Dephosphorylation of YB-1 is Required for Nuclear Localisation During G2 Phase of the Cell Cycle
Abstract
1. Introduction
2. Results
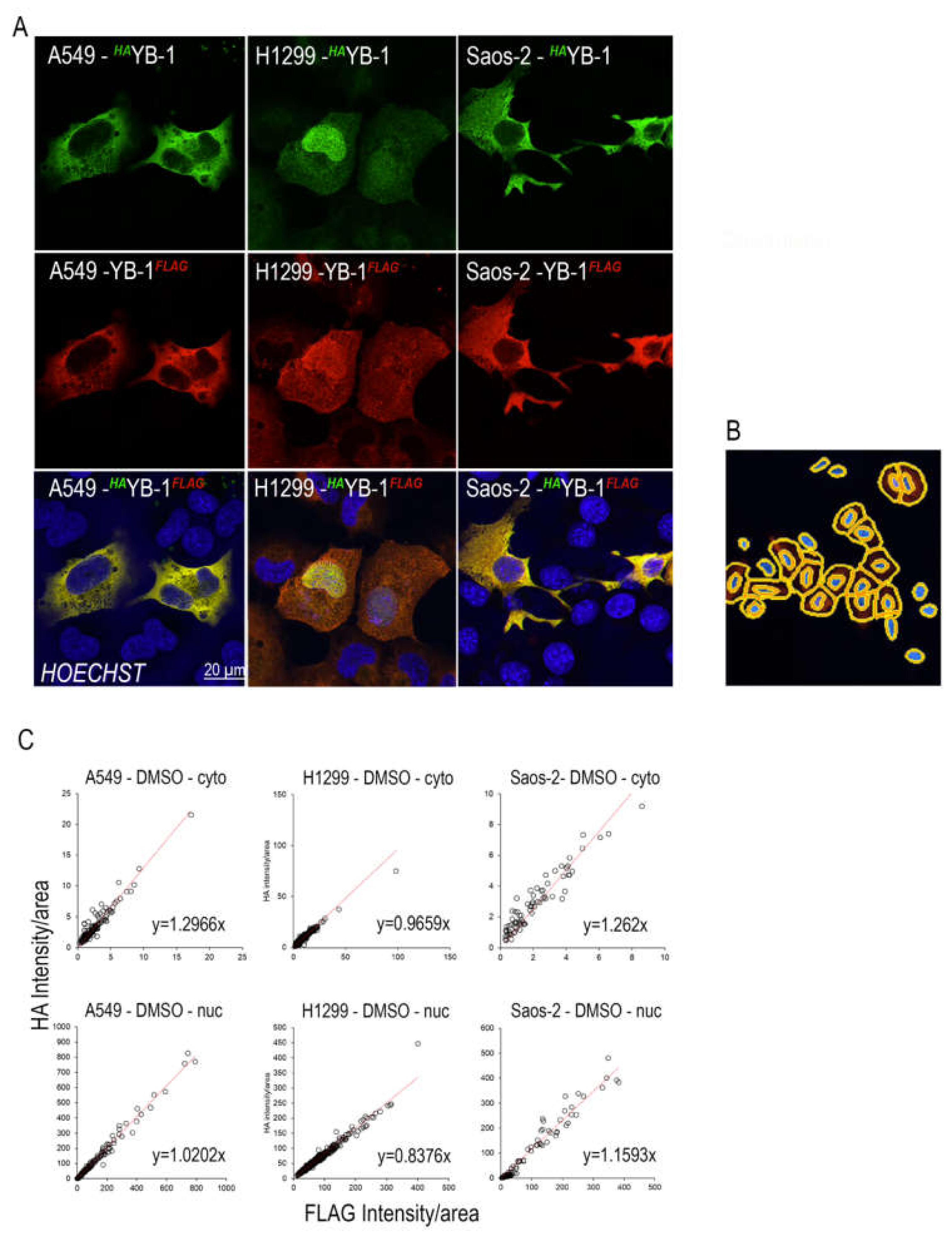
2.1. Full Length YB-1 is Present in Both Nuclear and Cytoplasmic Compartments
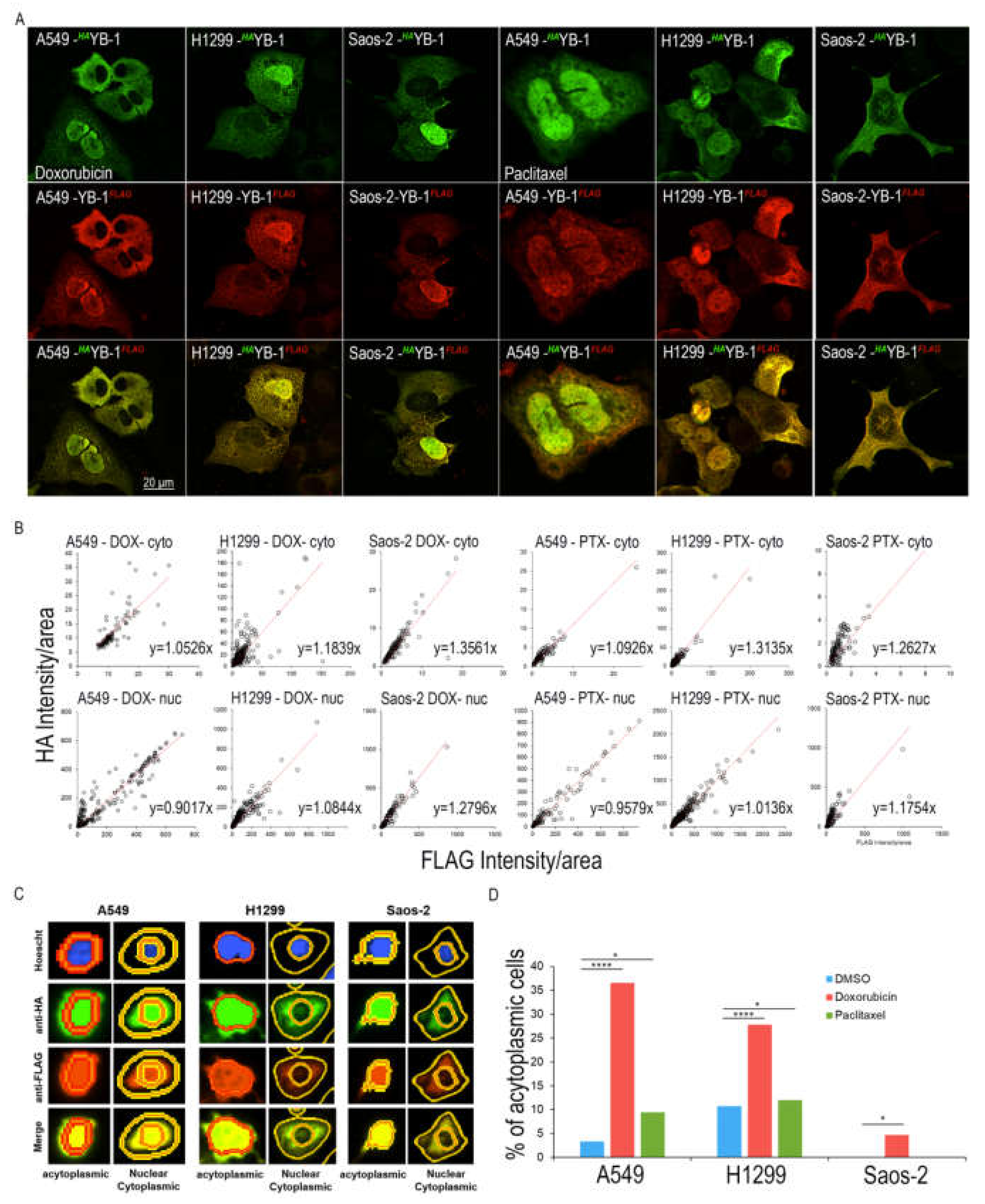
2.2. Specific Proteosomal Cleavage of YB-1 does not Occur in Response to Treatment with Doxorubicin or Paclitaxel
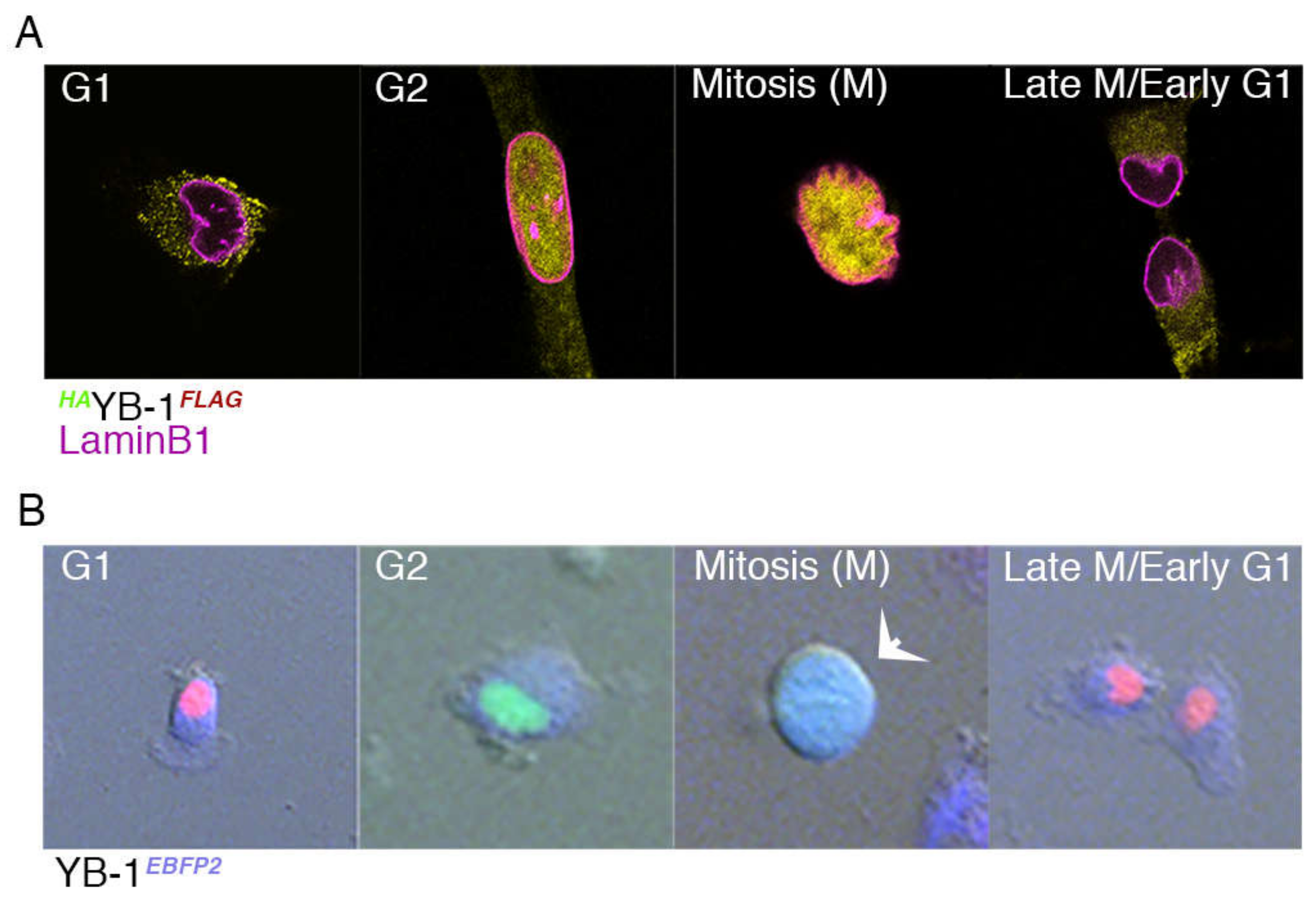
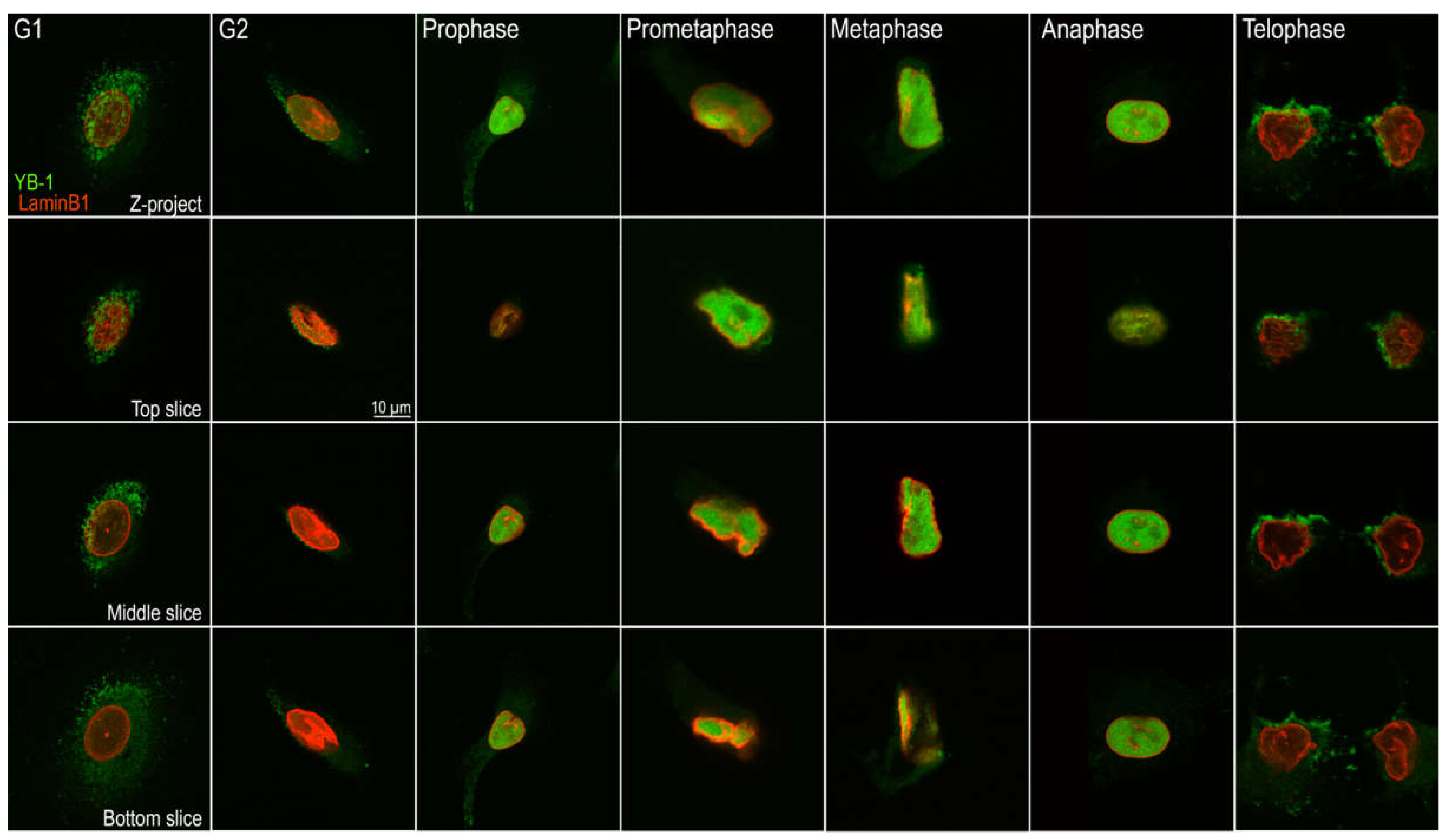
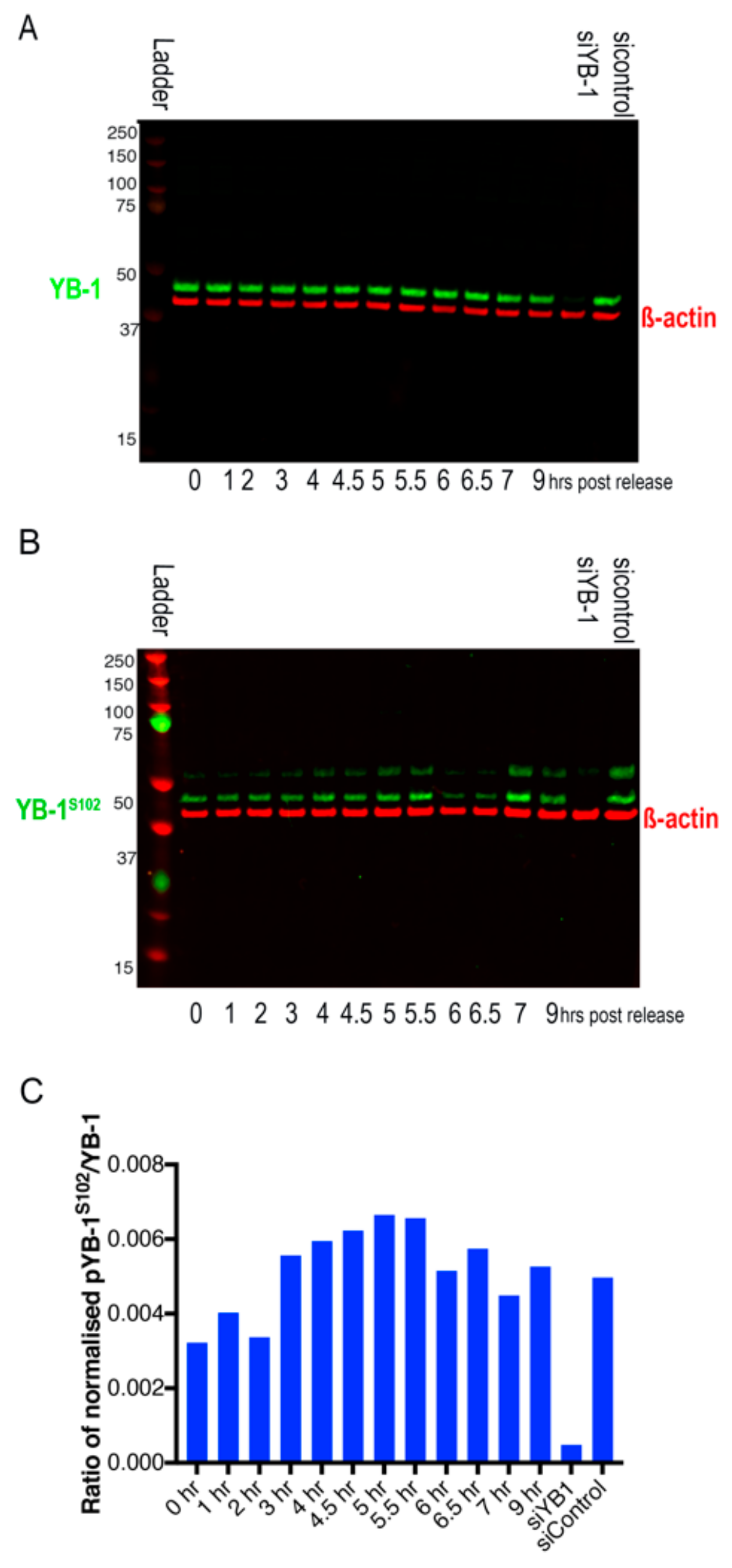
2.3. Nuclear Localisation of YB-1 Occurs during Late G2 Phase of the Cell Cycle
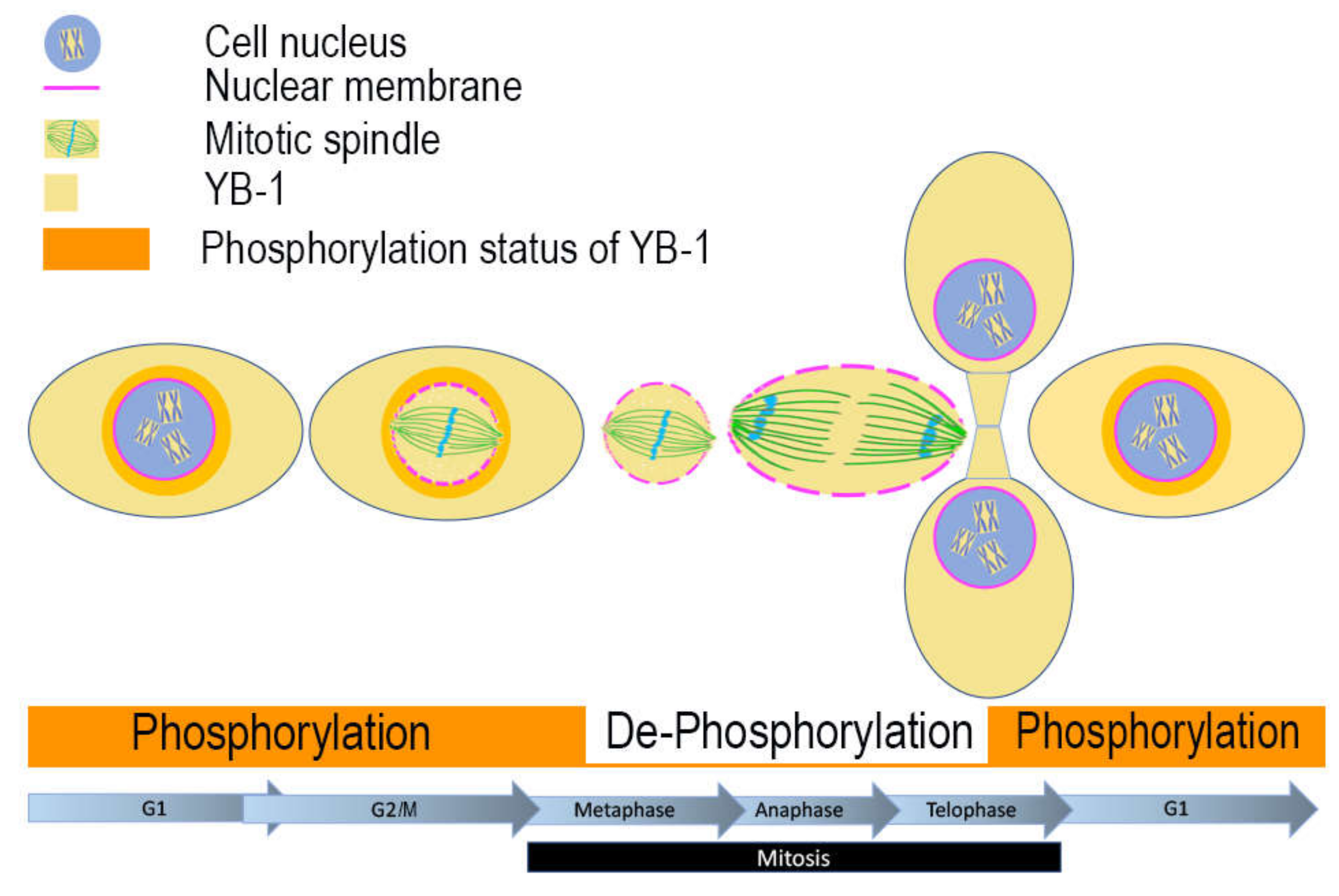
2.4. Nuclear Localisation of YB-1 is Dependent on Phosphorylation Status
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Generation of HAYB-1 FLAG and FLAGYB-1HA Expression Constructs
4.3. Generation of YB-1EBFP2 Expression Constructs
4.4. Generation of A549 FUCCI Cell Line
4.5. Plasmid Transfection and Drug Treatment
4.6. Live Imaging and Processing of Time-Lapse Data
4.7. Cell Fractionation and Western Blotting
4.8. Cell Synchronization and IF Labelling
4.9. Cell Proliferation Assay
4.10. Subcellular Fractionation of Cultured Cells for LC-MS/MS
4.11. Immunopurification of YB-1 and Mass Spectrometry
4.12. siRNA Transfection
4.13. Modelling the Structure of YB-1 Protein
4.14. MD Simulations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gessner, C.; Woischwill, C.; Schumacher, A.; Liebers, U.; Kuhn, H.; Stiehl, P.; Jurchott, K.; Royer, H.D.; Witt, C.; Wolff, G. Nuclear YB-1 expression as a negative prognostic marker in nonsmall cell lung cancer. Eur. Respir. J. 2004, 23, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Bonafe, P.; Fedoruk, M.N.; Whitmore, T.G.; Akbari, M.; Ralph, J.L.; Ettinger, S.; Gleave, M.E.; Nelson, C.C. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. The Prostate 2004, 59, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Janz, M.; Harbeck, N.; Dettmar, P.; Berger, U.; Schmidt, A.; Jurchott, K.; Schmitt, M.; Royer, H.D. Y-box factor YB-1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI-1. Int. J. Cancer 2002, 97, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Sakamoto, A.; Ohga, N.T.; Uchiumi, T.; Kohno, K.; Tsuneyoshi, M.; Kuwano, M.; Iwamoto, Y. Nuclear expression of YB-1 protein correlates with P-glycoprotein expression in human osteosarcoma. Clin. Cancer Res. 1998, 4, 2273–2277. [Google Scholar]
