1. Introduction
Prostate cancer (PCa) remains one of the leading causes of death among men in Western countries [
1]. Organ-confined PCa can often be cured predominantly either by radical prostatectomy or by primary radiation therapy, however about 15% of patients are diagnosed in a primary metastatic stage [
2]. In addition, it has been demonstrated that up to 50% of patients relapse after primary treatment over time claiming for an androgen deprivation therapy [
3,
4]. However, after a time of about 2–3 years, patients experience resistance to androgen deprivation therapy (metastatic castration-resistant prostate cancer, mCRPC). In the past five years, several new therapeutic options targeting the androgen receptor (AR)-signaling axis have been developed for this patient population, including the AR-inhibiting agent enzalutamide [
5,
6]. Response to these drugs, however, is only temporary, as the majority of treated patients develop drug resistance. Currently, the biological mechanisms of drug resistance are still under investigation.
The cullin-RING ligases (CRLs) comprise the largest class of E3 ubiquitin ligases. As part of the ubiquitin-proteasome system, CRLs mediate the ubiquitination of protein substrates destined for proteasomal degradation and are thus responsible for posttranslational regulation of numerous targets [
7,
8]. CRLs are multi-protein complexes with a common catalytic core consisting of a member of the cullin-family (CUL1-3, 4A, 4B, 5, 7 in human) and a RING (really interesting new gene) protein, also known as RBX1 (RING box protein-1) or ROC1 (regulator of cullins-1) [
9]. Adaptor proteins, which recruit a variety of different receptors, link specific substrates to the catalytic core, while the RING subunit provides a docking site to the ubiquitin conjugating enzyme E2. Besides the regulation of substrate recruitment by the variety of receptor subunits and the required post-translational modifications of the targets, the activity of CRLs is further modulated by the NEDD8/Cand1 cycle [
10]. In a process named neddylation, the NEDD8 protein is covalently bound to the CUL subunit of the enzymatic core by the NEDD8-conjugating enzyme UBC12, resulting in the activation of the CRL. By the intrinsic deneddylating activity of the constitutive photomorphogenesis 9 signalosome (CSN), NEDD8 is removed, leading to the dissociation of adapter protein and substrate-receptor. Deneddylated cullins are subsequently sequestered by the cullin-associated NEDD8-dissociated protein 1 (Cand1) and held in an inactive state. NEDD8 conjugation again causes the dissociation of Cand1, and thus activation of the ligase [
8]. It has been previously shown that suppression of Cand1 results in increased CRL activity
in vitro [
11,
12,
13]. Generally, aberrant regulation of the ubiquitin system is associated with the development and progression of several cancer entities, including urological malignancies [
14,
15]. Interestingly, depending on the CRL activity and the bound substrate-receptor, RING-ligases can have both oncogenic and tumor-suppressive properties [
16,
17,
18].
In the present study, we therefore aimed to elucidate the role of Cand1 in PCa patients’ samples as well as in PCa cell lines. In this context, we not only examined therapy-naive PCa cells, but also included cell lines resistant to the AR-inhibiting agent enzalutamide, as we speculated an involvement of Cand1 in enzalutamide resistance mechanisms.
2. Materials and Methods
2.1. Tissue Microarray and Immunohistochemistry
The use of archived tissue samples for this study was approved by the Ethics Committee of the Medical University Innsbruck (UN3174, AM 3174), informed consent of all patients included in the study is available.
To evaluate differences in Cand1 expression between malignant and benign prostate tissue, we constructed a tissue microarray (TMA) of PCa patients who underwent a radical prostatectomy due to biopsy confirmed localized PCa. In addition, punches of paraffin embedded metastatic PCa cell lines (PC3, DU145, PC3-DR and DU145-DR) were included as control. For each selected case, three cancer tissue cores and three benign cores were punched. The TMA was assembled using a manual tissue arrayer (Beecher Instruments, Sun Prairie, WI, USA). Hematoxilin/Eosin (HE) and p63/alpha-methylacyl-CoA racemase (AMACR) immunohistochemistry (IHC) double staining to control the histological diagnosis and Cand1 IHC were performed on a Discovery-XT staining device (Ventana, Tucson, AZ, USA) using the following antibodies: Cand1 (Cell Signaling Technology, 2316 ZA Leiden, The Netherlands), anti-p63 (Sigma Aldrich, Vienna, Austria), anti-AMACR (Dako, Vienna, Austria). Microscope images were taken with a Zeiss Imager Z2 microscope (Zeiss, Vienna, Austria) equipped with a Pixelink PLB622-CU camera (Canimpex Enterprises Ltd, Halifax, NS, Canada). IHC expression analysis was performed by the uro-pathologist G.S. multiplying the percentage of positive cells with the staining intensity (no staining: 0, weak light: 1, medium: 2, strong: 3).
2.2. Cell Lines and Cell Culture
The human PCa cell lines LNCaP, DU145 and PC3 were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA), whereas LAPC-4 cell line was a generous gift from Dr. A. Cato (University of Karlsruhe, Karlsruhe, Germany). The androgen independent cell subline LNCaP abl was established by Culig et al. by cultivating androgen sensitive LNCaP cells in steroid free medium for 87 passages [
19]. The enzalutamide-resistant cell lines (EnzaR) of LAPC-4 and LNCaP abl were also generated by our group as described before [
20]. The DUCaP cell line as well as the benign prostatic hyperplasia epithelial cell line BPH-1 were a generous gift from Dr. J. Schalken (Radboud University Nijmegen, 6525 XZ Nijmegen, Netherlands), whereas NAF PF179T (hTERT immortalized normal prostate tissue associated fibroblasts), CAF PF179T (hTERT immortalized prostate cancer associated fibroblasts) and EP156T (hTERT immortalized prostate epithelial cells) were established in collaboration with Dr. Varda Rotter (Weizmann Institute, Rehovot, Israel) [
21]. The RWPE-1 cell line established at the Michigan State University was provided by Dr. William Watson (University College Dublin, Ireland) [
22]. The identity of the used cancer cell lines was confirmed by forensic DNA fingerprinting methods using the AmpFlSTR
® SGM Plus
® PCR amplification kit (Applied Biosystems, Brunn am Gebirge, Austria).
