Simple Summary
The clinical relevance of Nuclear factor erythroid 2-Related Factor 2 (NRF2) in human breast cancer remains unclear. A total of 5443 breast cancer patients with transcriptomic profile were analyzed for the clinical relevance of NRF2 expression, including cancer aggressiveness, immune cell infiltration, patient survival, and drug response. We found that tumors with high NRF2 expression were associated with better survival in ER-positive/HER2-negative breast cancer. NRF2 expression was equivalent in immune, stromal, and cancer cells in tumor microenvironment. We found that high NRF2 expression was associated with enhanced tumor-infiltrating lymphocytes in ER-positive/HER2-negative breast cancer. NRF2 expression significantly correlated with drug sensitivity in multiple ER-positive breast cancer cell lines, but not associated with pathological complete response after neoadjuvant chemotherapy in breast cancer patients regardless of subtypes.
Abstract
Nuclear factor erythroid 2-related factor 2 (NRF2) is a key modifier in breast cancer. It is unclear whether NRF2 suppresses or promotes breast cancer progression. We studied the clinical relevance of NRF2 expression by conducting in silico analyses in 5443 breast cancer patients from several large patient cohorts (METABRIC, GSE96058, GSE25066, GSE20194, and GSE75688). NRF2 expression was significantly associated with better survival, low Nottingham pathological grade, and ER-positive/HER2-negative and triple negative breast cancer (TNBC). High NRF2 ER-positive/HER2-negative breast cancer enriched inflammation- and immune-related gene sets by GSEA. NRF2 expression was elevated in immune, stromal, and cancer cells. High NRF2 tumors were associated with high infiltration of immune cells (CD8+, CD4+, and dendritic cells (DC)) and stromal cells (adipocyte, fibroblasts, and keratinocytes), and with low fraction of Th1 cells. NRF2 expression significantly correlated with area under the curve (AUC) of several drug response in multiple ER-positive breast cancer cell lines, however, there was no significant association between NRF2 and pathologic complete response (pCR) rate after neoadjuvant chemotherapy in human samples. Finally, high NRF2 breast cancer was associated with high expression of immune checkpoint molecules. In conclusion, NRF2 expression was associated with enhanced tumor-infiltrating lymphocytes in ER-positive/HER2-negative breast cancer.
Keywords:
biomarker; breast cancer; gene expression; hormonal; metastasis; NRF2; survival; treatment response 1. Introduction
Breast cancer is the most common cancer among women worldwide. Estrogen receptor (ER)-positive breast cancer is both the most common (>70%) and least aggressive subtype of breast cancer [1]. Unfortunately, the main challenges with this subtype include late recurrence, which occurs in 40% of patients more than 10 years after diagnosis [2]. These tumors also have a poor response to neoadjuvant chemotherapy (NAC) [3,4]. Therefore, it is necessary to develop a prognostic biomarker for ER-positive breast cancer to anticipate who will recur.
In recent years, several publications have described the role of nuclear factor erythroid 2-related factor 2 (NRF2) in cancer progression. Data lack on whether NRF2 suppresses or promotes tumor progression [5,6,7]. Some of these findings lead to the question of whether NRF2 should be targeted as an anti-cancer therapy [8]. NRF2 is commonly known as a tumor suppressor because it protects cells from oxidative or electrophilic insults and is thus anti-carcinogenic. However, NRF2 promotes survival of both normal and malignant cells. NRF2 in normal cells activates broad defense mechanisms, such as elimination of reactive oxygen species (ROS), dampening of inflammation, drug and carcinogen detoxication, and intermediary metabolism [9,10], all of which protect cells from various carcinogenic stressors. There is cross-talk between NRF2 signaling and NF-kB, p53, and Notch1 signaling, which affects cell survival [11]. On the other hand, constitutively elevated NRF2 levels in cancer cells can enhance growth and develop chemotherapy resistance by creating a redox environment [12,13]. To this end, high levels of NRF2 are generally correlated with poor prognosis in multiple types of cancer [5,6].
Limited data exist on NRF2 activation in breast cancer. NRF2 promotes breast cancer progression by enhancing glycolysis through coactivation of HIF1α, which allows some to suggest NRF2 as a therapeutic target for breast cancer [14]. On the contrary, high NRF2 gene expression was reported to be associated with better outcomes in ER-positive breast cancer [15]. Given that estrogen is known as a ROS inducer, ER-positive tumors may upregulate NRF2 activity and accelerate their antioxidant response to resist oxidative stress [16]. Another study described a correlation between NRF2 activity and survival outcome in ER-positive breast cancer compared to triple negative breast cancer (TNBC) [17]. To date, there is no study that has elucidated the mechanism in which NRF2 activity in a bulk tumor is associated with better survival in ER-positive breast cancer patients.
Rapid evolution in microarray and sequencing technologies has revolutionized the depth and complexity at which molecular data are obtained and examined today in biomedical research. Although it is difficult to reproduce the human tumor microenvironment in vivo and vitro, it is possible to grasp several immune functions by using the gene expression of bulk tumor and algorithms. We previously reported on the relationship between transcriptome and clinical relevance with immunity and hallmark pathway by computational analysis using large patient cohorts using several algorithms, such as xCell and gene set variant analysis (GSVA) [18,19,20,21,22]. For example, we found the significant contribution of immune cells in a favorable survival outcome of high glucocorticoid receptor (GR) expression in ER-positive breast cancer [23]. We also established a 4-gene score based on genes differentially expressed between the parental and lung metastasis cell lines that predicts neoadjuvant chemotherapy (NAC) response in ER-positive/human epidermal growth factor receptor 2 (HER2)-negative breast cancer [24]. To this end, we are equipped with the necessary methods to analyze the clinical relevance of NRF2 activity and its associated features in the tumor microenvironment.
We hypothesized that both cancer cells and immune cells in the tumor microenvironment contribute to the expression of NRF2 in a bulk tumor. We also hypothesized that infiltration of immune cells may contribute to survival outcomes in breast cancer patients. The novelty of the study is to investigate the clinical relevance of NRF2 using a large amount of human sample data.