- Raffetseder, U.; Frye, B.; Rauen, T.; Jurchott, K.; Royer, H.D.; Jansen, P.L.; Mertens, P.R. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J. Biol. Chem. 2003, 278, 18241–18248. [Google Scholar] [CrossRef]
- Shibahara, K.; Sugio, K.; Osaki, T.; Uchiumi, T.; Maehara, Y.; Kohno, K.; Yasumoto, K.; Sugimachi, K.; Kuwano, M. Nuclear expression of the Y-box binding protein, YB-1, as a novel marker of disease progression in non-small cell lung cancer. Clin. Cancer Res. 2001, 7, 3151–3155. [Google Scholar]
- Sorokin, A.V.; Selyutina, A.A.; Skabkin, M.A.; Guryanov, S.G.; Nazimov, I.V.; Richard, C.; Th'ng, J.; Yau, J.; Sorensen, P.H.; Ovchinnikov, L.P.; et al. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 2005, 24, 3602–3612. [Google Scholar] [CrossRef]
- Stenina, O.I.; Poptic, E.J.; DiCorleto, P.E. Thrombin activates a Y box-binding protein (DNA-binding protein B) in endothelial cells. J. Clin. Investig. 2000, 106, 579–587. [Google Scholar] [CrossRef]
- Van Roeyen, C.R.; Scurt, F.G.; Brandt, S.; Kuhl, V.A.; Martinkus, S.; Djudjaj, S.; Raffetseder, U.; Royer, H.D.; Stefanidis, I.; Dunn, S.E.; et al. Cold shock Y-box protein-1 proteolysis autoregulates its transcriptional activities. CCS 2013, 11, 63. [Google Scholar] [CrossRef]
- Sakura, H.; Maekawa, T.; Imamoto, F.; Yasuda, K.; Ishii, S. Two human genes isolated by a novel method encode DNA-binding proteins containing a common region of homology. Gene 1988, 73, 499–507. [Google Scholar]
- Berquin, I.M.; Pang, B.; Dziubinski, M.L.; Scott, L.M.; Chen, Y.Q.; Nolan, G.P.; Ethier, S.P. Y-box-binding protein 1 confers EGF independence to human mammary epithelial cells. Oncogene 2005, 24, 3177–3186. [Google Scholar] [CrossRef] [PubMed]
