Deletion of Von Hippel–Lindau Interferes with Hyper Osmolality Induced Gene Expression and Induces an Unfavorable Gene Expression Pattern
Abstract
1. Introduction
2. Results
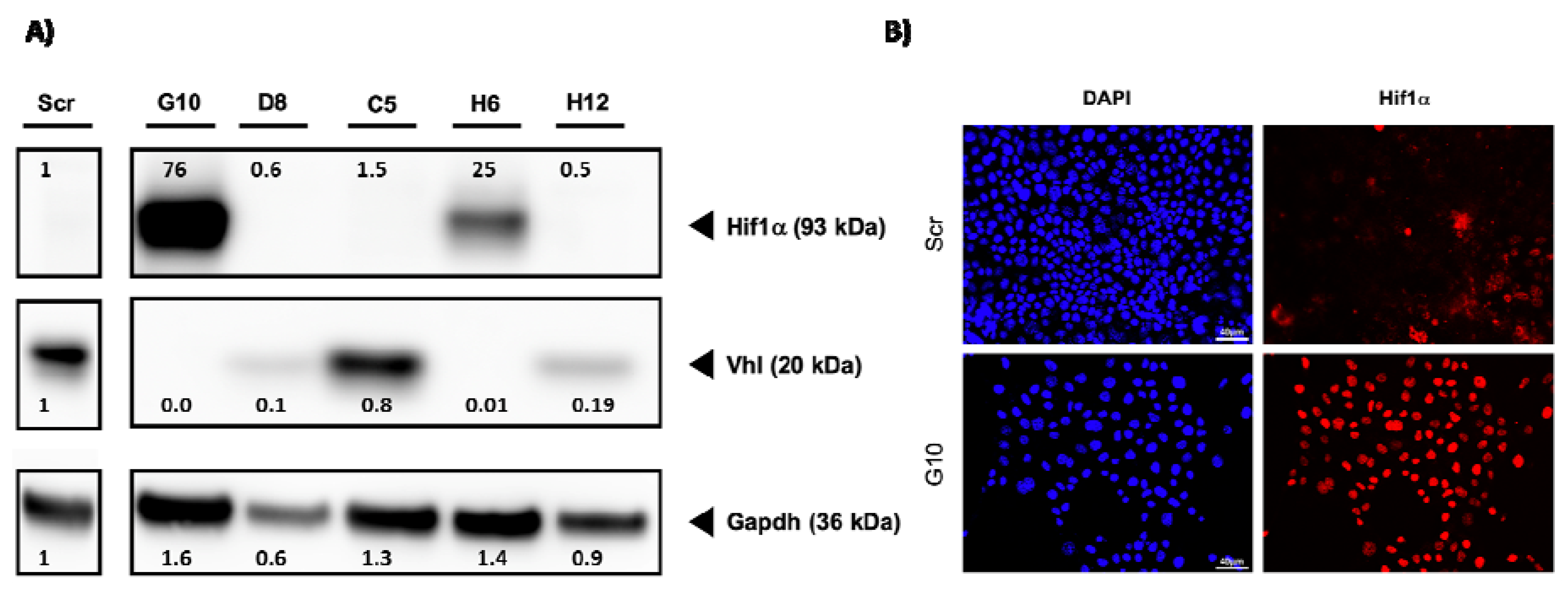
2.1. Generation of VHL-Deficient Cells
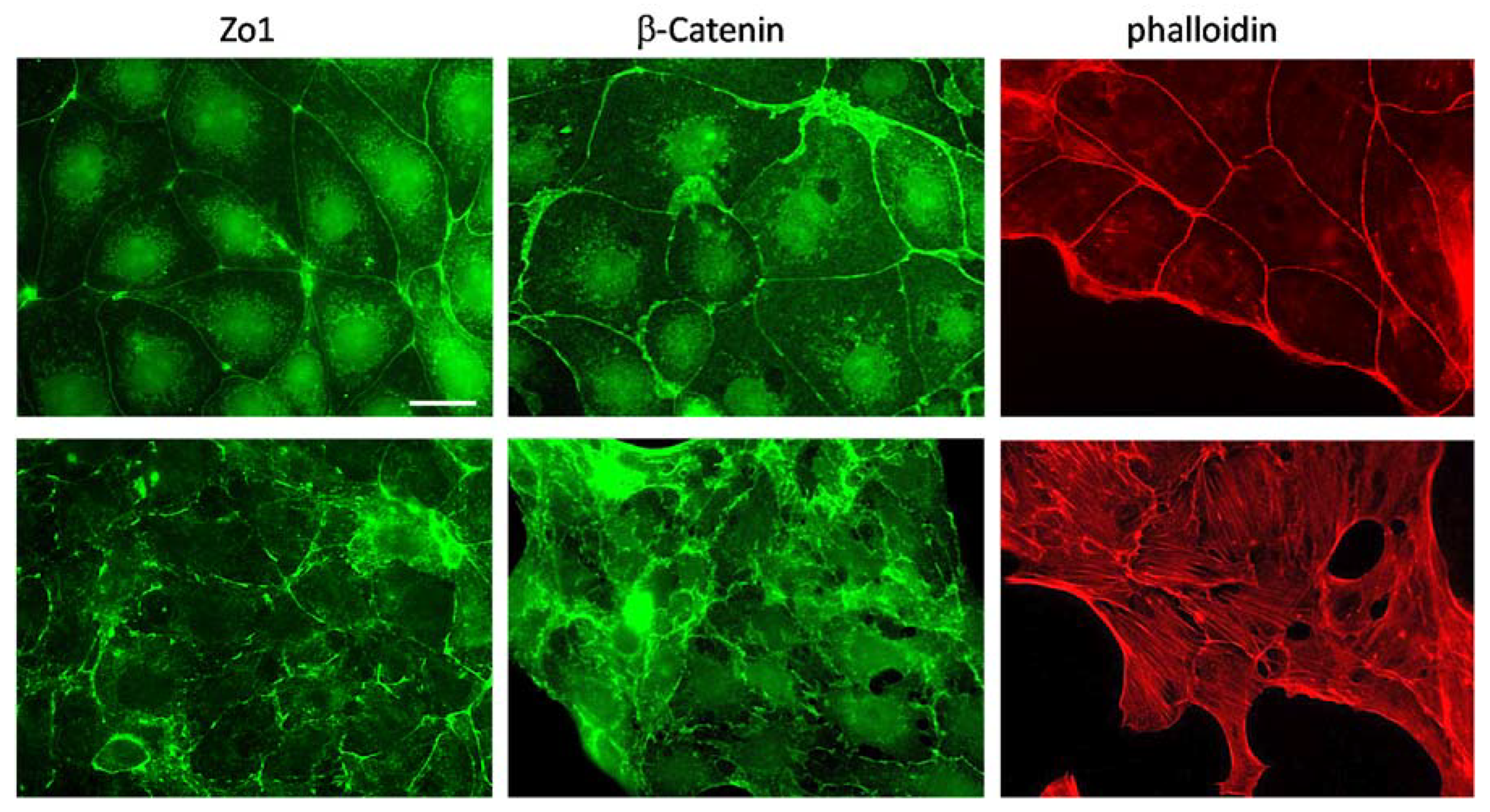
2.2. Vhl Deletion Induces Loss of Epithelial Structures
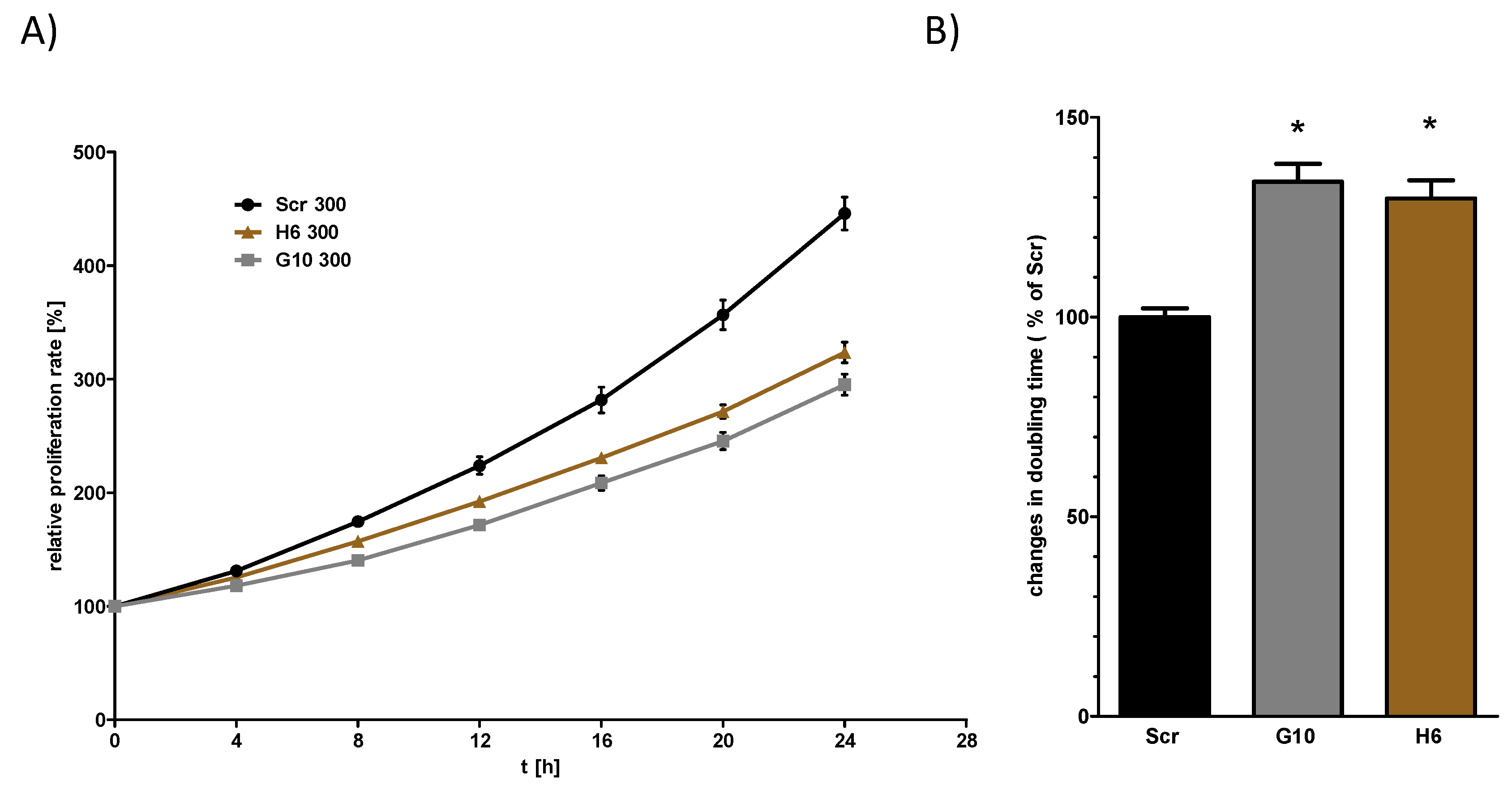
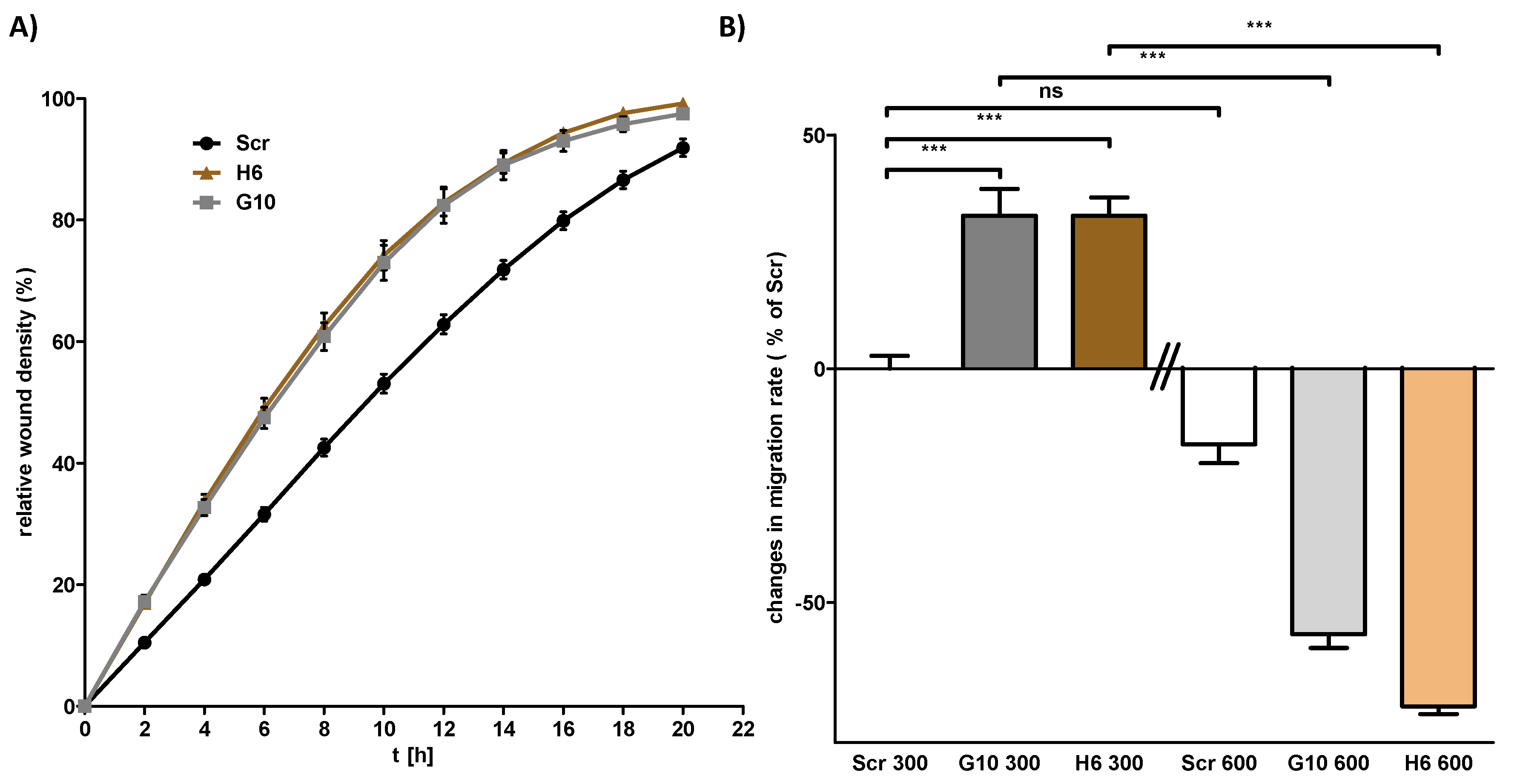
2.3. Vhl Deletion is Associated with Changes in Proliferation and Migration Behavior
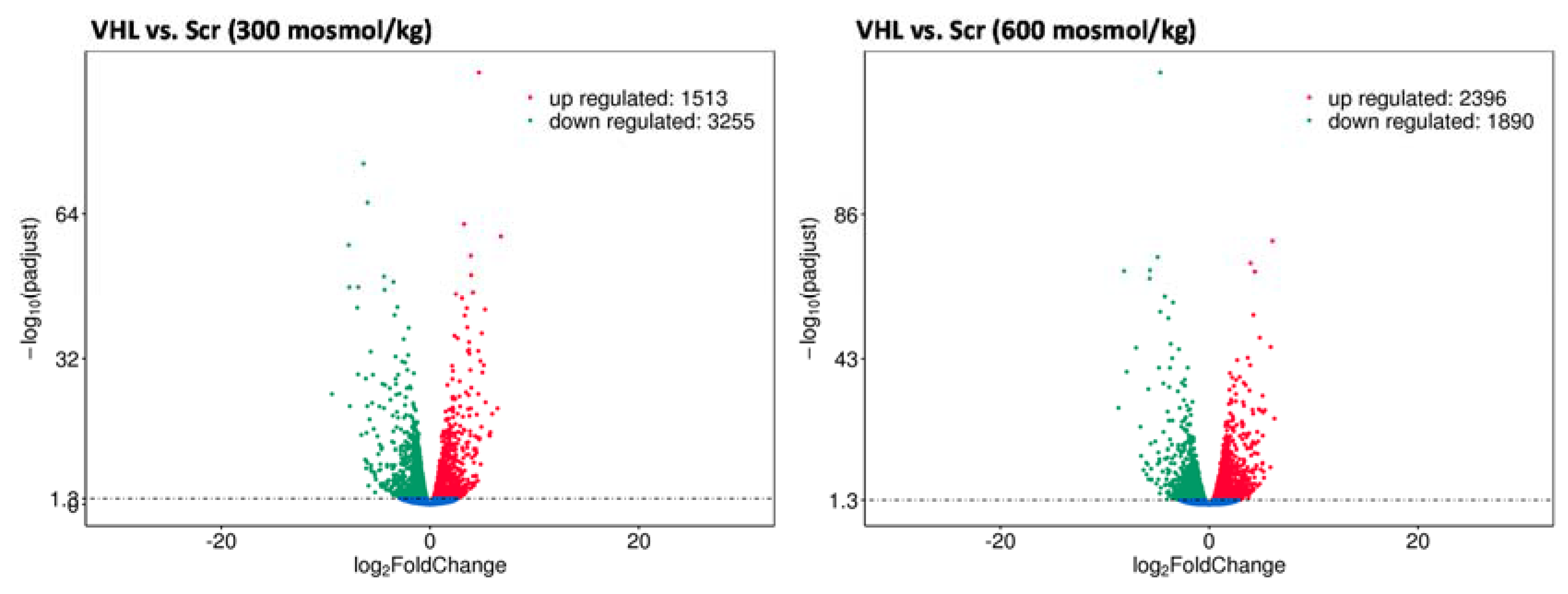
2.4. Vhl Deletion Affects Expression of Hyperosmolality Regulated Genes
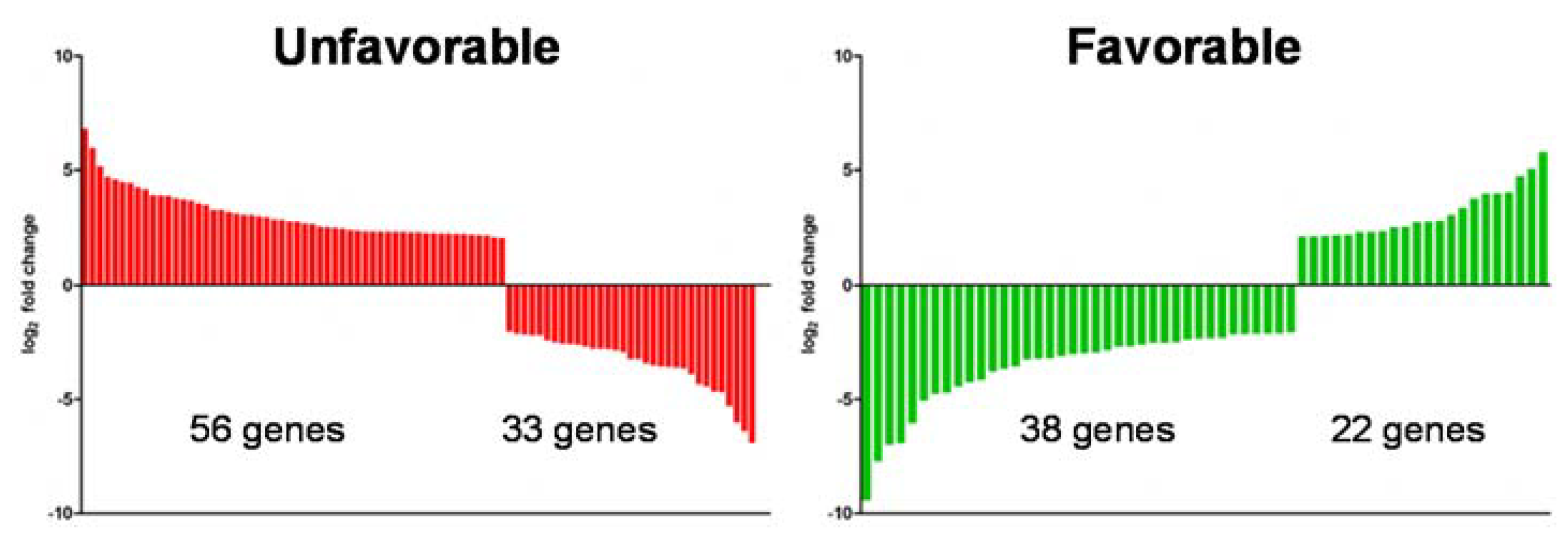
2.5. Loss of Vhl Function Leads to an Unfavorable Gene Expression Pattern
3. Discussion
4. Material and Methods
4.1. Cell Culture
4.2. Oligos and Primers
4.3. Cloning of gRNAs and Vector Production in Escherichia coli
4.4. Vectors
4.5. Viral Transfection of mpkCCD Cells and Sequencing of Vhl Locus
4.6. Isolation of Single-Cell Clones and Tracking of Indels (TIDE)
4.7. Western Blot
4.8. Immunofluorescence
4.9. Proliferation and Migration
4.10. Real-Time PCR (RT-PCR)
4.11. Preparation of Samples for Next-Generation Sequencing
4.12. Favorable and Unfavorable Gene Expression in Kidney Cancer
4.13. Induced Gene Expression by Osmolality in Kidney Cancer
4.14. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.I.; Campbell, S.C.; Escudier, B. Renal cell carcinoma. Lancet 2009, 373, 1119–1132. [Google Scholar] [CrossRef]
- McLaughlin, J.; Lipworth, L.; Tarone, R.; Blot, W. Cancer Epidemiology and Prevention; Oxford University Press: Oxford, UK, 2006; pp. 1087–1100. [Google Scholar]