2. Results
2.1. High NRF2 Expression Is Significantly Associated with Better Survival in ER-Positive/HER2-Negative Breast Cancer
Advances in technology and banking of transcriptomic data have been extremely rapid and robust in recent years. The large breast cancer patient cohort, METABRIC, was updated with clinical parameters, including longer survival follow-up. Another robust patient cohort (GSE96058) became available, which allows survival analyses by subtypes with stronger power. Thus, it was of interest to investigate how NRF2 gene expression impacts different breast cancer subtypes using the latest METABRIC and GSE96058 cohorts. The top tertile was used as a cut-off between high and low NRF2 groups within each cohort. High NRF2 expression was significantly associated with better disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS) in ER-positive/HER2-negative breast cancer in the METABRIC cohort (Figure 1; p = 0.039, p = 0.011, and p < 0.001, respectively). The OS results were validated in the GSE96058 cohort (p = 0.018). On the other hand, although high NRF2 expression in TNBC tends to be associated with better survival, there are no significant differences between high and low NRF2 group in TNBC nor HER2-positive breast cancer in both patient cohorts.
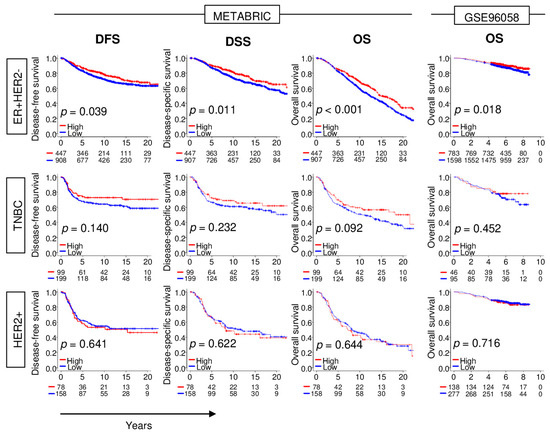
Figure 1.
Association between levels of expression of NRF2 and survival outcomes of breast cancer patients in the METABRIC and GSE96058 cohorts. Disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS) of NRF2 low (blue) and high (red) of NRF2 expression by breast cancer subtype.
2.2. High NRF2 Expression Is Significantly Associated with Lower Nottingham Pathological Grade, and ER-Positive/HER2-Negative and TNBC Subtype
Given the results of Figure 1, we anticipated that high NRF2 expression would be associated with less aggressive clinical features. There was no trend of NRF2 expression in the American Joint Commission on Cancer (AJCC) cancer staging. On the other hand, we found that higher NRF2 expression was associated with lower Nottingham pathological grade, which reflects less cancer cell proliferation. High NRF2 expression was associated with ER-positive/HER2-negative and TNBC subtypes. These results were consistent in both the METABRIC and GSE96058 cohorts (Figure 2).
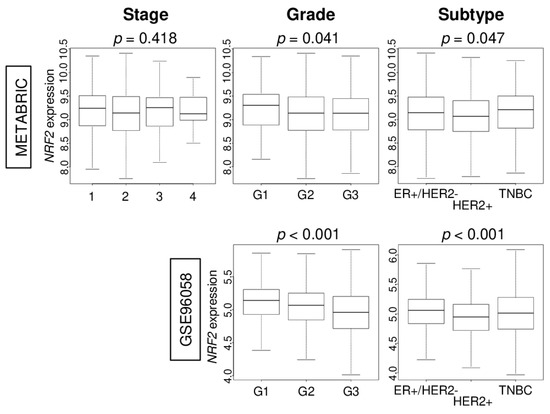
Figure 2.
Association of NRF2 levels of expression according to breast cancer clinical features in the METABRIC and GSE96058 cohorts. Boxplots of the NRF2 expression by breast cancer American Joint Committee on Cancer (AJCC) stage, Nottingham pathological grade, and breast cancer subtypes [estrogen receptor-positive/human epidermal growth factor receptor 2-negative (ER+/HER2), triple negative breast cancer (TNBC), and HER2-positive].
2.3. High NRF2 Tumors Are Enriched with Inflammation- and Immune-Related Gene Sets in ER-Positive/HER2-Negative Breast Cancer
Given that high NRF2 expression was associated with less cancer cell proliferation and with better survival outcomes in ER-positive/HER2-negative breast cancer, it was of interest to investigate the biological basis of this association in this subtype. Gene set enrichment analysis (GSEA) was used to study the enrichment of MSigDB Hallmark gene sets in breast cancers with high NRF2. We found that inflammation- and immune-related gene sets were significantly enriched in high NRF2 tumors consistently in both the METABRIC and GSE96058 cohorts, such as inflammatory response, IL6/JAK/STAT3 signaling, tumor necrosis factor (TNF)-α signaling, complement, coagulation, allograft rejection, IL2/STAT5 signaling, interferon (IFN)-γ response, and apoptosis (Figure 3). Tumors with high NRF2 expression also significantly expressed pro-cancerous gene sets, such as KRAS signaling up, TGF-β signaling, hypoxia, and angiogenesis, in both patient cohorts. Furthermore, tumors with high NRF2 expression was significantly associated with high expression of inflammatory-related genes; IL6, Lp-PLA2 (PLA2G7), and Myeloperoxidase (MPO), and apoptosis-related genes; FAS, TNF, and TNFR1, as well as high score of inflammatory-related gene sets; IL6/JAC/STAT3 signaling, and inflammatory response pathway, and apoptosis-related gene sets; Apoptosis and TNFα signaling Via NFkB pathway, which calculated by GSVA algorithm in both METABRIC and GSE96058 cohorts except for TNF and TNFR1 expression in the METABRIC cohort (Figure S1A,B). The METABRIC cohort did not have MPO expression data.
