Premature MicroRNA-Based Therapeutic: A “One-Two Punch” against Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
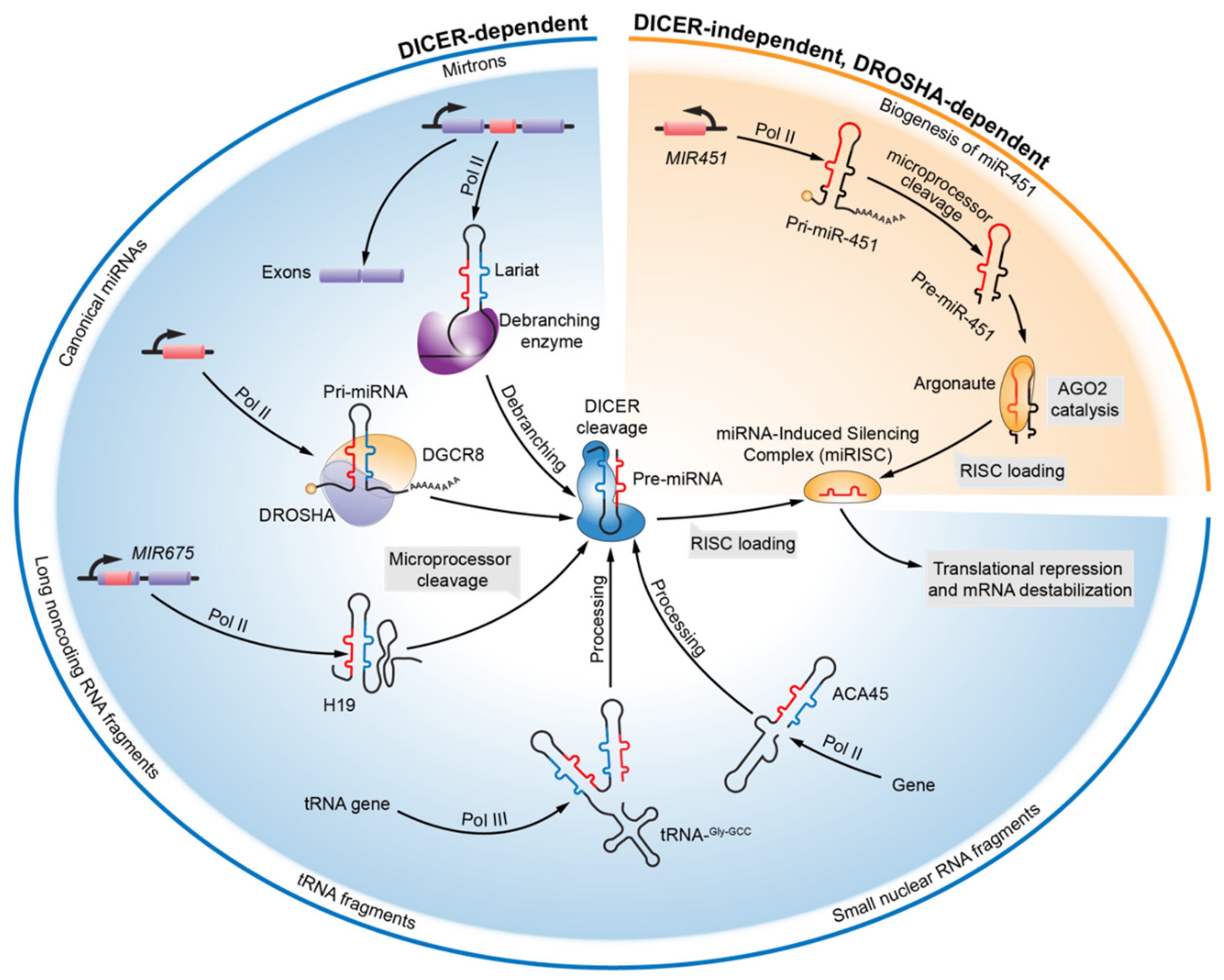
2. Evolving Knowledge of miRNA Biogenesis
3. Expanding miRNA–RNA Regulatory Networks
4. Regulatory Potential of miRNA* Species
5. Caveat and Opportunity over Premature miRNA Perturbance
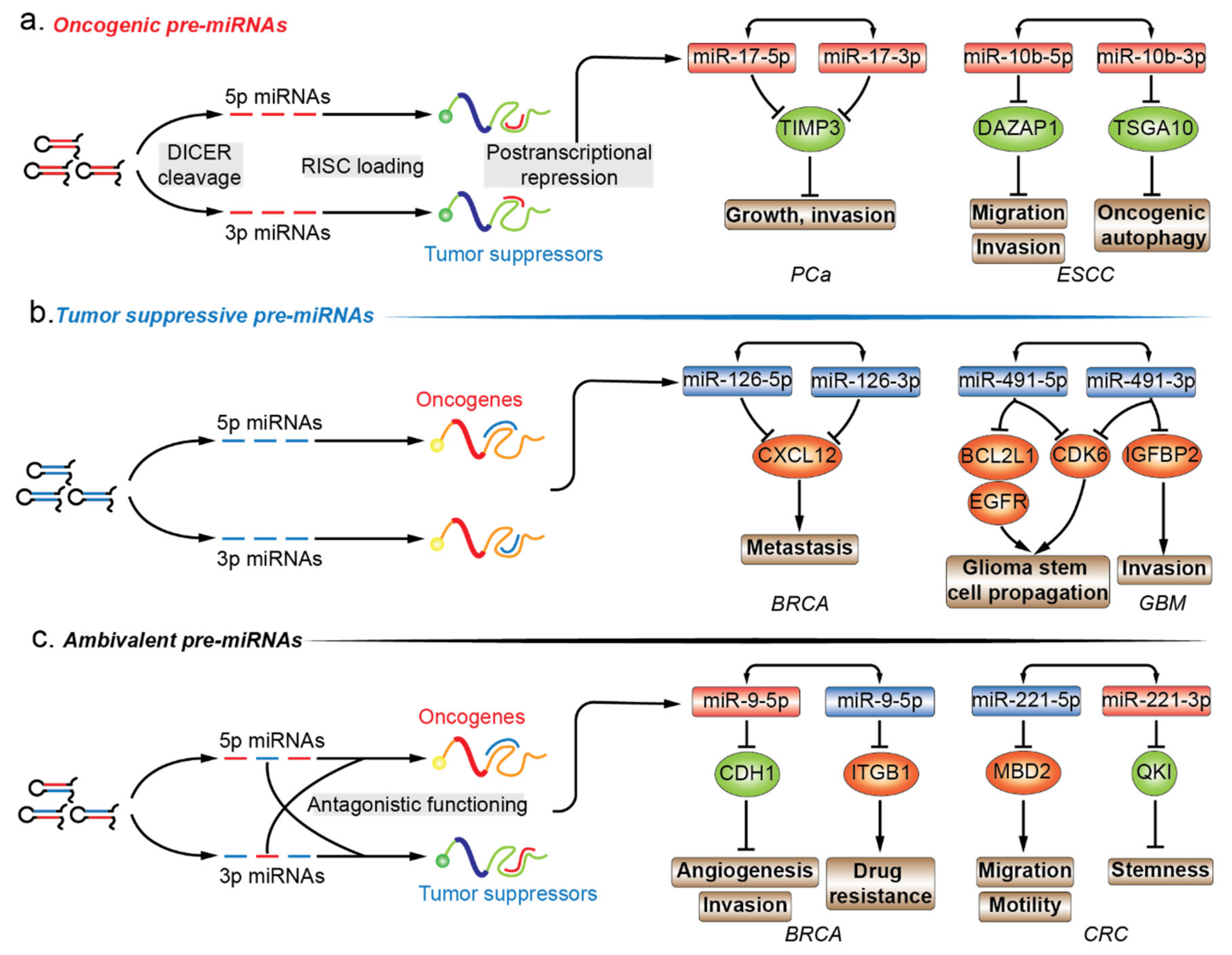
6. Functional Annotation of Premature miRNAs in Cancers
6.1. Oncogenic Pre-miRNAs
6.2. Tumor-Suppressive Pre-miRNAs
6.3. Ambivalent Pre-miRNAs
7. The “Ongoing” Premature miRNA Interventions
8. Conclusion and Future Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.; Tomari, Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. Roles for MicroRNAs in conferring robustness to biological processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef]
- Kosik, K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006, 7, 911–920. [Google Scholar] [CrossRef]
- Small, E.M.; Olson, E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [Green Version]
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–251. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Berindan-Neagoe, I.; Monroig, P.D.C.; Pasculli, B.; Calin, G.A. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014, 64, 311–336. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–221. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagos-Quintana, M. Identification of Novel Genes Coding for Small Expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, É.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [Green Version]
- Han, J. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.S. Functional anatomy of the human microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef] [Green Version]
- Yi, R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [Green Version]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Tambe, A.; Kidwell, M.A.; Noland, C.L.; Schneider, C.P.; Doudna, J.A. Dicer-TRBP complex formation ensures accurate mammalian MicroRNA biogenesis. Mol. Cell 2015, 57, 397–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar] [CrossRef]