- Lasham, A.; Moloney, S.; Hale, T.; Homer, C.; Zhang, Y.F.; Murison, J.G.; Braithwaite, A.W.; Watson, J. The Y-box-binding protein, YB1, is a potential negative regulator of the p53 tumor suppressor. J. Biol. Chem. 2003, 278, 35516–35523. [Google Scholar] [CrossRef]
- Lasham, A.; Lindridge, E.; Rudert, F.; Onrust, R.; Watson, J. Regulation of the human Fas promoter by YB-1, Puralpha and AP-1 transcription factors. Gene 2000, 252, 1–13. [Google Scholar] [CrossRef]
- Jurchott, K.; Bergmann, S.; Stein, U.; Walther, W.; Janz, M.; Manni, I.; Piaggio, G.; Fietze, E.; Dietel, M.; Royer, H.D. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J. Biol. Chem. 2003, 278, 27988–27996. [Google Scholar] [CrossRef]
- Kotake, Y.; Arikawa, N.; Tahara, K.; Maru, H.; Naemura, M. Y-box Binding Protein 1 Is Involved in Regulating the G2/M Phase of the Cell Cycle. Anticancer Res. 2017, 37, 1603–1608. [Google Scholar] [CrossRef]
- Guay, D.; Gaudreault, I.; Massip, L.; Lebel, M. Formation of a nuclear complex containing the p53 tumor suppressor, YB-1, and the Werner syndrome gene product in cells treated with UV light. Int. J. Biochem. Cell Biol. 2006, 38, 1300–1313. [Google Scholar] [CrossRef]
- Ohga, T.; Uchiumi, T.; Makino, Y.; Koike, K.; Wada, M.; Kuwano, M.; Kohno, K. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J. Biol. Chem. 1998, 273, 5997–6000. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Homer, C.; Edwards, S.J.; Hananeia, L.; Lasham, A.; Royds, J.; Sheard, P.; Braithwaite, A.W. Nuclear localization of Y-box factor YB1 requires wild-type p53. Oncogene 2003, 22, 2782–2794. [Google Scholar] [CrossRef][Green Version]
- Oda, Y.; Ohishi, Y.; Saito, T.; Hinoshita, E.; Uchiumi, T.; Kinukawa, N.; Iwamoto, Y.; Kohno, K.; Kuwano, M.; Tsuneyoshi, M. Nuclear expression of Y-box-binding protein-1 correlates with P-glycoprotein and topoisomerase II alpha expression, and with poor prognosis in synovial sarcoma. J. Pathol. 2003, 199, 251–258. [Google Scholar] [CrossRef]
- Habibi, G.; Leung, S.; Law, J.H.; Gelmon, K.; Masoudi, H.; Turbin, D.; Pollak, M.; Nielsen, T.O.; Huntsman, D.; Dunn, S.E. Redefining prognostic factors for breast cancer: YB-1 is a stronger predictor of relapse and disease-specific survival than estrogen receptor or HER-2 across all tumor subtypes. Breast Cancer Res. 2008, 10, R86. [Google Scholar] [CrossRef]