LNCaP, DU145, PC3, DUCaP and BPH-1 were grown in Roswell Park Memorial Institute (RPMI) 1640 medium without L-Glutamine (Lonza, Basel, Switzerland) containing 10% fetal bovine serum (FBS; Biowest, Nuaillé, France), 1% penicillin/streptomycin (Lonza, Basel, Switzerland) and 1% GlutaMAX (Gibco, Vienna, Austria). LAPC-4 cells were cultivated in the same medium but additionally supplemented with 1 nM dihydrotestosterone (DHT) (Sigma Aldrich, Vienna, Austria). NAF and CAF were cultivated in Minimal Essential Medium (MEM, Gibco, Vienna, Austria) with Earle´s Salts without L-glutamine supplemented with 10% FBS, 1% penicillin/streptomycin, 1% GlutaMAX, 1 mM sodium pyruvate (100 mM, Lonza, Basel, Switzerland) and 1% NEAA (Lonza, Basel, Switzerland). The epithelial cell line RWPE-1 was grown in Keratinocyte Serum Free Medium (K-SFM, Gibco, Vienna, Austria) without FBS but supplemented with 0.05 mg/mL Bovine Pituitary Extract (BPE) and 5 ng/mL epidermal growth factor (EGF). LNCaP abl were grown in RPMI 1640 without L-Glutamine containing 10% charcoal stripped fetal bovine serum (CS-FBS; PAN Biotech, Aidenbach, Germany), 1% penicillin/streptomycin, 1% GlutaMAX, 1 mM sodium pyruvate, 10 mM HEPES and 2.5 g/L D (+) glucose. EnzaR cell lines were treated with the corresponding medium plus 8 µM enzalutamide (MedChemExpress, Stockholm, Sweden) for LAPC-4 EnzaR and 13 µM enzalutamide for LNCaP abl EnzaR cells. The corresponding amount of ethanol was used for vehicle treated control cells.
2.3. Cand1 Overexpression
Depending on the corresponding PCa cell line, 1.2 × 105 to 2.0 × 105 cells were seeded in 6-well plates to 50%–60% confluence 24 hours before transfection. For overexpression of Cand1, ViaFectTM transfection reagent (Promega, Mannheim, Germany) was used according to the manufacture´s instruction as well as pCMV6-Entry-Cand1 (SC125317, Origene, Hiddenhausen, Germany) overexpression plasmid and pCMV6-Entry (PS100001, Origene, Hiddenhausen, Germany) control plasmid. Then, 72 hours after transfection, cells were used for immunofluorescence staining.
2.4. Cand1 Downregulation
For downregulation of Cand1 in PCa cells, a pool of four targeting siRNAs (On-Targetplus Human Cand1siRNA smart pool, L-015562-01, Dharmacon, Vienna, Austria) was used: GACUUUAGGUUUAUGGCUA (J-015562-09), CGUGCAACAUGUACAACUA (J-015562-10), CAACAAGAACCUACAUACA (J-015562-11), CAUAACAAGCCAUCAUUAA (J-015562-12). A pool of 4 non-targeting siRNAs (ON-Targetplus Non-targeting Control Pool, D-001810-10-20, Dharmacon, Vienna, Austria) served as negative control (siCtrl). Both, siCand1 and siCtrl were resuspended in 1× siRNA Buffer (B-002000-UB-100, Dharmacon, Vienna, Austria). Transfection of targeting or control siRNAs (50 nM) was performed using Lipofectamine2000 transfection reagent (Invitrogen, Lofer, Austria) according to the manufacturer´s instruction. Target gene downregulation was confirmed by qRT-PCR and Western blot analysis. When performing functional experiments (p21 expression levels and SKP Elisa), the cell cycle was synchronized with a thymidine block using 2 mM thymidine (Sigma Aldrich, Vienna, Austria) for 18 hours.
2.5. Western Blot Analysis
Total protein was extracted from cells using RIPA lysis buffer (1% Triton X-100 (Serva, Heidelberg, Germany), 0.5% sodium deoxicholate (Sigma Aldrich, Vienna, Austria), 150 mM sodium chloride (Lactan, Graz, Austria), 50 mM Tris (Sanova, Vienna, Austria)) without sodium dodecyl sulphate (SDS) containing 5 mM sodium fluoride (NaF, Sigma Aldrich, Vienna, Austria), 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma Aldrich, Vienna, Austria), 1% Triton X-100, 1% Phosphatase Inhibitor Cocktail 2 (Sigma Aldrich, Vienna, Austria) and 0.5% Protease Inhibitor Cocktail Set III (Calbiochem, Vienna, Austria). After addition of lysis buffer to the cell pellets, the samples were shaken at 4 °C for 1 hour. Cell debris were then removed by centrifugation (maximum speed, 4 °C,10 minutes), samples were diluted in lysis buffer and 4× NuPAGE LDS Sample Buffer (Invitrogen, Lofer, Austria) and 50 µg protein per lane were loaded on a 4%–12% Bis-Tris protein (Invitrogen, Lofer, Austria) sodium dodecyl sulfate polyacrylamide gel. Electrophoresis (SDS-PAGE) was performed in a XCell SureLock Mini-Cell Electrophoresis System at 150 V using 1× NuPAGE MOPS running buffer (Invitrogen, Lofer, Austria).
Western blot was performed as previously described by our group [
20,
23]. Nitrocellulose membranes were incubated with the primary antibodies overnight at 4 °C. The following primary antibodies were used: Cand1 (1:500, Rabbit monoclonal Antibody (RmAb), Cell Signaling Technology, 2316 ZA Leiden, The Netherlands), PARP p85 Fragment (1:500, Rabbit polyclonal Antibody (RpAb), Promega, Mannheim, Germany), p21 (1:500, Mouse monoclonal Antibody (MmAb), Cell Signaling, 2316 ZA Leiden, The Netherlands) and GAPDH (1:50000, Mouse monoclonal Antibody (MmAb), Millipore, Vienna, Austria). Afterwards the membranes were incubated with infrared fluorescent dye labeled secondary antibodies (LiCor Biosciences, Bad Homburg, Germany) for 1 hour at room temperature and scanned using the Odyssey infrared imaging system (LiCor Biosciences, Bad Homburg, Germany). Densitometric analysis was performed using Odyssey application software (LiCor Biosciences, Bad Homburg, Germany).
2.6. Bradford Assay
The protein concentration was determined using the Bradford assay. For this, a standard curve was generated using bovine serum albumin (BSA, Sigma Aldrich, Vienna, Austria) with concentrations ranging from 0 to 40 µg/mL. The samples were diluted in phosphate buffered saline (PBS) and 1 mL of 1× Bradford reagent was added to each cuvette. The absorbance was measured at 595 nm using the Spectrophotometer U 2000 (Hitachi, Metrohm Inula GmbH, Vienna, Austria) and the protein concentrations were calculated based on the standard curve.