- Hakimi, A.A.; Chen, Y.B.; Wren, J.; Gonen, M.; Abdel-Wahab, O.; Heguy, A.; Liu, H.; Takeda, S.; Tickoo, S.K.; Reuter, V.E.; et al. Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. Eur. Urol. 2013, 63, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Gnarra, J.R.; Tory, K.; Weng, Y.; Schmidt, L.; Wei, M.H.; Li, H.; Latif, F.; Liu, S.; Chen, F.; Duh, F.M.; et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 1994, 7, 85–90. [Google Scholar] [CrossRef]
- Nickerson, M.L.; Jaeger, E.; Shi, Y.; Durocher, J.A.; Mahurkar, S.; Zaridze, D.; Matveev, V.; Janout, V.; Kollarova, H.; Bencko, V.; et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin. Cancer Res. 2008, 14, 4726–4734. [Google Scholar] [CrossRef]
- Rankin, E.B.; Tomaszewski, J.E.; Haase, V.H. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006, 66, 2576–2583. [Google Scholar] [CrossRef]
- Frew, I.J.; Thoma, C.R.; Georgiev, S.; Minola, A.; Hitz, M.; Montani, M.; Moch, H.; Krek, W. pVHL and PTEN tumour suppressor proteins cooperatively suppress kidney cyst formation. EMBO J. 2008, 27, 1747–1757. [Google Scholar] [CrossRef]
- Schönenberger, D.; Rajski, M.; Harlander, S.; Frew, I.J. Vhl deletion in renal epithelia causes HIF-1α-dependent, HIF-2α-independent angiogenesis and constitutive diuresis. Oncotarget 2016, 7, 60971–60985. [Google Scholar] [CrossRef]
- Edemir, B.; Pavenstadt, H.; Schlatter, E.; Weide, T. Mechanisms of cell polarity and aquaporin sorting in the nephron. Pflug. Arch. 2011, 461, 607–621. [Google Scholar] [CrossRef]
- Nielsen, S.; Frøkiær, J.; Marples, D.; Kwon, T.H.; Agre, P.; Knepper, M.A. Aquaporins in the kidney: From molecules to medicine. Physiol. Rev. 2002, 82, 205–244. [Google Scholar] [CrossRef]