- Lasham, A.; Samuel, W.; Cao, H.; Patel, R.; Mehta, R.; Stern, J.L.; Reid, G.; Woolley, A.G.; Miller, L.D.; Black, M.A.; et al. YB-1, the E2F Pathway, and Regulation of Tumor Cell Growth. J. NatI. Cancer Inst. 2012, 104, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Woolley, A.G.; Algie, M.; Samuel, W.; Harfoot, R.; Wiles, A.; Hung, N.A.; Tan, P.H.; Hains, P.; Valova, V.A.; Huschtscha, L.; et al. Prognostic Association of YB-1 Expression in Breast Cancers: A Matter of Antibody. PloS ONE 2011, 6, e20603. [Google Scholar] [CrossRef] [PubMed]
- Stenina, O.I.; Shaneyfelt, K.M.; DiCorleto, P.E. Thrombin induces the release of the Y-box protein dbpB from mRNA: a mechanism of transcriptional activation. Proc. NatI. Acad. Sci. USA 2001, 98, 7277–7282. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.G.; Vogt, P.K. Inhibition of protein synthesis by Y box-binding protein 1 blocks oncogenic cell transformation. Mol. Cell Biol. 2005, 25, 2095–2106. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.B.; Ma, W.; Valova, V.A.; Algie, M.; Harfoot, R.; Woolley, A.G.; Robinson, P.J.; Braithwaite, A.W. Genotoxic stress-induced nuclear localization of oncoprotein YB-1 in the absence of proteolytic processing. Oncogene 2010, 29, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.A.; Randolph, J.K.; Yalowich, J.C.; Ritke, M.K.; Gewirtz, D.A. Interference by doxorubicin with DNA unwinding in MCF-7 breast tumor cells. Mol. Pharmacol. 1994, 45, 649–656. [Google Scholar]
- Arnal, I.; Wade, R.H. How does taxol stabilize microtubules? CB 1995, 5, 900–908. [Google Scholar] [CrossRef]
- Wang, T.H.; Wang, H.S.; Soong, Y.K. Paclitaxel-induced cell death: where the cell cycle and apoptosis come together. Cancer 2000, 88, 2619–2628. [Google Scholar] [CrossRef]
- Tada, S. Cdt1 and geminin: role during cell cycle progression and DNA damage in higher eukaryotes. Front. Biosci. 2007, 12, 1629–1641. [Google Scholar] [CrossRef]