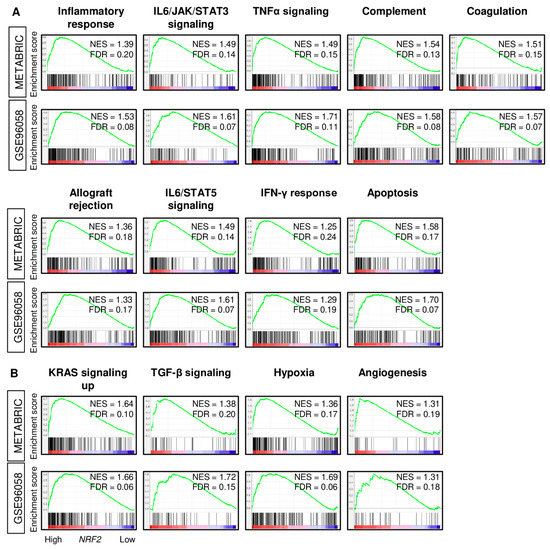
Figure 3.
Gene Set Enrichment Assay (GSEA) with enrichment gene sets in the NRF2 high expression group of ER-positive/HER2-negative patients from the METABRIC and GSE96058 cohorts. (A) Inflammation- and immune-related gene sets (inflammatory response, IL6/JAK/STAT3 signaling, TNF-α signaling, complement, coagulation, allograft rejection, IL2/STAT5 signaling, interferon (IFN)-γ response, and apoptosis and (B) Pro-cancerous-related gene sets (KRAS signaling up, TGF-β signaling, hypoxia, and angiogenesis) with normalized enrichment score (NES) and false discovery rate (FDR). Statistical significance was defined by an FDR of 0.25, as recommended by the GSEA software.
2.4. NRF2 Is Expressed in Immune Cells as well as Cancer Cells, and High NRF2 Tumors Are Infiltrated with Anti-Cancer Immune Cells and Stromal Cells
Given that tumors with high NRF2 expression were associated with better survival outcomes and enriched inflammation- and immune-related gene sets in ER-positive/HER2-negative subtype, we hypothesized that non-cancer cells in the tumor microenvironment may be involved in NRF2 expression of a bulk tumor. Single-cell sequencing technology allows transcriptomic profile and a better understanding of the function of an individual cell in the tumor microenvironment. Hence, a single-cell sequencing cohort of primary breast cancer (GSE75688) was used to analyze NRF2 expression differences between immune (T cells, B cells, and myeloid cells), tumor, and stromal cells. Immune cells expressed NRF2 in equivalent levels as tumor cells (Figure 4A). In immune cells, NRF2 expression levels were lower in B cells than in T cells or myeloid cells.
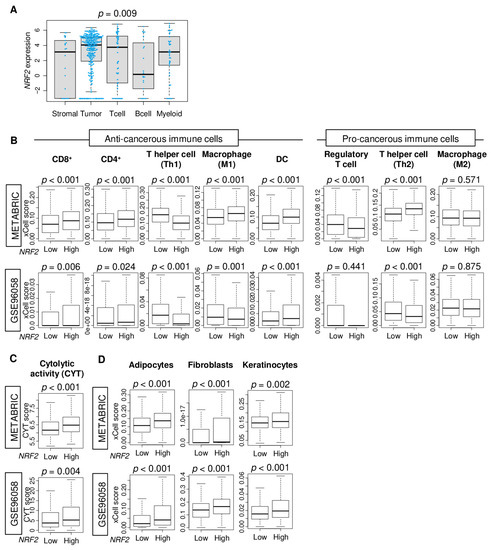
Figure 4.
Association of NRF2 expression level with fraction of immune cells and cytolytic activity (CYT). (A) Boxplots comparing NRF2 expression levels by tumor cells, T cells, B cells, myeloid cells, and stromal cells in the GSE75688 cohort. Boxplots depicting the fraction of (B) immune cells, (C) CYT, and (D) stromal cells by NRF2 low and high groups in METABRIC and GSE96058 cohorts. The cut-off of top tertile of NRF2 expression was considered as NRF2 high and low within each cohort. One-way ANOVA test was used for comparison.
Therefore, it was of interest to investigate which immune cell in the tumor microenvironment was associated with NRF2 gene expression in the bulk tumor. Immune and stromal cell compositions in bulk tumors were estimated using the xCell algorithm and compared between NRF2 low and high groups in ER-positive/HER2-negative subtype of the METABRIC and GSE96058 cohorts. We found that NRF2 high tumors had a significantly high fraction of CD8, CD4, and dendritic cells (DC), and low fraction of T helper 1 type cells (Th1) consistently in both cohorts (Figure 4B). M1 macrophages, natural killer, regulatory T cells, and T helper 2 type cells (Th2) were highly infiltrated in high NRF2 tumors in the METABRIC cohort but not validated by the GSE96058 cohort.
Cytolytic activity score (CYT), defined as the sum of expression of granzyme A (GZMA) and perforin (PRF1), was used to evaluate overall anti-cancer immune cell killing in the tumor microenvironment (25594174). We found that NRF2 high group was significantly associated with a high level of CYT consistently in both patient cohorts (Figure 4C, p < 0.001, and p = 0.004, respectively).
The high NRF2 group was also significantly associated with a high fraction of stromal cells, including adipocytes, fibroblasts, and keratinocytes, consistently in both patient cohorts (Figure 4D). These results suggested that high NRF2 expression was associated with a high fraction of anti-cancer immune cells with cytolytic activity as well as infiltration of stromal cells.
2.5. NRF2 Expression Was Significantly Associated with Treatment Response In Vitro, but Association Was Noted with Pathological Complete Response (pCR) after Neoadjuvant Chemotherapy (NAC)
Next, we investigated the relationship between NRF2 expression and treatment response using in vitro data obtained from DepMap portal and patient cohorts that underwent NAC (GSE25066 and GSE20194). The levels of NRF2 expression correlated with levels of area of under the curve (AUC) for paclitaxel, 5-FU (fluorouracil), tamoxifen, and fulvestrant in ER-positive/HER2-negative breast cancer cell lines (CAMA1, EFM19, HCC1428, HCC1500, KPL1, MCF7, MDAMB175VII, MDAMB415, T47D, and ZR751) (Figure 5A, r = 0.88 [p = 0.02], r = 0.86 [p < 0.01], r = 0.75 [p < 0.05], and r = 0.96 [p < 0.01], respectively). There were no significant differences between low and high NRF2 groups in pCR rate after NAC in the GSE25066 and GSE20194 cohorts (Figure 5B). These results suggest that in vitro results of NRF2 expression may not be directly translatable to the clinical setting.