2.7. Caspase-Glo 3/7 Assay
Depending on the cell line, 7.5 × 104 to 3.0 × 105 cells were seeded in 6 well plates, transfected with 50 nM siCand1 or siCtrl and harvested after 96 hours of incubation. Cell lysis and protein extraction were performed as described above for SDS-PAGE samples. The measurement was carried out in black 96-well plates using lysis buffer as blank. 20 µL of PBS and 5 µL of well vortexed sample were incubated with 25 µL of Caspase-Glo 3/7 Substrate (Promega, Mannheim, Germany) on a horizontal shaker for 5 minutes and other 30 minutes at room temperature in the dark. Luminescence was measured using the Chameleon V Multitechnology Plate Reader (HVD Life Sciences, Vienna, Austria) and the results (relative light units, RLU) were normalized to the inserted amount of protein determined via Bradford assay.
2.8. Cell Viability Assay (WST-1)
Cell viability upon downregulation of Cand1 was assessed using the Cell Proliferation Reagent WST-1 (Roche Diagnostics GmbH, Vienna, Austria). Depending on the cell line, 3000–5000 cells were grown in five replicates in 96-well plates and transfected the next day as indicated. After 96 hours, WST-1 reagent was added 1:10 to the cells in each well and further incubated at 37 °C. The absorbance was measured after 2 hours at 420–480 nm using the Chameleon V Multitechnology Plate Reader (HVD Life Sciences, Vienna, Austria).
2.9. Proliferation Assay (3H-Thymidine Incorporation Assay)
Cell proliferation upon downregulation of Cand1 was determined by the 3H-thymidine incorporation assay. Thereby, 24 hours before transfection with 50 nM siRNA, depending on the cell line, 3000–5000 cells were seeded in five replicates onto 96-well plates. Then, 72 hours after transfection, 1 µCi of 3H-thymidine (Hanke Laboratory Products, Vienna, Austria) was added to each well and the cells were further incubated at 37 °C for 24 hours. After freezing the plate at −20 °C, cell DNA was harvested on 96-well filter plates (PerkinElmer Life Sciences Austria, Vienna, Austria) and 50 µL of scintillation fluid (PerkinElmer Life Sciences Austria, Vienna, Austria) were added. Incorporated radioactivity was quantified using the Chameleon V Multitechnology Plate Reader.
2.10. SKP1 and SKP2 ELISA
Cell pellets were re-suspended in ice-cold PBS and cells lysis was performed by ultra-sonification. Cell lysates were then centrifuged at 1500× g, 10 minutes at 4 °C and supernatants were used in the ELISA for SKP1 and SKP2, which were performed according to the manual of the manufactory instructions (Abbexa, Cambridge, UK; SKP1: Catalog No.: abx153099, SKP2: Catalog No.: abx153100). The SKP1 and SKP2 concentrations of the samples were interpolated from each standard curve and the obtained results were normalized to the amount of protein determined via Bradford Assay.
2.11. Immunofluorescence
Depending on the cell line, 1.2 × 105 to 2.0 × 105 cells were grown on sterile glass coverslips in 6-well plates for 24 hours and transfected with 50 nM siCtrl, 50 nM siCand1 or 4 µg pCMV6-Entry-Cand1 as described above. After 72 hours of incubation, cells were washed with PBS and fixed in 4% paraformaldehyde (PFA) for 10 minutes at room temperature. After a second washing procedure, cells were permeabilized with 0.2% Triton X-100 in PBS with 1% BSA for 5 minutes on ice. Blocking was carried out with 1% BSA in PBS for 30 minutes at room temperature, exchanging the solution three times. The cells on the coverslips were incubated for 1 hour at 37 °C in a wet chamber with 20 µL of primary antibody diluted in blocking solution. The following antibodies were used: Cand1 (1:50, RmAb, Cell Signaling Technology, 2316 ZA Leiden, The Netherlands), Rabbit IgG Isotype Control (1:250, RmAb, Cell Signaling Technology, 2316 ZA Leiden, The Netherlands) and Cytokeratin-8/KRTB (1:100, Chicken polyclonal Antibody (CpAb), Sigma Aldrich, Vienna, Austria) for isotype control. After incubation, cells were rinsed in PBS four times, dried on a paper towel and probed for 1 hour at 37 °C in a wet chamber in the dark with 100 µL of respective secondary antibody (goat anti rabbit IgG (Alexa Fluor 555), goat anti-chicken IgG (Alexa Fluor 488), both Invitrogen, Lofer, Austria) diluted 1:500 in blocking solution. Again, cells were then rinsed four times in PBS and twice in aqua dest., dried on a paper towel and embedded using VECTASHIELD Antifade Mounting Medium containing DAPI (Linaris (VectorLabs), Szabo Scandic, Vienna, Austria), which stains DNA blue. Finally, cells were visualized by fluorescent microscopy using the Axio Imager Z2 microscope (Zeiss, Vienna, Austria) and the TissueFaxs Cell Analysis System (TissueGnostics, Vienna, Austria).
2.12. Next Generation Sequencing of Cell Lines
Briefly, 200 ng of genomic DNA per sample was sheared using a Covaris M220 instrument (Covaris, Brighton, United Kingdom) resulting in an average fragment size of 183 base pairs as determined by Agilent Bioanalyzer 2100 (Agilent Technologies Austria, Vienna, Austria), analysis. Whole exome sequencing libraries were constructed using Agilent SureSelectXT Human All ExonV6+COSMIC kit (#5190-9307, Agilent Technoligies, Vienna, Austria) and sequenced paired-end (2×125 base pairs) on a HiSeq 2500 (Illumina, San Diego, U.S.A.) instrument with an average yield of 2 × 98.7 million reads per sample. Mutation calling was performed using the GATK best practices pipeline. Specifically, raw sequencing reads were aligned to the human genome (hg19) using Burrows-Wheeler Aligner and then processed using Picard tools (version 2.1.1) and GATK (version 3.6); mutations were called with the HaplotypeCaller.
2.13. Statistical Analyses
Statistical analyses were performed using GraphPad Prism 5.0 and R Platform for Statistical Computing. For cell culture experiments, data are shown as mean with standard error of mean (SEM) from at least three independent experiments. For patient studies, data are presented as boxplots (box denoting for median with interquartile range (IQR) and whiskers depicting median ± 150% IQR) with points coding for single patient samples layered on them. Statistical significance for differences between two independent groups were analyzed with two tailed T test or Mann–Whitney U test for normally and non-normally distributed data, respectively. Statistical significance for more than two independent groups was assessed, dependent on number of factors, with one- or two-way ANOVA. Significances are encoded as follows: *p < 0.05; **p < 0.01; ***p < 0.001.