- Zielke, N.; Edgar, B.A. FUCCI sensors: Powerful new tools for analysis of cell proliferation. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 469–487. [Google Scholar] [CrossRef]
- Bargou, R.C.; Jurchott, K.; Wagener, C.; Bergmann, S.; Metzner, S.; Bommert, K.; Mapara, M.Y.; Winzer, K.J.; Dietel, M.; Dorken, B.; et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nature Med. 1997, 3, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Reipas, K.M.; Law, J.H.; Couto, N.; Islam, S.; Li, Y.; Li, H.; Cherkasov, A.; Jung, K.; Cheema, A.S.; Jones, S.J.; et al. Luteolin is a novel p90 ribosomal S6 kinase (RSK) inhibitor that suppresses Notch4 signaling by blocking the activation of Y-box binding protein-1 (YB-1). Oncotarget 2013, 4, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Sinnberg, T.; Sauer, B.; Holm, P.; Spangler, B.; Kuphal, S.; Bosserhoff, A.; Schittek, B. MAPK and PI3K/AKT mediated YB-1 activation promotes melanoma cell proliferation which is counteracted by an autoregulatory loop. Exp. Dermatol. 2012, 21, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Stratford, A.L.; Habibi, G.; Astanehe, A.; Jiang, H.; Hu, K.; Park, E.; Shadeo, A.; Buys, T.P.; Lam, W.; Pugh, T.; et al. Epidermal growth factor receptor (EGFR) is transcriptionally induced by the Y-box binding protein-1 (YB-1) and can be inhibited with Iressa in basal-like breast cancer, providing a potential target for therapy. Breast Cancer Res. 2007, 9, R61. [Google Scholar] [CrossRef] [PubMed]
- Stratford, A.L.; Reipas, K.; Hu, K.; Fotovati, A.; Brough, R.; Frankum, J.; Takhar, M.; Watson, P.; Ashworth, A.; Lord, C.J.; et al. Targeting p90 Ribosomal S6 Kinase Eliminates Tumor-Initiating Cells by Inactivating Y-Box Binding Protein-1 in Triple-Negative Breast Cancers. Stem Cells 2012, 30, 1338–1348. [Google Scholar] [CrossRef]
- Martin, M.; Hua, L.; Wang, B.; Wei, H.; Prabhu, L.; Hartley, A.V.; Jiang, G.; Liu, Y.; Lu, T. Novel Serine 176 Phosphorylation of YBX1 Activates NF-kappaB in Colon Cancer. J. Biol. Chem. 2017, 292, 3433–3444. [Google Scholar] [CrossRef]