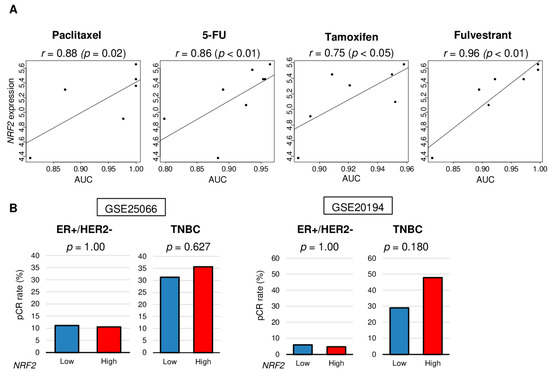
Figure 5.
Association of NRF2 expression level with treatment response of cell lines and human tumors. (A) Correlation plots between NRF2 expression level and area under the curve (AUC) of several drug sensitivity for ER-positive/HER2-negative breast cancer cell lines. Spearman correlation statistics are depicted. (B) Bar plots comparing the pathologic complete response (pCR) rates between low and high NRF2 groups among patients with ER-positive/HER2-negative tumors and triple negative breast cancer (TNBC) in the GSE25066 (n = 508), and GSE20194 (n = 278) cohorts. Fisher’s exact test was used to compare groups.
2.6. High NRF2 Tumors Are Significantly Associated with High Expression of Immune Checkpoint Molecules in ER-Positive/HER2-Negative Breast Cancer
Immunotherapy using immune checkpoint inhibitors is drawing attention as a new modality to treat cancer, however, it is only approved for TNBC. Therefore, it is necessary to develop a biomarker to identify which patients have a high expression of immune checkpoint molecules. We found that NRF2 high tumors were significantly associated with higher expression of major immune checkpoint molecules, namely programmed death-1 (PD-1), programmed death ligand 2 (PD-L2), indoleamine dioxygenase 1 (IDO1), and B- and T-lymphocyte attenuator (BTLA). These findings were consistently noted in both patient cohorts (Figure 6). NRF2 high tumors were also significantly associated with high expression of programmed death ligand 1 (PD-L1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA4) in GSE96058 cohort.
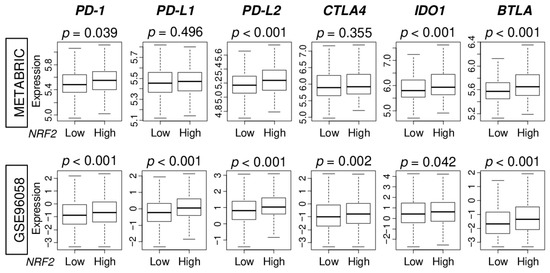
Figure 6.
Association of NRF2 expression with immune checkpoint molecules in the METABRIC and GSE96058 cohorts. Boxplots comparing low and high NRF2 group with expression levels of immune checkpoint molecules [programmed death-1; PD-1, programmed death ligand 1; PD-L1, programmed death ligand 2; PD-L2, cytotoxic T-lymphocyte-associated protein 4; CTLA4, indoleamine dioxygenase 1; IDO1, B- and T-lymphocyte attenuator; BTLA] in ER-positive/HER2-negative breast cancer patients.
3. Discussion
In this study, we found that high NRF2 expression was significantly associated with better DFS, DSS, and OS in ER-positive/HER2-negative, but not in other subtypes across two large breast cancer patient cohorts (METABRIC and GSE96058). High NRF2 expression was associated with low Nottingham pathological grade as well as ER-positive/HER2-negative and TNBC subtypes, but not with AJCC cancer staging. We found that high NRF2 expression ER-positive/HER2-negative breast cancer significantly enriched inflammation- and immune-related gene sets as well as pro-cancerous gene sets by GSEA. Interestingly, NRF2 expression was elevated not only in cancer cells but also in T cells, myeloid cells, and stromal cells. High NRF2 expression ER-positive/HER2-negative breast cancer was associated with increased tumor-infiltrating lymphocytes (CD8+ T cell, CD4+ T cell, and DC) and low fraction of Th1 cells. Several stromal cells, including adipocyte, fibroblasts, and keratinocytes, highly infiltrated tumors with high NRF2 expression levels. CYT, which assesses overall immune cytolytic activity, was elevated in high NRF2 expression tumors. NRF2 expression levels correlated with AUC of several drug response in vitro, however, there was no association between NRF2 expression and pCR rate after NAC in two cohorts. Finally, high NRF2 expression ER-positive/HER2-negative breast cancer was associated with high expression of immune checkpoint molecules.