2.14. Survival and Differential Gene Expression
For survival and differential gene expression, published studies available from Gene Expression Omnibus (GEO) were re-analyzed: GSE16560 (
n = 280), GSE70768 (
n = 110), GSE70769 (
n = 91), GSE40272 (
n = 87) and TCGA (
n = 419) [
24,
25,
26,
27]. For characteristics of the study cohorts, see
Supplementary Table S1. In brief, clinical information such as survival or Gleason score as well as normalized whole-genome and
Cand1 gene expression data provided by the study authors were extracted. In each study, patients were stratified according to tumor tissue Cand1 expression into Cand1
lo and Cand1
hi expressers. The stratification threshold was iteratively calculated with an in-house-written algorithm maximizing differences in survival tested with Mantel–Haenszel test. For analysis of survival differences between Cand1
lo and Cand1
hi individuals, Kaplan-Meier plots were generated and statistical significance for survival differences assessed with Mantel–Haenszel and Wilcoxon test and univariate Cox proportional hazard modeling (R packages survival and survminer). Identification of differentially expressed genes was performed with two-tailed T tests, p value correction was done with the Benjamini–Hochberg method (False Discovery Rate, p
FDR). Genes with pFDR < 0.05 and fold-regulation between Cand1
lo and Cand1
hi samples exceeding 1.25 were deemed significantly regulated. Genes significantly up- and downregulated in at least two out of four analyzed studies were further investigated for Gene Ontology (GO) term enrichment (biological process and molecular function GOs) with DAVID [
28,
29].
4. Discussion
The deregulation of CRL components such as of substrate-receptors or Cand1 have been described in various cancer entities, however there is conflicting evidence concerning the impact of Cand1 in prostatic malignancies. For example, Rulina et al. previously showed that the knockdown of Cand1 induces apoptosis in PCa cell lines, while Murata et al. provided evidence that Cand1 downregulation enhances cell growth in LNCaP PCa cells [
35,
36]. Therefore, the present study elucidated Cand1 in PCa using not only a preclinical model, but also patient samples to better clarify its clinical impact.
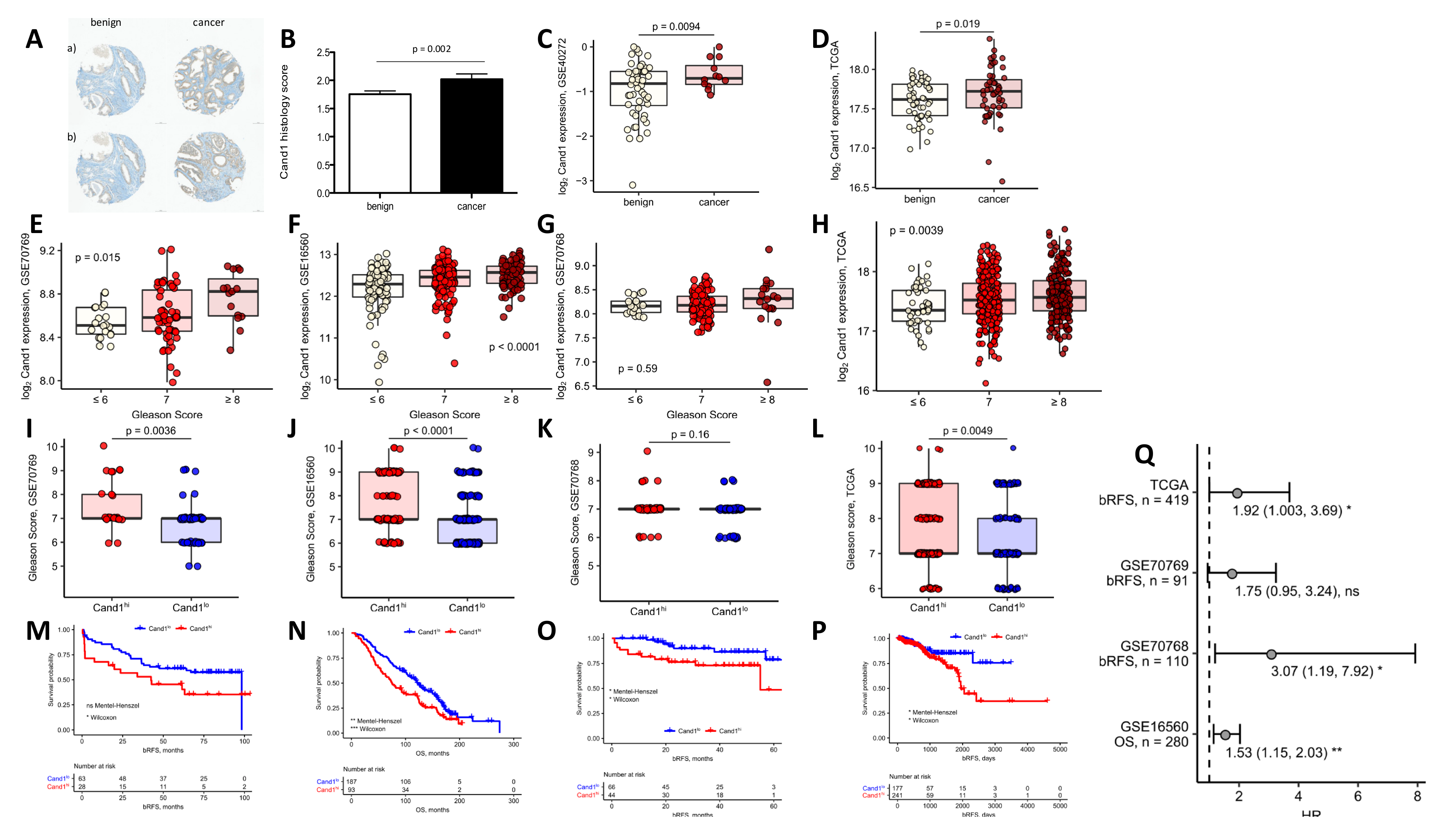
Using a TMA of patients with localized PCa, we found that Cand1 is significantly upregulated in PCa tissue compared to benign controls. These findings are in line with a previous small study [
37] as well as two publicly available datasets (GSE40272 and TGCA). In addition, we found that high Cand1 expression was associated with more aggressive PCa in three out of four GEO datasets. The next step was to investigate the potential impact of Cand1 in tumor recurrence and OS. From our patients, who donated samples for IHC analyses, only a small number of patients relapsed yet and fewer are deceased. Therefore, we relied on publicly available datasets to investigate this aspect. Thereby, data revealed that high expression of Cand1 was associated with poor prognosis (GSE 70769, GSE 70768, TCGA) and OS rates (GSE 16560). To summarize our clinical findings: for the first time we could show that in patient samples an upregulated expression of Cand1 yields a more aggressive PCa phenotype.