- Kwak, P.B.; Tomari, Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012, 19, 145–151. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.S.; Lai, E.C. Alternative miRNA Biogenesis Pathways and the Interpretation of Core miRNA Pathway Mutants. Mol. Cell 2011, 43, 892–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. MiRNA nomenclature: A view incorporating genetic origins, biosynthetic pathways, and sequence variants. Trends Genet. 2015, 31, 613–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berezikov, E.; Chung, W.J.; Willis, J.; Cuppen, E.; Lai, E.C. Mammalian mirtron genes. Mol. Cell 2007, 28, 328–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladewig, E.; Okamura, K.; Flynt, A.S.; Westholm, J.O.; Lai, E.C. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Res. 2012, 22, 1634–1645. [Google Scholar] [CrossRef] [Green Version]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. TRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [Green Version]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with MicroRNA-Like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Cai, X.; Cullen, B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. Rna 2007, 13, 313–316. [Google Scholar] [CrossRef] [Green Version]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D.; et al. A novel miRNA processing pathway independent of dicer requires argonaute2 catalytic activity. Science 2010, 328, 1694–1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.W.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.-S.; Maurin, T.; Robine, N.; Rasmussen, K.D.; Jeffrey, K.L.; Chandwani, R.; Papapetrou, E.P.; Sadelain, M.; O’Carroll, D.; Lai, E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 15163–15168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Sumazin, P.; Yang, X.; Chiu, H.-S.; Chung, W.-J.; Iyer, A.; Llobet-Navas, D.; Rajbhandari, P.; Bansal, M.; Guarnieri, P.; Silva, J.; et al. An extensive MicroRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 2011, 147, 370–381. [Google Scholar] [CrossRef] [Green Version]
- Tay, Y.; Kats, L.; Salmena, L.; Weiss, D.; Tan, S.M.; Ala, U.; Karreth, F.; Poliseno, L.; Provero, P.; Di Cunto, F.; et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011, 147, 344–357. [Google Scholar] [CrossRef] [Green Version]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [Green Version]