- Prabhu, L.; Mundade, R.; Wang, B.; Wei, H.; Hartley, A.V.; Martin, M.; McElyea, K.; Temm, C.J.; Sandusky, G.; Liu, Y.; et al. Critical role of phosphorylation of serine 165 of YBX1 on the activation of NF-kappaB in colon cancer. Oncotarget 2015, 6, 29396–29412. [Google Scholar] [CrossRef]
- Kloks, C.P.; Spronk, C.A.; Lasonder, E.; Hoffmann, A.; Vuister, G.W.; Grzesiek, S.; Hilbers, C.W. The solution structure and DNA-binding properties of the cold-shock domain of the human Y-box protein YB-1. J. Mol. Biol. 2002, 316, 317–326. [Google Scholar] [CrossRef]
- Chatterjee, M.; Rancso, C.; Stuhmer, T.; Eckstein, N.; Andrulis, M.; Gerecke, C.; Lorentz, H.; Royer, H.D.; Bargou, R.C. The Y-box binding protein YB-1 is associated with progressive disease and mediates survival and drug resistance in multiple myeloma. Blood 2008, 111, 3714–3722. [Google Scholar] [CrossRef]
- Basaki, Y.; Taguchi, K.; Izumi, H.; Murakami, Y.; Kubo, T.; Hosoi, F.; Watari, K.; Nakano, K.; Kawaguchi, H.; Ohno, S.; et al. Y-box binding protein-1 (YB-1) promotes cell cycle progression through CDC6-dependent pathway in human cancer cells. Eur. J. Cancer 2010, 46, 954–965. [Google Scholar] [CrossRef]
- Davies, A.H.; Barrett, I.; Pambid, M.R.; Hu, K.; Stratford, A.L.; Freeman, S.; Berquin, I.M.; Pelech, S.; Hieter, P.; Maxwell, C.; et al. YB-1 evokes susceptibility to cancer through cytokinesis failure, mitotic dysfunction and HER2 amplification. Oncogene 2011, 30, 3649–3660. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pagano, C.; di Martino, O.; Ruggiero, G.; Guarino, A.M.; Mueller, N.; Siauciunaite, R.; Reischl, M.; Foulkes, N.S.; Vallone, D.; Calabro, V. The tumor-associated YB-1 protein: new player in the circadian control of cell proliferation. Oncotarget 2017, 8, 6193–6205. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Lowe, M.; Herliczek, T.W.; Keyomarsi, K. Lovastatin mediated G1 arrest in normal and tumor breast cells is through inhibition of CDK2 activity and redistribution of p21 and p27, independent of p53. Oncogene 1998, 17, 2393–2402. [Google Scholar] [CrossRef] [PubMed]
- Terada, Y.; Inoshita, S.; Nakashima, O.; Yamada, T.; Kuwahara, M.; Sasaki, S.; Marumo, F. Lovastatin inhibits mesangial cell proliferation via p27Kip1. JASN 1998, 9, 2235–2243. [Google Scholar]