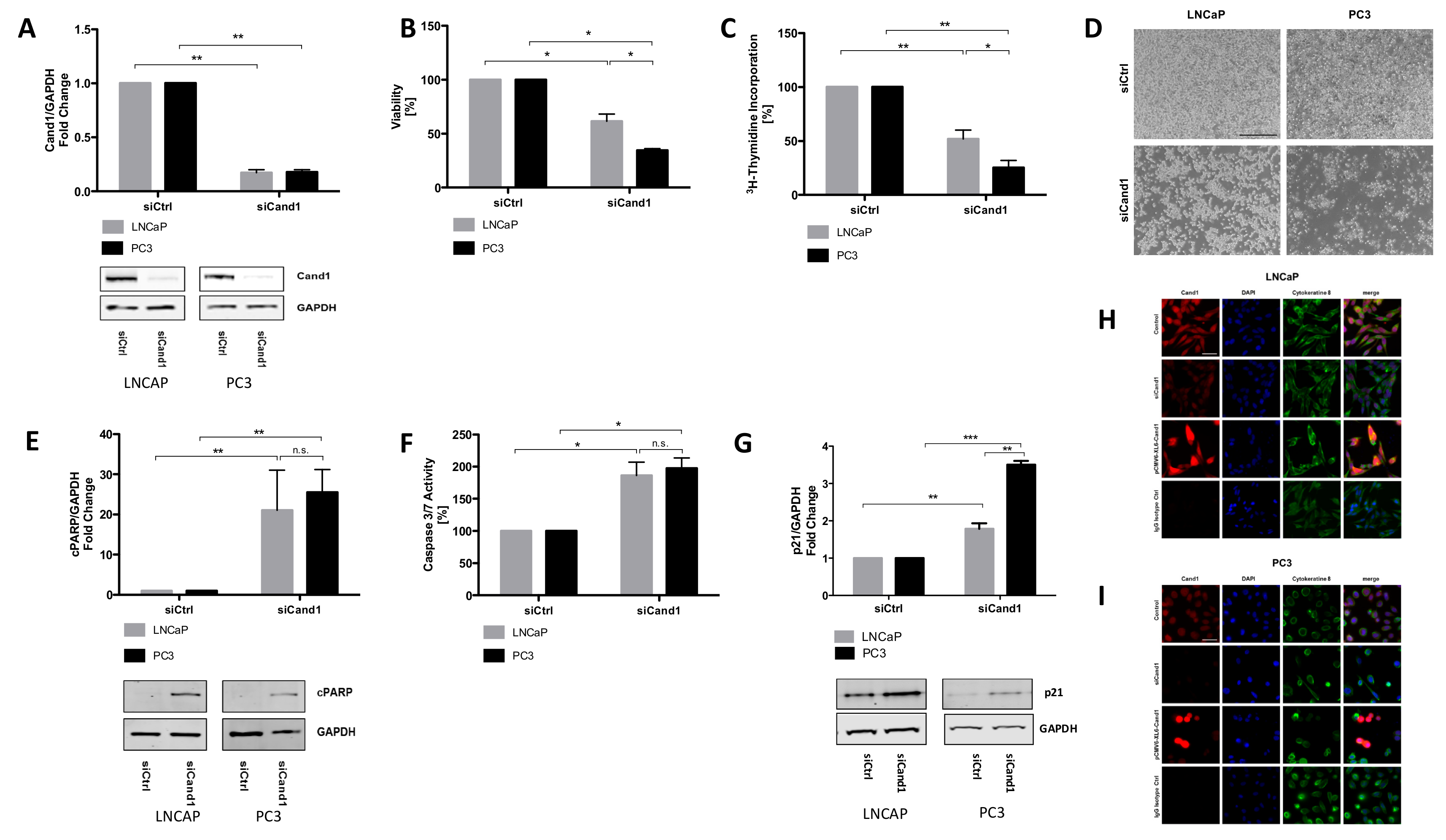
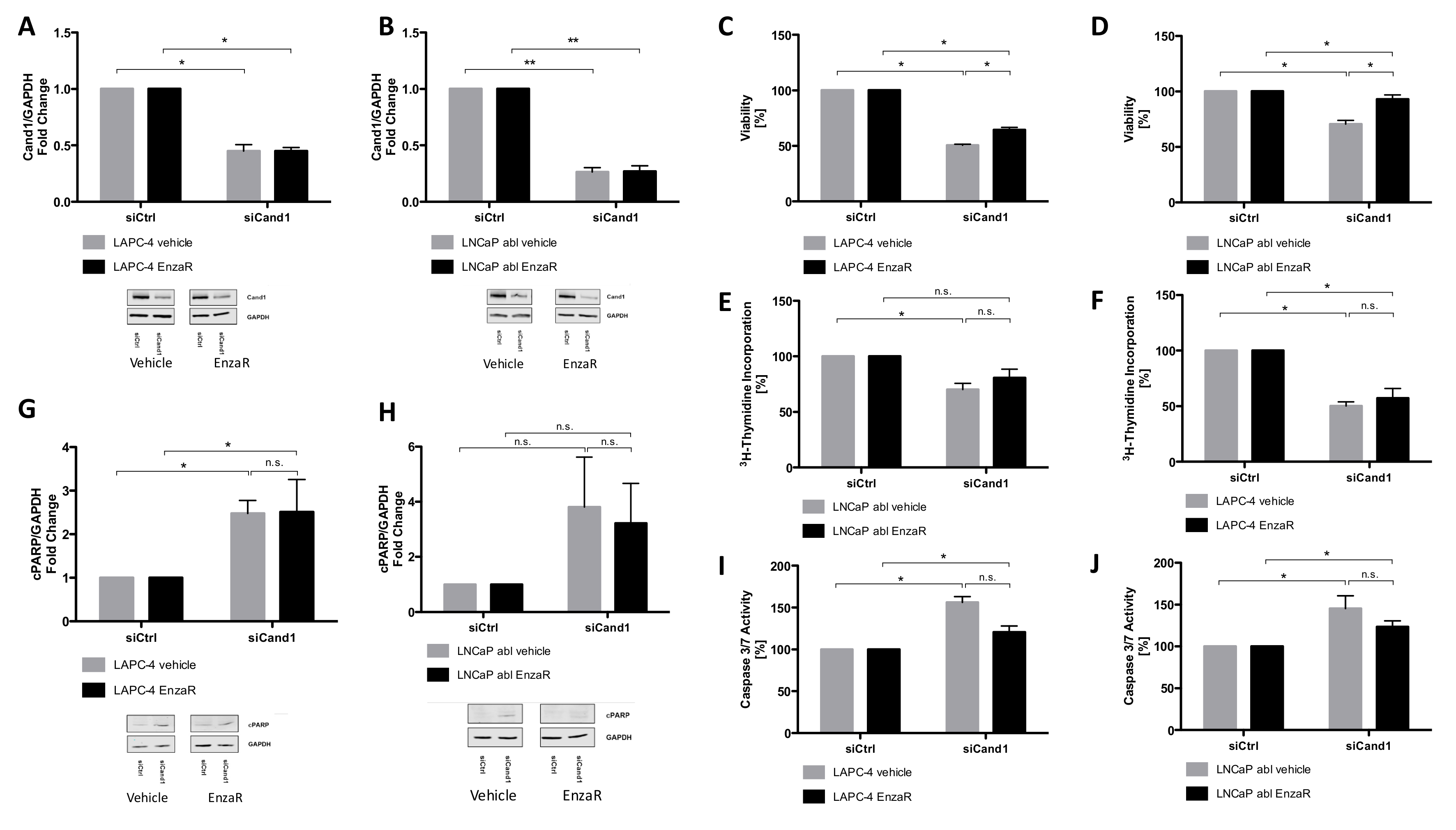
To understand the biological mechanisms of Cand1 expression in PCa, we modulated its expression in metastatic PCa cell lines and analyzed the resulting functional effects. Briefly, we observed that downregulation of Cand1 caused a significant decrease in cell viability and proliferation in both AR positive LNCaP and AR negative PC3 cells. Further functional experiments and investigations of crucial targets both in vitro but also in vivo of Cand1 in PCa would be highly interesting to further strengthen the findings of this study.
Our findings contrast the data of Murata et al., showing that Cand1 knockdown using miR-148a promotes proliferation of LNCaP cells [
36]. However, according to their published results, Cand1 knockdown in LNCaP cells, which was the only cell line used in this study, was quite moderate, compromising their conclusions. Generally, miRNA´s as well as siRNA´s can result in unspecific effects as the results can be biased by off-target effects. In the present study, this aspect was taken into account using an siRNA pool consisting of four targeting siRNA´s (On-Targetplus Human Cand1 siRNA smart pool, Dharmacon) to reduce off-target effects while still providing guaranteed gene silencing of gene targets. Moreover, one has to consider that the growth promoting effects of miR-148a overexpression described by Murata and colleagues may not only involve Cand1, but conceivably, other signaling pathways like PTEN-inhibited PI-3K/AKT (data not shown), also known to have pleotropic effects in cancer [