The novelty of the study is to investigate the clinical relevance of NRF2 using a large amount of human sample data. Recent advances in high-throughput technology resulted in accumulation of tumor transcriptome data of large sample size cohorts. There has been a dramatic advancement in the use of genetic analysis for cancer research due to the availability of data collected from around the world, including projects such as METABRIC and data repositories such as Gene Expression Omnibus (GEO). The clinical outcomes of these databases are updated and allow researchers to analyze long-term follow-up data. For example, the currently available median clinical follow-up is 14 years [25]. This is particularly relevant for ER-positive breast cancer that often relapses more than a decade after diagnosis. Recent advances in gene analysis technology allow us to obtain robust information from transcriptome data in bulk tumor by using algorithms which are released on a monthly basis. The tumor immune microenvironment, which plays a significant role in cancer progression and treatment response, has been analyzed traditionally by flow cytometry or immunohistochemistry. These approaches are highly labor-intensive and expensive when they are used to analyze thousands of patient samples, whereas computational algorithms can estimate the quantity of immune cells from tens of thousands of samples with less cost and much quicker [26,27,28,29] (as long as transcriptomic data are available). Furthermore, analyses such as GSEA explore the biological activity of a signaling pathway of interest and allow investigators to grasp the big picture of the intricately intertwined gene signaling pathways and to identify the mechanism that is in place. The clinical relevance of NRF2 expression in tumors remains controversial in breast cancer. Wofl et al. suggested that NRF2 expression of a bulk tumor may be useful as a predictive biomarker for ER-positive breast cancer using the METABRIC cohort in 2016 [15]. In this study, we have used the updated METABRIC cohort and obtained a similar result that patients with high NRF2 expression ER-positive breast tumor have a better prognosis. Importantly, this result was validated in a larger sample size cohort (GSE96058). Somewhat surprisingly, we found that not only cancer cells but also immune cells and stromal cells were expressing high levels of NRF2 gene in tumor microenvironment. NRF2 activation has also been reported to play a critical role in proper immune function. NRF2 suppresses macrophage inflammatory response by blocking the transcription of proinflammatory cytokines, including IL-6 and IL-1b. This was found to be independent of ROS and the canonical NRF2-binding motif in cytokine genes [30]. The regulation of the immune microenvironment also extends beyond macrophages to other myeloid populations and regulatory T cells [31,32], which can influence tumor progression and metastasis. NRF2 activation within the tumor microenvironment suppressed the progression of lung tumors [33]. Hayashi et al. reported that microenvironmental NRF2 activation suppresses the progression of malignant NRF2-acitvated tumors and that NRF2 activation in immune cells at least partially contributes to these suppressive effects [33]. Rong et al. reported that sulforaphane blocked prostaglandin E2 synthesis in parental and doxorubicin-resistant breast cancer 4T1 cell lines by activating NRF2, and triggered MDSCs to switch to an immunogenic phenotype, enhancing the anti-tumor activities of CD8+ T cells [34]. High immune activity is known to be associated with a better prognosis in many cancers, including breast cancer [35,36,37]. In vivo/vitro preclinical models are essential tools to elucidate cancer biology. With that said, we are also aware that no model can perfectly replicate human cancer. Therefore, in this study, we investigated the association between NRF2 expression and immunity in tumor microenvironment using large amounts of human sample data to see if what has been reported actually occurs in human tumors. Considering that high NRF2 expression ER-positive breast tumor was significantly associated with increased tumor-infiltrating lymphocytes, especially anti-cancer immune cells, CD8+ T cell, CD4+ T cell, and DC cells, we speculate that the better survival of high NRF2 expression ER-positive/HER2-negative breast cancer may be at least partly due to the reflection of high infiltration of these immune cells. This notion is in agreement with our other results that high NRF2 expression tumor enriched immune-related gene sets and was associated with overall cytolytic activity.
NRF2 is generally known as a tumor suppressor and several NRF2 activators are currently being tested as chemopreventive compounds in clinical trials. On the other hand, there are serious clinical concerns on enhanced NRF2 activity because it may also protect cancer cells from chemotherapeutic agents and facilitate cancer progression as NRF2 protects normal cells. These studies have included a diverse range of drugs, such as cisplatin, carboplatin, 5-fluorouracil, paclitaxel, bleomycin, doxorubicin, and etoposide [38,39,40]. Thus, the role of NRF2 in cancer is subject of controversial discussion, as it has been reported to have both pro- and anti-tumorigenic functions [15]. Furthermore, several papers reported that the role of NRF2 may be context-dependent, because of complex NRF2-related pathways [41,42]. One of the reasons for this is that the tumor environment is not constant in the in vivo and in vitro settings. Moreover, it is also difficult to reproduce the human tumor environment in vivo and in vitro. Several efforts have been made or are ongoing to develop novel therapeutics, but they have been hampered by the lack of preclinical models that reliably reproduce the human tumor microenvironment. We previously reported the utility of patient-derived xenograft mouse model using human breast cancer patient samples, however, it was not possible to replicate human tumors. Our results, which showed the different result of the association of NRF2 expression with treatment response between in vitro and neoadjuvant human patient data, indicate that the role of non-tumor cells in a bulk tumor, such as immune cells, may have clinical relevance. Additionally, this further emphasizes the importance of analyzing clinical specimens. We believe that our in silico approach is a useful tool to obtain a comprehensive view of human cancers in the clinical setting.
Although immune checkpoint inhibitors are approved for advanced breast cancer, their indication is very limited, and patient selection remains a major challenge. Given that our results show that the majority of immune checkpoint molecules were associated with high NRF2 expression in ER-positive/HER2-negative breast cancer, we cannot help but speculate that patients whose tumors show high NRF2 expression could be the population who may respond to immune checkpoint inhibitors even though they may not respond to chemotherapy.
Although we found clinical relevance of NRF2 expression in breast cancer using several algorithms, this study is not free from limitations. The biggest limitation is that our analysis is a retrospective study and limited in the measurement of gene expressions alone. Experimental approaches are needed to elucidate the mechanism in the future. Especially the relationship between the NRF2 expression and immunity should be directly quantified in tumoral immune cells using a gold standard such as flow cytometry or immunohistochemistry. Furthermore, to use NRF2 expression as a predictive biomarker in ER-positive/HER2-negative management, we need to conduct a prospective clinical trial.
In conclusion, we demonstrated that immune cells in addition to tumor cells express high levels of NRF2, and high NRF2 expression enriched inflammation- and immune-related gene sets and was associated with enhanced tumor-infiltrating lymphocytes in ER-positive/HER2-negative breast cancer, which may at least partly explain why high NRF2 expression was associated with better survival in that subtype.
4. Materials and Methods
4.1. Cohorts Used for Analyses
For the main analysis, we used the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (n = 1903) [25,43], and the GSE96058 (n = 3234) [44] cohorts. Both these cohorts have a large number of breast cancer samples with tumor transcriptome and clinical data. The METABRIC cohort data were obtained from the cBio Cancer Genomic portal [45]. The GSE96058 cohort data were obtained from the Gene Expression Omnibus (GEO) repository. We also obtained other GEO data from the GSE25066 (n = 508) [46] andGSE20194 (n = 248) [47] cohorts. Single cell sequencing data were obtained from primary breast cancer tumor from the GSE75688 cohort [48].
4.2. Data of Gene Expression and Treatment Response of Cell Lines
Gene expression and level of AUC of several drugs (paclitaxel, 5-FU, tamoxifen, and fulvestrant) of ER-positive/HER2-negative breast cancer cell lines were obtained through DepMap portal (https://depmap.org/portal/). Cell lines containing AUC data for each drug were used. ER-positive/HER2-negative breast cancer cell lines included CAMA1, EFM19, HCC1428, HCC1500, KPL1, MCF7, MDAMB175VII, MDAMB415, T47D, and ZR751.