- Rutnam, Z.J.; Du, W.W.; Yang, W.; Yang, X.; Yang, B.B. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat. Commun. 2014, 5, 1–15. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Huang, X.; Tan, W.; Yu, D.; Du, Z.; Chang, J.; Wei, L.; Han, Y.; Wang, C.; Che, X.; et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat. Genet. 2016, 48, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Liu, C.G.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Vannini, I.; Wise, P.M.; Challagundla, K.B.; Plousiou, M.; Raffini, M.; Bandini, E.; Fanini, F.; Paliaga, G.; Crawford, M.; Ferracin, M.; et al. Transcribed ultraconserved region 339 promotes carcinogenesis by modulating tumor suppressor microRNAs. Nat. Commun. 2017, 8, 1–19. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Cheetham, S.W.; Faulkner, G.J.; Dinger, M.E. Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat. Rev. Genet. 2020, 21, 191–201. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Terracciano, D.; Terreri, S.; De Nigris, F.; Costa, V.; Calin, G.A.; Cimmino, A. The role of a new class of long noncoding RNAs transcribed from ultraconserved regions in cancer. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 449–455. [Google Scholar] [CrossRef]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ro, S.; Park, C.; Young, D.; Sanders, K.M.; Yan, W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007, 35, 5944–5953. [Google Scholar] [CrossRef] [PubMed]
- Schulte, J.H.; Marschall, T.; Martin, M.; Rosenstiel, P.; Mestdagh, P.; Schlierf, S.; Thor, T.; Vandesompele, J.; Eggert, A.; Schreiber, S.; et al. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res. 2010, 38, 5919–5928. [Google Scholar] [CrossRef] [Green Version]
- Okamura, K.; Phillips, M.D.; Tyler, D.M.; Duan, H.; Chou, Y.T.; Lai, E.C. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat. Struct. Mol. Biol. 2008, 15, 354–363. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Phillips, M.D.; Betel, D.; Mu, P.; Ventura, A.; Siepel, A.C.; Chen, K.C.; Lai, E.C. Widespread regulatory activity of vertebrate microRNA* species. RNA 2011, 17, 312–326. [Google Scholar] [CrossRef] [Green Version]
- Medley, J.C.; Panzade, G.; Zinovyeva, A.Y. microRNA strand selection: Unwinding the rules. WIREs RNA 2020, 1–22. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.Y.; Yan, Z.; Xu, Y.; Hu, H.; Menzel, C.; Zhou, Y.H.; Chen, W.; Khaitovich, P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics 2009, 10, 413. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, 68–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Du, W.W.; Li, H.; Liu, F.; Khorshidi, A.; Rutnam, Z.J.; Yang, B.B. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013, 41, 9688–9704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, S.W.; Fang, L.; Shatseva, T.; Rutnam, Z.J.; Yang, X.; Du, W.; Lu, W.