38,
39].
Another argument speaking in favor of the model presented in our work is that low Cand1 expression in primary prostate carcinoma tissue could be linked to better patient survival in four out of five investigated study cohorts. Besides the findings of Rulina that knockdown of Cand1 induces apoptosis in PCa, our findings of reduced cell proliferation due to Cand1 knockdown are also in line with a recent study in liver cancer where Che et al. showed that Cand1 was overexpressed in cancerous issues compared to the corresponding adjacent benign ones and that high expression of Cand1 was associated with poor OS [
35,
40].
Besides impaired cell viability and proliferation, we were able to show that downregulation of Cand1 resulted in significantly increased apoptosis, reflected by increased caspase 3/7 activity and elevated levels of cPARP in LNCaP and PC3 cells. Consistent with this finding, Rulina et al. demonstrated that Cand1 knockdown induces apoptosis in PCa cell lines [
35]. Chua and colleagues hypothesized that Cand1 differentially regulates CRLs depending on their cellular localization, as they found Cand1 in human embryonic kidney cells (HEK293) to be predominantly present in the cytoplasm [
41].
In the present study, we provide evidence using immunofluorescence staining and IHC analyses that Cand1 is localized in both, nucleoplasm and cytosol of LNCaP and PC3 cells and PCa tissue areas, respectively. Moreover, our findings are supported by validated data from the Human Protein Atlas
As CRLs target a variety of cell cycle regulatory proteins including p21, p27 and cyclin E, we measured p21protein levels in PCa cell lines upon Cand1 downregulation and found as expected a significant upregulation of p21. However, in contrast to Dubiel et al., who showed that Cand1 knockdown increases the level of the FBP SKP2 in cervical cancer cells, we did not find any alterations of SKP1 or SKP2 upon Cand1 downregulation [
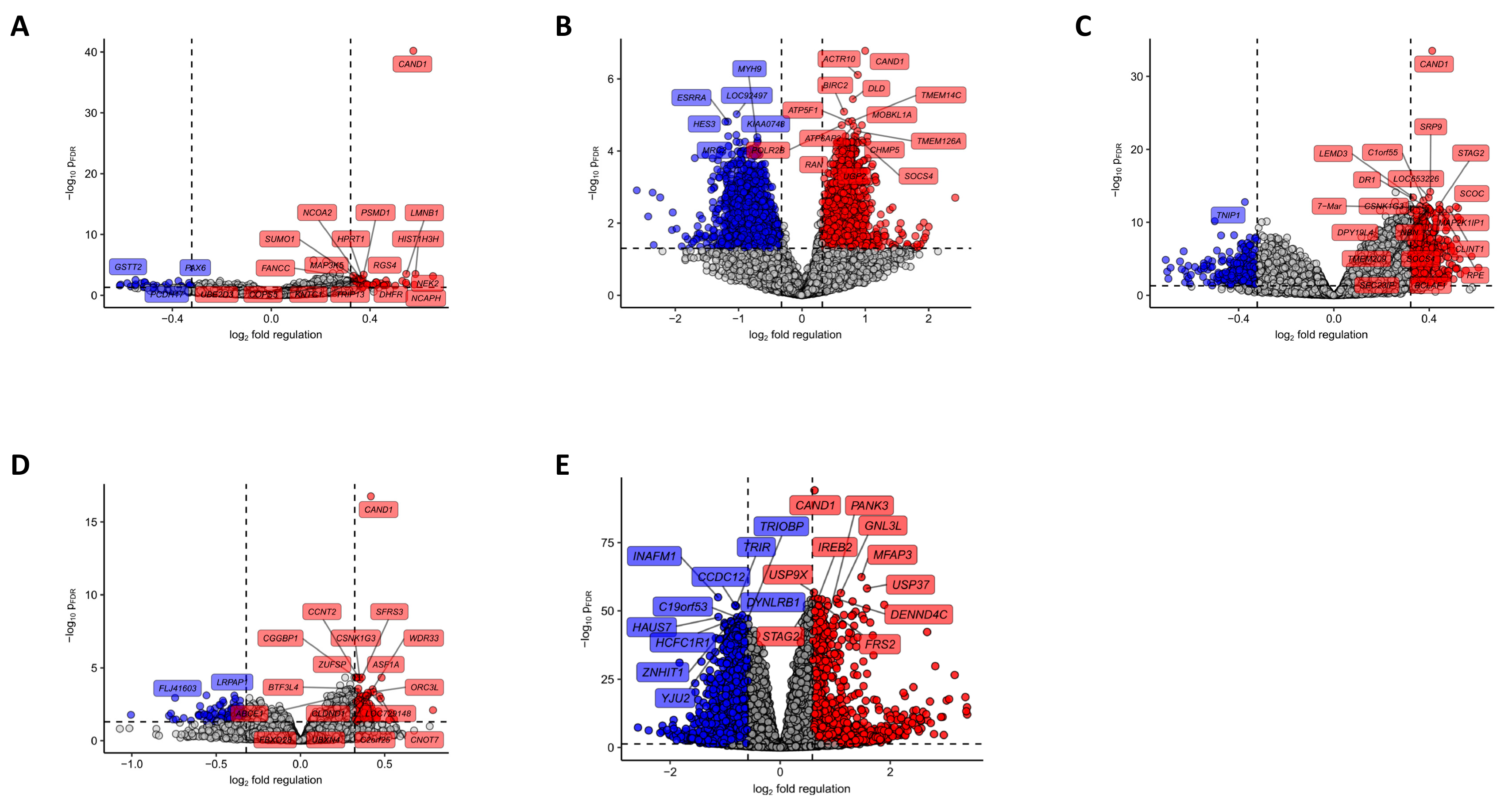
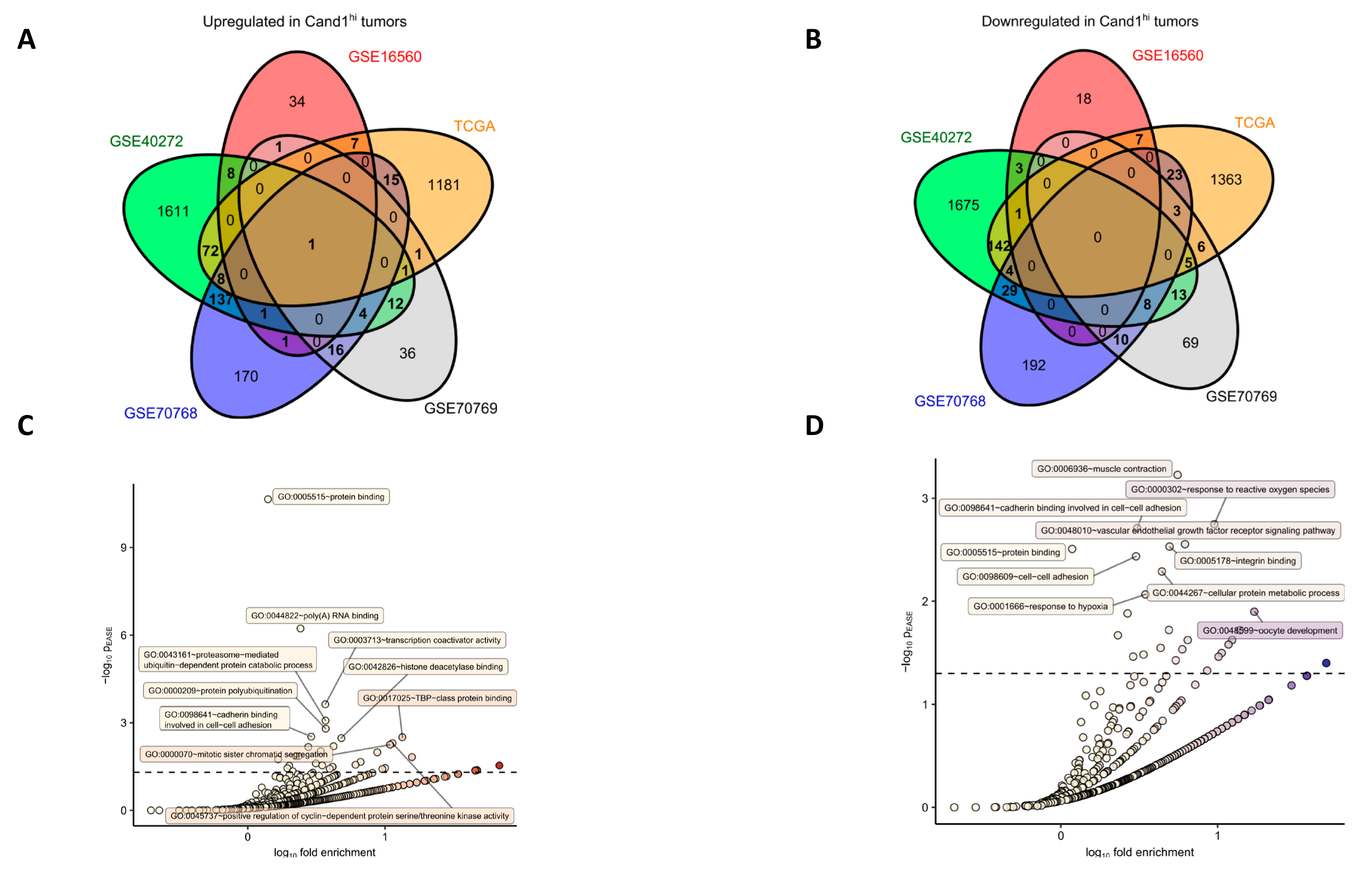
42]. Using bioinformatics tools, we detected genes significantly upregulated in Cand1
hi tumors like genes coding for proteins linked to mRNA turnover, protein polyubiquitination, and proteasomal degradation as well as positive regulation of the cell cycle, fitting to anti-proliferative effects of the Cand1 knockdown in cell culture experiments. Cand1 is a central modulator/regulator in protein ubiquitinylation and degradation and unlikely to directly regulate gene transcription. We therefore expect that the different gene expressions associated with Cand1
Hi/Cand1
Lo is an indirect effect.