- Hasler, U.; Jeon, U.S.; Kim, J.A.; Mordasini, D.; Kwon, H.M.; Feraille, E.; Martin, P.Y. Tonicity-responsive enhancer binding protein is an essential regulator of aquaporin-2 expression in renal collecting duct principal cells. J. Am. Soc. Nephrol. 2006, 17, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, H.; Woo, S.K.; Dahl, S.C.; Handler, J.S.; Kwon, H.M. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc. Natl. Acad. Sci. USA 1999, 96, 2538–2542. [Google Scholar] [CrossRef] [PubMed]
- Bogusławska, J.; Popławski, P.; Alseekh, S.; Koblowska, M.; Iwanicka-Nowicka, R.; Rybicka, B.; Kędzierska, H.; Głuchowska, K.; Hanusek, K.; Tański, Z.; et al. MicroRNA-Mediated Metabolic Reprograming in Renal Cancer. Cancers 2019, 11, 1825. [Google Scholar] [CrossRef] [PubMed]
- Schulze Blasum, B.; Schroter, R.; Neugebauer, U.; Hofschroer, V.; Pavenstadt, H.; Ciarimboli, G.; Schlatter, E.; Edemir, B. The kidney-specific expression of genes can be modulated by the extracellular osmolality. FASEB J. 2016, 30, 3588–3597. [Google Scholar] [CrossRef]
- Kandabarau, S.; Leiz, J.; Krohn, K.; Winter, S.; Bedke, J.; Schwab, M.; Schaeffeler, E.; Edemir, B. Hypertonicity-Affected Genes Are Differentially Expressed in Clear Cell Renal Cell Carcinoma and Correlate with Cancer-Specific Survival. Cancers 2019, 12, 6. [Google Scholar] [CrossRef]
- Lapinskas, E.J.; Svobodova, S.; Davis, I.D.; Cebon, J.; Hertzog, P.J.; Pritchard, M.A. The Ets transcription factor ELF5 functions as a tumor suppressor in the kidney. Twin Res. Hum. Genet. 2011, 14, 316–322. [Google Scholar] [CrossRef]