4.3. Tumor-Infiltrating Cells Scoring Using xCell Algorithm and Cytolytic Activity Score (CYT)
The xCell, a bioinformatics algorithm [29], was used to predict immune composition in the METABRIC and GSE96058 cohort samples. A set of 64 cell reference profiles were used. A signature to predict their absolute levels within each sample was developed, as we described previously [49,50,51,52,53,54,55,56,57]. The cytolytic activity score (CYT) as defined by Rooney et al. was used in the algorithm using the gene expression levels of granzyme A (GZMA) and perforin (PRF1) published in Cell, 2015 [58].
4.4. Gene Set Expression Analyses
Gene set enrichment analyses were performed using Gene Set Enrichment Analyses (GSEA) software (Java version 4.0) [59] with MSigDB Hallmark gene sets [60]. Statistical significance was defined by a false discovery rate (FDR) of 0.25, as recommended by the GSEA software.
4.5. Other
The R software (version 4.0.1, R Project for Statistical Computing) was used for all analyses. The top tertile for tumor NRF2 expression was divided into high and low NRF2 groups within cohorts. For group comparison, a one-way analysis of variance (ANOVA) and Fisher’s exact test were used. The Kaplan–Meier method and log-rank test were used for survival analyses. Boxplots were used to depict median and inter-quartile level values.
5. Conclusions
We found that immune cells, in addition to tumor cells, express high levels of NRF2 in the tumor microenvironment. High NRF2 expression levels enriched inflammation- and immune-related gene sets and were associated with enhanced tumor-infiltrating lymphocytes in ER-positive/HER2-negative breast cancer. These results may explain why high NRF2 expression was associated with better survival outcomes in this subtype.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/12/3856/s1, Figure S1: Association of the NRF2 with gene expression and pathway score of inflammatory- and apoptosis-related markers in ER+/HER2- breast cancer in the METABRIC and GSE96058 cohorts, Table S1: Correlation of the NRF2 expression with gene expressions of Hallmark_apoptosis gene set in ER-positive/HER2-negative breast cancer in the METABRIC and GSE96058 cohorts.
Author Contributions
Conceptualization, I.E., K.T., M.O., R.M.; methodology, K.T., M.O., Y.T.; formal analysis, M.O.; writing—original draft preparation, M.O.; writing—review & editing, F.A.A., K.T., L.Y.; supervision, K.T.; project administration, K.T. All authors have read and agree to the published version of the manuscript.
Funding
This work was supported by National Institutes of Health (NIH), USA grant R01CA160688 to K.T., and National Cancer Institute, USA cancer center support grant P30-CA016056 to Roswell Park Comprehensive Cancer Center (RPCCC).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
AJCC, American Joint Committee on Cancer; DFS, disease-free survival; DSS, disease-specific survival; ER, estrogen receptor; FDR, false discovery rate; GSVA, gene set variation analysis; HER2, human epidermal growth factor receptor 2-negative; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; neoadjuvant chemotherapy (NAC); NES, normalized enrichment score; OS, overall survival; pCR, pathological complete response; PFS, progression-free survival; TNBC, triple negative breast cancer.
References
- Dodson, A.; Parry, S.; Ibrahim, M.; Bartlett, J.M.; Pinder, S.; Dowsett, M.; Miller, K. Breast cancer biomarkers in clinical testing: Analysis of a UK national external quality assessment scheme for immunocytochemistry and in situ hybridisation database containing results from 199,300 patients. J. Pathol. Clin. Res. 2018, 4, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, T.; Yan, L.; Asaoka, M.; Rashid, O.; Takabe, K. Late recurrence of breast cancer is associated with pro-cancerous immune microenvironment in the primary tumor. Sci. Rep. 2019, 9, 16942. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.A.; Cirrincione, C.; Henderson, I.C.; Citron, M.L.; Budman, D.R.; Goldstein, L.J.; Martino, S.; Perez, E.A.; Muss, H.B.; Norton, L.; et al. Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA 2006, 295, 1658–1667. [Google Scholar] [CrossRef]
- Bonnefoi, H.; Litière, S.; Piccart, M.; MacGrogan, G.; Fumoleau, P.; Brain, E.; Petit, T.; Rouanet, P.; Jassem, J.; Moldovan, C.; et al. Pathological complete response after neoadjuvant chemotherapy is an independent predictive factor irrespective of simplified breast cancer intrinsic subtypes: A landmark and two-step approach analyses from the EORTC 10994/BIG 1-00 phase III trial. Ann. Oncol. 2014, 25, 1128–1136. [Google Scholar] [CrossRef] [PubMed]
- Sporn, M.B.; Liby, K.T. NRF2 and cancer: The good, the bad and the importance of context. Nat. Rev. Cancer 2012, 12, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Motohashi, H.; Yamamoto, M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells Devoted Mol. Cell. Mech. 2011, 16, 123–140. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N. Nrf2: Friend or foe for chemoprevention? Carcinogenesis 2010, 31, 90–99. [Google Scholar] [CrossRef]
- Agyeman, A.S.; Chaerkady, R.; Shaw, P.G.; Davidson, N.E.; Visvanathan, K.; Pandey, A.; Kensler, T.W. Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Res. Treat. 2012, 132, 175–187. [Google Scholar] [CrossRef]