In this study, we used AR positive (LNCaP) and AR negative (PC3) cell lines to address the issue if the effect of Cand1 is in regard of the androgen receptor. The results obtained in our study clearly demonstrate that Cand1 acts independently of the AR as we observed that in AR negative PC3 cells cell proliferation and cell viability were affected to a greater extent by the downregulation of Cand1 compared to AR positive LNCaP cells. Furthermore, all pathway analyses conducted in our study did not identify the AR-pathway to be involved in Cand1 signalling and therefore we conclude that Cand1 does not influence the AR.
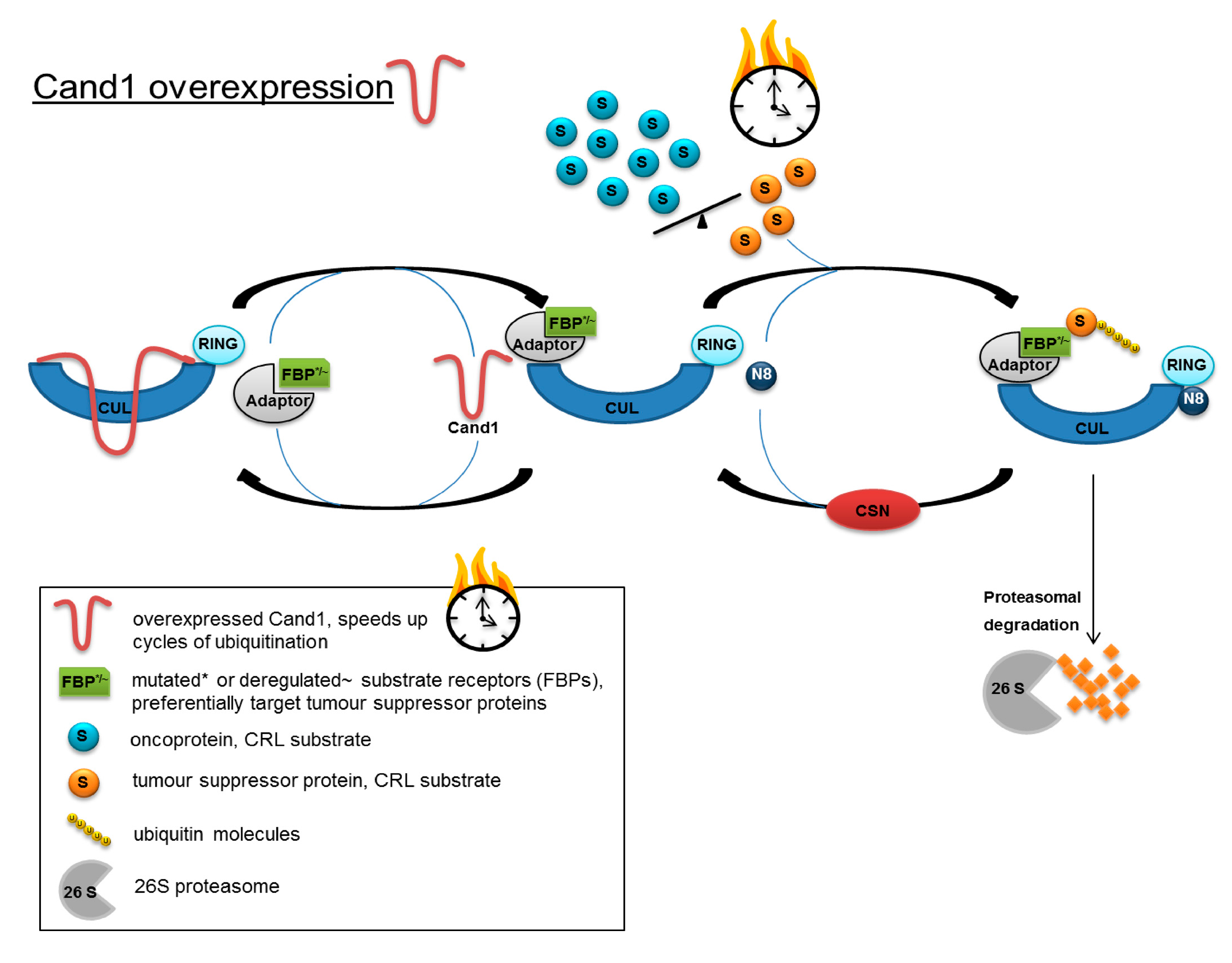
When integrating the results obtained in the present study into the model of CRL control proposed by Schmidt et al. [
43], Cand1 may adopt the role of a time-limiting factor in the two regulatory cycles of Cand1 and CSN: intracellular availability of Cand1 combined with a deregulated balance or mutation-dependent change in binding affinity of the different substrate-receptors (FBPs in SCF) might influence the fate of a prostatic cell. In fact, overexpression and deletions of FBPs as well as point mutations have been detected in cancer (reviewed in [
30]). High Cand1 expression could accelerate the turnover of CRLs, thus advancing ubiquitination and degradation of tumor suppressor proteins, which are preferentially targeted by the deregulated/mutated FBPs (
Figure 6). By lowering the level of Cand1, proteasomal degradation would be slowed down, resulting in an accumulation of tumor suppressor proteins and the induction of apoptosis.
Enzalutamide is an AR inhibitor approved for patients with non-metastatic hormone and metastatic castration-resistant PCa. In addition, it recently has been FDA approved in patients with hormone-naïve metastatic PCa due to a positive phase III study where enzalutamide plus ADT has shown to increase OS and progression free survival (PFS) compared to ADT plus standard androgen ablation [
44]. Generally, enzalutamide binds competitively to the AR, inhibits nuclear translocation, and impairs binding of the receptor to the DNA [
45]. Response to this drug, however, is only temporary, as the majority of treated patients develop drug resistance after 9–15 months. Thus, the identification of resistance mechanisms to the drug is of important need.
Interestingly, DNA sequencing performed on our recently generated EnzaR PCa cells revealed Cand1 to be commonly mutated compared to corresponding parental cell lines. The identified mutations were all single nucleotide missense mutations not present in the parental cell lines associated with an amino acid substitution, however only with a small impact on the 3D structure and a low level of the mutant alleles of around 15%. To assess a potential functional involvement of Cand1 in the development of drug resistance, we tested the effect of Cand1 knock-down in the EnzaR sublines of LAPC-4 and LNCaP abl. Thereby, we could show that Cand1 knockdown negatively affected cell viability and proliferation and induced apoptosis in both vehicle-treated control cells as well as EnzaR cells. Hence, we conclude that the mutations found in Cand1 of EnzaR cells are not likely to play a major role in conferring resistance to the drug.