- Yu, M.J.; Miller, R.L.; Uawithya, P.; Rinschen, M.M.; Khositseth, S.; Braucht, D.W.; Chou, C.L.; Pisitkun, T.; Nelson, R.D.; Knepper, M.A. Systems-level analysis of cell-specific AQP2 gene expression in renal collecting duct. Proc. Natl. Acad. Sci. USA 2009, 106, 2441–2446. [Google Scholar] [CrossRef]
- Hasler, U.; Vinciguerra, M.; Vandewalle, A.; Martin, P.Y.; Feraille, E. Dual effects of hypertonicity on aquaporin-2 expression in cultured renal collecting duct principal cells. J. Am. Soc. Nephrol. 2005, 16, 1571–1582. [Google Scholar] [CrossRef]
- Brinkman, E.K.; Chen, T.; Amendola, M.; van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014, 42, e168. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, X.; Wang, B.; Zhang, K.; Liu, S.; Yue, K.; Zhang, L.; Wang, X. Loss of VHL expression contributes to epithelial-mesenchymal transition in oral squamous cell carcinoma. Oral Oncol. 2014, 50, 809–817. [Google Scholar] [CrossRef]
- Guo, H.; German, P.; Bai, S.; Barnes, S.; Guo, W.; Qi, X.; Lou, H.; Liang, J.; Jonasch, E.; Mills, G.B.; et al. The PI3K/AKT Pathway and Renal Cell Carcinoma. J. Genet. Genom. 2015, 42, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Gurzu, S.; Turdean, S.; Kovecsi, A.; Contac, A.O.; Jung, I. Epithelial-mesenchymal, mesenchymal-epithelial, and endothelial-mesenchymal transitions in malignant tumors: An update. World J. Clin. Cases 2015, 3, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed]
- Schokrpur, S.; Hu, J.; Moughon, D.L.; Liu, P.; Lin, L.C.; Hermann, K.; Mangul, S.; Guan, W.; Pellegrini, M.; Xu, H.; et al. CRISPR-Mediated VHL Knockout Generates an Improved Model for Metastatic Renal Cell Carcinoma. Sci. Rep. 2016, 6, 29032. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, T.; Ibe, J.C.F.; Usha Raj, J.; Zhou, G. Knockdown of von Hippel–Lindau protein decreases lung cancer cell proliferation and colonization. FEBS Lett. 2012, 586, 1510–1515. [Google Scholar] [CrossRef]