- Malhotra, D.; Portales-Casamar, E.; Singh, A.; Srivastava, S.; Arenillas, D.; Happel, C.; Shyr, C.; Wakabayashi, N.; Kensler, T.W.; Wasserman, W.W.; et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010, 38, 5718–5734. [Google Scholar] [CrossRef]
- Wakabayashi, N.; Slocum, S.L.; Skoko, J.J.; Shin, S.; Kensler, T.W. When NRF2 talks, who’s listening? Antioxid. Redox Signal. 2010, 13, 1649–1663. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Sun, Z.; Villeneuve, N.F.; Zhang, S.; Zhao, F.; Li, Y.; Chen, W.; Yi, X.; Zheng, W.; Wondrak, G.T.; et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 2008, 29, 1235–1243. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T.; Iijima, K.; Miyamoto, M.; Nakahara, I.; Tanaka, H.; Ohtsuji, M.; Suzuki, T.; Kobayashi, A.; Yokota, J.; Sakiyama, T.; et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008, 68, 1303–1309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.S.; Du, G.Y.; Zhang, Z.G.; Zhou, Z.; Sun, H.L.; Yu, X.Y.; Shi, Y.T.; Xiong, D.N.; Li, H.; Huang, Y.H. NRF2 facilitates breast cancer cell growth via HIF1α-mediated metabolic reprogramming. Int. J. Biochem. Cell Biol. 2018, 95, 85–92. [Google Scholar] [CrossRef]
- Wolf, B.; Goebel, G.; Hackl, H.; Fiegl, H. Reduced mRNA expression levels of NFE2L2 are associated with poor outcome in breast cancer patients. BMC Cancer 2016, 16, 821. [Google Scholar] [CrossRef]
- Okoh, V.; Deoraj, A.; Roy, D. Estrogen-induced reactive oxygen species-mediated signalings contribute to breast cancer. Biochim. Biophys. Acta 2011, 1815, 115–133. [Google Scholar] [CrossRef]
- Karihtala, P.; Kauppila, S.; Soini, Y.; Arja Jukkola, V. Oxidative stress and counteracting mechanisms in hormone receptor positive, triple-negative and basal-like breast carcinomas. BMC Cancer 2011, 11, 262. [Google Scholar] [CrossRef]
- Asaoka, M.; Ishikawa, T.; Takabe, K.; Patnaik, S.K. APOBEC3-Mediated RNA Editing in Breast Cancer is Associated with Heightened Immune Activity and Improved Survival. Int. J. Mol. Sci. 2019, 20, 5621. [Google Scholar] [CrossRef]
- Okano, M.; Oshi, M.; Butash, A.L.; Katsuta, E.; Tachibana, K.; Saito, K.; Okayama, H.; Peng, X.; Yan, L.; Kono, K.; et al. Triple-Negative Breast Cancer with High Levels of Annexin A1 Expression Is Associated with Mast Cell Infiltration, Inflammation, and Angiogenesis. Int. J. Mol. Sci. 2019, 20, 4197. [Google Scholar] [CrossRef]
- Takahashi, H.; Asaoka, M.; Yan, L.; Rashid, O.M.; Oshi, M.; Ishikawa, T.; Nagahashi, M.; Takabe, K. Biologically Aggressive Phenotype and Anti-cancer Immunity Counterbalance in Breast Cancer with High Mutation Rate. Sci. Rep. 2020, 10, 1852. [Google Scholar] [CrossRef]
- Asaoka, M.; Patnaik, S.K.; Zhang, F.; Ishikawa, T.; Takabe, K. Lymphovascular invasion in breast cancer is associated with gene expression signatures of cell proliferation but not lymphangiogenesis or immune response. Breast Cancer Res. Treat. 2020, 181, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Oshi, M.; Asaoka, M.; Ishikawa, T.; Endo, I.; Takabe, K. ASO Author Reflections: Transitioning From Morphology to Transcriptomics in Capturing Tumor Biology. Ann. Surg. Oncol. 2020, 27, 4486–4487. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Elkhanany, A.; Oshi, M.; Dai, T.; Opyrchal, M.; Mohammadpour, H.; Repasky, E.A.; Takabe, K. Contribution of Immune Cells to Glucocorticoid Receptor Expression in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 4635. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Katsuta, E.; Yan, L.; Ebos, J.M.L.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. A Novel 4-Gene Score to Predict Survival, Distant Metastasis and Response to Neoadjuvant Therapy in Breast Cancer. Cancers 2020, 12, 1148. [Google Scholar] [CrossRef]
- Rueda, O.M.; Sammut, S.J.; Seoane, J.A.; Chin, S.F.; Caswell-Jin, J.L.; Callari, M.; Batra, R.; Pereira, B.; Bruna, A.; Ali, H.R.; et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019, 567, 399–404. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Miao, Y.R.; Zhang, Q.; Lei, Q.; Luo, M.; Xie, G.Y.; Wang, H.; Guo, A.Y. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv. Sci. 2020, 7, 1902880. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef]
- Hiramoto, K.; Satoh, H.; Suzuki, T.; Moriguchi, T.; Pi, J.; Shimosegawa, T.; Yamamoto, M. Myeloid lineage-specific deletion of antioxidant system enhances tumor metastasis. Cancer Prev. Res. 2014, 7, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Maj, T.; Wang, W.; Crespo, J.; Zhang, H.; Wang, W.; Wei, S.; Zhao, L.; Vatan, L.; Shao, I.; Szeliga, W.; et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 2017, 18, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Kuga, A.; Suzuki, M.; Panda, H.; Kitamura, H.; Motohashi, H.; Yamamoto, M. Microenvironmental Activation of Nrf2 Restricts the Progression of Nrf2-Activated Malignant Tumors. Cancer Res. 2020, 80, 3331–3344. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.; Huang, L.; Yi, K.; Chen, H.; Liu, S.; Zhang, W.; Yuan, C.; Song, X.; Wang, F. Co-administration of sulforaphane and doxorubicin attenuates breast cancer growth by preventing the accumulation of myeloid-derived suppressor cells. Cancer Lett. 2020, 493, 189–196. [Google Scholar] [CrossRef]
- Oshi, M.; Asaoka, M.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Ishikawa, T.; Endo, I.; Takabe, K. CD8 T Cell Score as a Prognostic Biomarker for Triple Negative Breast Cancer. Int. J. Mol. Sci. 2020, 21, 6968. [Google Scholar] [CrossRef]