- Stratford, A.L.; Fry, C.J.; Desilets, C.; Davies, A.H.; Cho, Y.Y.; Li, Y.; Dong, Z.; Berquin, I.M.; Roux, P.P.; Dunn, S.E. Y-box binding protein-1 serine 102 is a downstream target of p90 ribosomal S6 kinase in basal-like breast cancer cells. Breast Cancer Res. 2008, 10, R99. [Google Scholar] [CrossRef]
- Kahn, K.; Plaxco, K.W. Principles of Biomolecular Recognition. In Recognition Receptors in Biosensors; Zoureb, M., Ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- He, Y.; Chen, Y.; Mooney, S.M.; Rajagopalan, K.; Bhargava, A.; Sacho, E.; Weninger, K.; Bryan, P.N.; Kulkarni, P.; Orban, J. Phosphorylation-induced Conformational Ensemble Switching in an Intrinsically Disordered Cancer/Testis Antigen. J. Biol. Chem. 2015, 290, 25090–25102. [Google Scholar] [CrossRef]
- Eliseeva, I.; Kim, E.; Guryanov, S.; Ovchinnikov, L.; Lyabin, D. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Moscow) 2011, 76, 1402–1433. [Google Scholar] [CrossRef]
- Evdokimova, V.; Ovchinnikov, L.P.; Sorensen, P.H. Y-box binding protein 1: providing a new angle on translational regulation. Cell Cycle 2006, 5, 1143–1147. [Google Scholar] [CrossRef]
- Wolffe, A.P.; Tafuri, S.; Ranjan, M.; Familari, M. The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biologist 1992, 4, 290–298. [Google Scholar]
- Wu, J.; Stratford, A.L.; Astamehe, A.; Dunn, S.E. YB-1 is a transcription/translation factor that orchestrates the oncogenome by hardwiring signal transduction to gene expression. Transl. Oncogenomics 2007, 2, 49–65. [Google Scholar]
- Mordovkina, D.A.; Kim, E.R.; Buldakov, I.A.; Sorokin, A.V.; Eliseeva, I.A.; Lyabin, D.N.; Ovchinnikov, L.P. Transportin-1-dependent YB-1 nuclear import. Biochem. Biophys. Res. Commun. 2016, 480, 629–634. [Google Scholar] [CrossRef]
- Reid, G.; Wallant, N.C.; Patel, R.; Antonic, A.; Saxon-Aliifaalogo, F.; Cao, H.; Webster, G.; Watson, J.D. Potent subunit-specific effects on cell growth and drug sensitivity from optimised siRNA-mediated silencing of ribonucleotide reductase. J. RNAi Gene Silenc. 2009, 5, 321–330. [Google Scholar]