- Ozcan, A.; Zhai, J.; Hamilton, C.; Shen, S.S.; Ro, J.Y.; Krishnan, B.; Truong, L.D. PAX-2 in the diagnosis of primary renal tumors: Immunohistochemical comparison with renal cell carcinoma marker antigen and kidney-specific cadherin. Am. J. Clin. Pathol. 2009, 131, 393–404. [Google Scholar] [CrossRef]
- Kraus, S.; Abel, P.D.; Nachtmann, C.; Linsenmann, H.J.; Weidner, W.; Stamp, G.W.; Chaudhary, K.S.; Mitchell, S.E.; Franke, F.E.; El Lalani, N. MUC1 mucin and trefoil factor 1 protein expression in renal cell carcinoma: Correlation with prognosis. Hum. Pathol. 2002, 33, 60–67. [Google Scholar] [CrossRef]
- Khurana, K.K.; Truong, L.D.; Verani, R.R. Image analysis of proliferating cell nuclear antigen expression and immunohistochemical profiles in renal cell carcinoma associated with acquired cystic kidney disease: Comparison with classic renal cell carcinoma. Mod. Pathol. 1998, 11, 339–346. [Google Scholar]
- Shen, S.S.; Krishna, B.; Chirala, R.; Amato, R.J.; Truong, L.D. Kidney-specific cadherin, a specific marker for the distal portion of the nephron and related renal neoplasms. Mod. Pathol. 2005, 18, 933–940. [Google Scholar] [CrossRef]
- Pritchett, T.L.; Bader, H.L.; Henderson, J.; Hsu, T. Conditional inactivation of the mouse von Hippel-Lindau tumor suppressor gene results in wide-spread hyperplastic, inflammatory and fibrotic lesions in the kidney. Oncogene 2015, 34, 2631–2639. [Google Scholar] [CrossRef]