- Narayanan, S.; Kawaguchi, T.; Yan, L.; Peng, X.; Qi, Q.; Takabe, K. Cytolytic Activity Score to Assess Anticancer Immunity in Colorectal Cancer. Ann. Surg. Oncol. 2018, 25, 2323–2331. [Google Scholar] [CrossRef]
- Wakiyama, H.; Masuda, T.; Motomura, Y.; Hu, Q.; Tobo, T.; Eguchi, H.; Sakamoto, K.; Hirakawa, M.; Honda, H.; Mimori, K. Cytolytic Activity (CYT) Score Is a Prognostic Biomarker Reflecting Host Immune Status in Hepatocellular Carcinoma (HCC). Anticancer Res. 2018, 38, 6631–6638. [Google Scholar] [CrossRef]
- Homma, S.; Ishii, Y.; Morishima, Y.; Yamadori, T.; Matsuno, Y.; Haraguchi, N.; Kikuchi, N.; Satoh, H.; Sakamoto, T.; Hizawa, N.; et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin. Cancer Res. 2009, 15, 3423–3432. [Google Scholar] [CrossRef]
- Jiang, T.; Chen, N.; Zhao, F.; Wang, X.J.; Kong, B.; Zheng, W.; Zhang, D.D. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 2010, 70, 5486–5496. [Google Scholar] [CrossRef]
- Shibata, T.; Kokubu, A.; Gotoh, M.; Ojima, H.; Ohta, T.; Yamamoto, M.; Hirohashi, S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 2008, 135, 1358–1368. [Google Scholar] [CrossRef]
- Sun, K.A.; Li, Y.; Meliton, A.Y.; Woods, P.S.; Kimmig, L.M.; Cetin-Atalay, R.; Hamanaka, R.B.; Mutlu, G.M. Endogenous itaconate is not required for particulate matter-induced NRF2 expression or inflammatory response. eLife 2020, 9. [Google Scholar] [CrossRef]
- Yi, Z.; Deng, M.; Scott, M.J.; Fu, G.; Loughran, P.A.; Lei, Z.; Li, S.; Sun, P.; Yang, C.; Li, W.; et al. Immune-Responsive Gene 1/Itaconate Activates Nuclear Factor Erythroid 2-Related Factor 2 in Hepatocytes to Protect Against Liver Ischemia-Reperfusion Injury. Hepatology 2020. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Brueffer, C.; Vallon-Christersson, J.; Grabau, D.; Ehinger, A.; Häkkinen, J.; Hegardt, C.; Malina, J.; Chen, Y.; Bendahl, P.-O.; Malmberg, M.; et al. Clinical Value of RNA Sequencing-Based Classifiers for Prediction of the Five Conventional Breast Cancer Biomarkers: A Report From the Population-Based Multicenter Sweden Cancerome Analysis Network—Breast Initiative. JCO Precis. Oncol. 2018, 2, 1–18. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, l1. [Google Scholar] [CrossRef] [PubMed]
- Hatzis, C.; Pusztai, L.; Valero, V.; Booser, D.J.; Esserman, L.; Lluch, A.; Vidaurre, T.; Holmes, F.; Souchon, E.; Wang, H.; et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA 2011, 305, 1873–1881. [Google Scholar] [CrossRef]
- Popovici, V.; Chen, W.; Gallas, B.G.; Hatzis, C.; Shi, W.; Samuelson, F.W.; Nikolsky, Y.; Tsyganova, M.; Ishkin, A.; Nikolskaya, T.; et al. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. BCR 2010, 12, R5. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.; Eum, H.H.; Lee, H.O.; Lee, K.M.; Lee, H.B.; Kim, K.T.; Ryu, H.S.; Kim, S.; Lee, J.E.; Park, Y.H.; et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017, 8, 15081. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Oshi, M.; Katsuta, E.; Yan, L.; Satyananda, V.; Matsuhashi, N.; Futamura, M.; Akao, Y.; Yoshida, K.; Takabe, K. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am. J. Cancer Res. 2020, 10, 897–907. [Google Scholar]
- Oshi, M.; Takahashi, H.; Tokumaru, Y.; Yan, L.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. G2M Cell Cycle Pathway Score as a Prognostic Biomarker of Metastasis in Estrogen Receptor (ER)-Positive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 2921. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Katsuta, E.; Oshi, M.; Sporn, J.C.; Yan, L.; Le, L.; Matsuhashi, N.; Futamura, M.; Akao, Y.; Yoshida, K.; et al. High Expression of miR-34a Associated with Less Aggressive Cancer Biology but Not with Survival in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 45. [Google Scholar] [CrossRef] [PubMed]
- Tokumaru, Y.; Asaoka, M.; Oshi, M.; Katsuta, E.; Yan, L.; Narayanan, S.; Sugito, N.; Matsuhashi, N.; Futamura, M.; Akao, Y.; et al. High Expression of microRNA-143 is Associated with Favorable Tumor Immune Microenvironment and Better Survival in Estrogen Receptor Positive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 3213. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Oshi, M.; Asaoka, M.; Yan, L.; Endo, I.; Takabe, K. Molecular Biological Features of Nottingham Histological Grade 3 Breast Cancers. Ann. Surg. Oncol. 2020, 27, 4475–4485. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; Rashid, O.M.; Takabe, K. Fibroblasts as a Biological Marker for Curative Resection in Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 3890. [Google Scholar] [CrossRef]
- Oshi, M.; Takahashi, H.; Tokumaru, Y.; Yan, L.; Rashid, O.M.; Nagahashi, M.; Matsuyama, R.; Endo, I.; Takabe, K. The E2F Pathway Score as a Predictive Biomarker of Response to Neoadjuvant Therapy in ER+/HER2- Breast Cancer. Cells 2020, 9, 1643. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Oshi, M.; Katsuta, E.; Yan, L.; Huang, J.L.; Nagahashi, M.; Matsuhashi, N.; Futamura, M.; Yoshida, K.; Takabe, K. Intratumoral Adipocyte-High Breast Cancer Enrich for Metastatic and Inflammation-Related Pathways but Associated with Less Cancer Cell Proliferation. Int. J. Mol. Sci. 2020, 21, 5744. [Google Scholar] [CrossRef]
- Oshi, M.; Newman, S.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Nagahashi, M.; Takabe, K. Intra-Tumoral Angiogenesis Is Associated with Inflammation, Immune Reaction and Metastatic Recurrence in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 6708. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).