- Kall, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008, 9, 40. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Homeyer, N.; Horn, A.H.; Lanig, H.; Sticht, H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J. Mol. Model. 2006, 12, 281–289. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald—An N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle—An Analytical Version of the Shake and Rattle Algorithm for Rigid Water Models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Hubbard, S.J.; Thornton, J.M. NACCESS; University College London: London, UK, 1993. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. Model 1996, 14, 33–38. [Google Scholar] [CrossRef]
- De Lano, W. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehta, S.; McKinney, C.; Algie, M.; Verma, C.S.; Kannan, S.; Harfoot, R.; Bartolec, T.K.; Bhatia, P.; Fisher, A.J.; Gould, M.L.; et al. Dephosphorylation of YB-1 is Required for Nuclear Localisation During G2 Phase of the Cell Cycle. Cancers 2020, 12, 315. https://doi.org/10.3390/cancers12020315
Mehta S, McKinney C, Algie M, Verma CS, Kannan S, Harfoot R, Bartolec TK, Bhatia P, Fisher AJ, Gould ML, et al. Dephosphorylation of YB-1 is Required for Nuclear Localisation During G2 Phase of the Cell Cycle. Cancers. 2020; 12(2):315. https://doi.org/10.3390/cancers12020315
Chicago/Turabian StyleMehta, Sunali, Cushla McKinney, Michael Algie, Chandra S. Verma, Srinivasaraghavan Kannan, Rhodri Harfoot, Tara K. Bartolec, Puja Bhatia, Alistair J. Fisher, Maree L. Gould, and et al. 2020. "Dephosphorylation of YB-1 is Required for Nuclear Localisation During G2 Phase of the Cell Cycle" Cancers 12, no. 2: 315. https://doi.org/10.3390/cancers12020315
APA StyleMehta, S., McKinney, C., Algie, M., Verma, C. S., Kannan, S., Harfoot, R., Bartolec, T. K., Bhatia, P., Fisher, A. J., Gould, M. L., Parker, K., Cesare, A. J., Cunliffe, H. E., Cohen, S. B., Kleffmann, T., Braithwaite, A. W., & Woolley, A. G. (2020). Dephosphorylation of YB-1 is Required for Nuclear Localisation During G2 Phase of the Cell Cycle. Cancers, 12(2), 315. https://doi.org/10.3390/cancers12020315