- Harlander, S.; Schönenberger, D.; Toussaint, N.C.; Prummer, M.; Catalano, A.; Brandt, L.; Moch, H.; Wild, P.J.; Frew, I.J. Combined mutation in Vhl, Trp53 and Rb1 causes clear cell renal cell carcinoma in mice. Nat. Med. 2017, 23, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Chou, C.L.; Knepper, M.A. Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J. Am. Soc. Nephrol. 2015, 26, 2669–2677. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Schrödter, S.; Braun, M.; Syring, I.; Klümper, N.; Deng, M.; Schmidt, D.; Perner, S.; Müller, S.C.; Ellinger, J. Identification of the dopamine transporter SLC6A3 as a biomarker for patients with renal cell carcinoma. Mol. Cancer 2016, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Rafael, D.; Doktorovova, S.; Florindo, H.F.; Gener, P.; Abasolo, I.; Schwartz, S., Jr.; Videira, M.A. EMT blockage strategies: Targeting Akt dependent mechanisms for breast cancer metastatic behaviour modulation. Curr. Gene Ther. 2015, 15, 300–312. [Google Scholar] [CrossRef]
- Wetzel, R.K.; Pascoa, J.L.; Arystarkhova, E. Stress-induced expression of the gamma subunit (FXYD2) modulates Na,K-ATPase activity and cell growth. J. Biol. Chem. 2004, 279, 41750–41757. [Google Scholar] [CrossRef]
- Izumi, Y.; Yang, W.; Zhu, J.; Burg, M.B.; Ferraris, J.D. RNA-Seq analysis of high NaCl-induced gene expression. Physiol. Genom. 2015, 47, 500–513. [Google Scholar] [CrossRef]
- Maeoka, Y.; Wu, Y.; Okamoto, T.; Kanemoto, S.; Guo, X.P.; Saito, A.; Asada, R.; Matsuhisa, K.; Masaki, T.; Imaizumi, K.; et al. NFAT5 up-regulates expression of the kidney-specific ubiquitin ligase gene Rnf183 under hypertonic conditions in inner-medullary collecting duct cells. J. Biol. Chem. 2019, 294, 101–115. [Google Scholar] [CrossRef]
