug Metab. Pharmacokinet. 2012, 27, 85–105. [Google Scholar] [CrossRef]
- Michaelis, M.; Rothweiler, F.; Wurglics, M.; Aniceto, N.; Dittrich, M.; Zettl, H.; Wiese, M.; Wass, M.N.; Ghafourian, T.; Schubert-Zsilavecz, M.; et al. Substrate-specific effects of pirinixic acid derivatives on ABCB1-mediated drug transport. Oncotarget 2016, 7, 11664–11676. [Google Scholar] [CrossRef]
- Nakahara, T.; Kita, A.; Yamanaka, K.; Mori, M.; Amino, N.; Takeuchi, M.; Tominaga, F.; Kinoyama, I.; Matsuhisa, A.; Kudou, M.; et al. Broad spectrum and potent antitumor activities of YM155, a novel small-molecule survivin suppressant, in a wide variety of human cancer cell lines and xenograft models. Cancer Sci. 2011, 102, 614–621. [Google Scholar] [CrossRef]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Meresse, P.; Dechaux, E.; Monneret, C.; Bertounesque, E. Etoposide: Discovery and medicinal chemistry. Curr. Med. Chem. 2004, 11, 2443–2466. [Google Scholar] [CrossRef] [PubMed]
- Broxterman, H.J.; Gotink, K.J.; Verheul, H.M. Understanding the causes of multidrug resistance in cancer: A comparison of doxorubicin and sunitinib. Drug Resist. Updat. 2009, 12, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Dilruba, S.; Kalayda, G.V. Platinum-based drugs: Past, present and future. Cancer Chemother. Pharmacol. 2016, 77, 1103–1124. [Google Scholar] [CrossRef]
- Kotchetkov, R.; Cinatl, J.; Blaheta, R.; Vogel, J.U.; Karaskova, J.; Squire, J.; Hernáiz Driever, P.; Klingebiel, T.; Cinatl, J., Jr. Development of resistance to vincristine and doxorubicin in neuroblastoma alters malignant properties and induces additional karyotype changes: A preclinical model. Int. J. Cancer 2003, 104, 36–43. [Google Scholar] [CrossRef]
- Kotchetkov, R.; Driever, P.H.; Cinatl, J.; Michaelis, M.; Karaskova, J.; Blaheta, R.; Squire, J.A.; Von Deimling, A.; Moog, J.; Cinatl, J., Jr. Increased malignant behavior in neuroblastoma cells with acquired multi-drug resistance does not depend on P-gp expression. Int. J. Oncol. 2005, 27, 1029–1037. [Google Scholar] [CrossRef]
- Giaccone, G.; Zatloukal, P.; Roubec, J.; Floor, K.; Musil, J.; Kuta, M.; van Klaveren, R.J.; Chaudhary, S.; Gunther, A.; Shamsili, S. Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non-small-cell lung cancer. J. Clin. Oncol. 2009, 27, 4481–4486. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Quinn, D.I.; Ferrari, A.; Ahmann, F.; Giaccone, G.; Drake, T.; Keating, A.; de Bono, J.S. A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxane-pretreated prostate cancer. Ann. Oncol. 2012, 23, 968–973. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michaelis, M.; Voges, Y.; Rothweiler, F.; Weipert, F.; Zia-Ahmad, A.; Cinatl, J.; von Deimling, A.; Westermann, F.; Rödel, F.; Wass, M.N.; et al. Testing of the Survivin Suppressant YM155 in a Large Panel of Drug-Resistant Neuroblastoma Cell Lines. Cancers 2020, 12, 577. https://doi.org/10.3390/cancers12030577
Michaelis M, Voges Y, Rothweiler F, Weipert F, Zia-Ahmad A, Cinatl J, von Deimling A, Westermann F, Rödel F, Wass MN, et al. Testing of the Survivin Suppressant YM155 in a Large Panel of Drug-Resistant Neuroblastoma Cell Lines. Cancers. 2020; 12(3):577. https://doi.org/10.3390/cancers12030577
Chicago/Turabian StyleMichaelis, Martin, Yvonne Voges, Florian Rothweiler, Fabian Weipert, Amara Zia-Ahmad, Jaroslav Cinatl, Andreas von Deimling, Frank Westermann, Franz Rödel, Mark N. Wass, and et al. 2020. "Testing of the Survivin Suppressant YM155 in a Large Panel of Drug-Resistant Neuroblastoma Cell Lines" Cancers 12, no. 3: 577. https://doi.org/10.3390/cancers12030577
APA StyleMichaelis, M., Voges, Y., Rothweiler, F., Weipert, F., Zia-Ahmad, A., Cinatl, J., von Deimling, A., Westermann, F., Rödel, F., Wass, M. N., & Cinatl, J., Jr. (2020). Testing of the Survivin Suppressant YM155 in a Large Panel of Drug-Resistant Neuroblastoma Cell Lines. Cancers, 12(3), 577. https://doi.org/10.3390/cancers12030577