- Jeon, U.S.; Kim, J.A.; Sheen, M.R.; Kwon, H.M. How tonicity regulates genes: Story of TonEBP transcriptional activator. Acta Physiol. 2006, 187, 241–247. [Google Scholar] [CrossRef]
- Burg, M.B.; Ferraris, J.D.; Dmitrieva, N.I. Cellular response to hyperosmotic stresses. Physiol. Rev. 2007, 87, 1441–1474. [Google Scholar] [CrossRef]
- Tellechea, M.; Buxade, M.; Tejedor, S.; Aramburu, J.; Lopez-Rodriguez, C. NFAT5-Regulated Macrophage Polarization Supports the Proinflammatory Function of Macrophages and T Lymphocytes. J. Immunol. 2018, 200, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Boland, B.S.; Widjaja, C.E.; Banno, A.; Zhang, B.; Kim, S.H.; Stoven, S.; Peterson, M.R.; Jones, M.C.; Su, H.I.; Crowe, S.E.; et al. Immunodeficiency and autoimmune enterocolopathy linked to NFAT5 haploinsufficiency. J. Immunol. 2015, 194, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Suh, J.H.; Choi, S.Y.; Kang, H.J.; Lee, H.H.; Ye, B.J.; Lee, G.R.; Jung, S.W.; Kim, C.J.; Lee-Kwon, W.; et al. Tonicity-responsive enhancer-binding protein promotes hepatocellular carcinogenesis, recurrence and metastasis. Gut 2019, 68, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Wang, Y.; Li, J.; Xiao, Y.; Liu, Z. NFAT5 inhibits invasion and promotes apoptosis in hepatocellular carcinoma associated with osmolality. Neoplasma 2017, 64, 502–510. [Google Scholar] [CrossRef]
- Kuper, C.; Beck, F.X.; Neuhofer, W. NFAT5-mediated expression of S100A4 contributes to proliferation and migration of renal carcinoma cells. Front. Physiol. 2014, 5, 293. [Google Scholar] [CrossRef]
- Leisz, S.; Schulz, K.; Erb, S.; Oefner, P.; Dettmer, K.; Mougiakakos, D.; Wang, E.; Marincola, F.M.; Stehle, F.; Seliger, B. Distinct von Hippel-Lindau gene and hypoxia-regulated alterations in gene and protein expression patterns of renal cell carcinoma and their effects on metabolism. Oncotarget 2015, 6, 11395–11406. [Google Scholar] [CrossRef][Green Version]
- Rinschen, M.M.; Yu, M.J.; Wang, G.; Boja, E.S.; Hoffert, J.D.; Pisitkun, T.; Knepper, M.A. Quantitative phosphoproteomic analysis reveals vasopressin V2-receptor-dependent signaling pathways in renal collecting duct cells. Proc. Natl. Acad. Sci. USA 2010, 107, 3882–3887. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Groß, A.; Chernyakov, D.; Gallwitz, L.; Bornkessel, N.; Edemir, B. Deletion of Von Hippel–Lindau Interferes with Hyper Osmolality Induced Gene Expression and Induces an Unfavorable Gene Expression Pattern. Cancers 2020, 12, 420. https://doi.org/10.3390/cancers12020420
Groß A, Chernyakov D, Gallwitz L, Bornkessel N, Edemir B. Deletion of Von Hippel–Lindau Interferes with Hyper Osmolality Induced Gene Expression and Induces an Unfavorable Gene Expression Pattern. Cancers. 2020; 12(2):420. https://doi.org/10.3390/cancers12020420
Chicago/Turabian StyleGroß, Alexander, Dmitry Chernyakov, Lisa Gallwitz, Nicola Bornkessel, and Bayram Edemir. 2020. "Deletion of Von Hippel–Lindau Interferes with Hyper Osmolality Induced Gene Expression and Induces an Unfavorable Gene Expression Pattern" Cancers 12, no. 2: 420. https://doi.org/10.3390/cancers12020420
APA StyleGroß, A., Chernyakov, D., Gallwitz, L., Bornkessel, N., & Edemir, B. (2020). Deletion of Von Hippel–Lindau Interferes with Hyper Osmolality Induced Gene Expression and Induces an Unfavorable Gene Expression Pattern. Cancers, 12(2), 420. https://doi.org/10.3390/cancers12020420