-Y.; Xuan, J.W.; Deng, Z.; Yang, B.B. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J. Cell Sci. 2013, 126, 1517–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Lu, Y.; Ren, Y.; Yuan, J.; Zhang, N.; Kimball, H.; Zhou, L.; Yang, M. Starvation-induced suppression of DAZAP1 by miR-10b integrates splicing control into TSC2-regulated oncogenic autophagy in esophageal squamous cell carcinoma. Theranostics 2020, 10, 4983–4996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, J.; Fu, Z.; Dong, L.; Tang, Y.; Xu, C.; Wang, H.; Zhang, T.; Wu, Y.; Dong, C.; et al. Hypoxia-induced microRNA-10b-3p promotes esophageal squamous cell carcinoma growth and metastasis by targeting TSGA10. Aging 2019, 11, 10374–10384. [Google Scholar] [CrossRef]
- Shao, N.; Ma, G.; Zhang, J.; Zhu, W. MIR-221-5p enhances cell proliferation and metastasis through post-transcriptional regulation of SOCS1 in human prostate cancer. BMC Urol. 2018, 18, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Wang, X.; He, H.H.; Sweeney, C.J.; Liu, S.X.; Brown, M.; Balk, S.; Lee, G.S.; Kantoff, P.W. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene 2014, 33, 2790–2800. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yang, P.; Sun, T.; Li, D.; Xu, X.; Rui, Y.; Li, C.; Chong, M.; Ibrahim, T.; Mercatali, L.; et al. MiR-126 and miR-126 * repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat. Cell Biol. 2013, 15, 284–294. [Google Scholar] [CrossRef] [Green Version]
- Uchida, A.; Seki, N.; Mizuno, K.; Misono, S.; Yamada, Y.; Kikkawa, N.; Sanada, H.; Kumamoto, T.; Suetsugu, T.; Inoue, H. Involvement of dual-strand of the miR-144 duplex and their targets in the pathogenesis of lung squamous cell carcinoma. Cancer Sci. 2019, 110, 420–432. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Wang, Q.; Wang, Y.; Huang, K.; Yang, C.; Li, Y.; Yi, K.; Kang, C. EGFR/c-myc axis regulates TGFβ/Hippo/Notch pathway via epigenetic silencing miR-524 in gliomas. Cancer Lett. 2017, 406, 12–21. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Granberg, K.J.; Wang, Q.; Moore, L.M.; Ji, P.; Gumin, J.; Sulman, E.P.; Calin, G.A.; Haapasalo, H.; et al. Two mature products of MIR-491 coordinate to suppress key cancer hallmarks in glioblastoma. Oncogene 2015, 34, 1619–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.J.; Tsai, M.M.; Hung, P.S.; Kao, S.Y.; Liu, T.Y.; Wu, K.J.; Chiou, S.H.; Lin, S.C.; Chang, K.W. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010, 70, 1635–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, K.W.; Kao, S.Y.; Wu, Y.H.; Tsai, M.M.; Tu, H.F.; Liu, C.J.; Lui, M.T.; Lin, S.C. Passenger strand miRNA miR-31 regulates the phenotypes of oral cancer cells by targeting RhoA. Oral Oncol. 2013, 49, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. MiR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zawistowski, J.S.; Nakamura, K.; Parker, J.S.; Granger, D.A.; Golitz, B.T.; Johnson, G.L. MicroRNA 9-3p targets 1 integrin to sensitize claudin-low breast cancer cells to MEK inhibition. Mol. Cell. Biol. 2013, 33, 2260–2274. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef]
- Biagioni, F.; Bossel Ben-Moshe, N.; Fontemaggi, G.; Canu, V.; Mori, F.; Antoniani, B.; Di Benedetto, A.; Santoro, R.; Germoni, S.; De Angelis, F.; et al. miR-10b *, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol. Med. 2012, 4, 1214–1229. [Google Scholar] [CrossRef]
- Yuan, K.; Xie, K.; Fox, J.; Zeng, H.; Gao, H.; Huang, C.; Wu, M. Decreased levels of mir-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology 2013, 145, 853–864.e9. [Google Scholar] [CrossRef] [Green Version]
- Mukohyama, J.; Isobe, T.; Hu, Q.; Hayashi, T.; Watanabe, T.; Maeda, M.; Yanagi, H.; Qian, X.; Yamashita, K.; Minami, H.; et al. MiR-221 targets QKI to enhance the tumorigenic capacity of human colorectal cancer stem cells. Cancer Res. 2019, 79, 5151–5158. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef]
- Chen, L.; Kang, C. miRNA interventions serve as ‘magic bullets’ in the reversal of glioblastoma hallmarks. Oncotarget 2015, 6, 38628–38642. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 2007, 26, 745–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Rooij, E.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Hill, J.; Olson, E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef] [Green Version]
- Xiao, C.; Calado, D.P.; Galler, G.; Thai, T.H.; Patterson, H.C.; Wang, J.; Rajewsky, N.; Bender, T.P.; Rajewsky, K. MiR-150 controls B Cell differentiation by targeting the transcription factor c-Myb. Cell 2007, 131, 146–159. [Google Scholar] [CrossRef] [Green Version]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Hirata, Y.; Murai, N.; Yanaihara, N.; Saito, M.; Saito, M.; Urashima, M.; Murakami, Y.; Matsufuji, S.; Okamoto, A. MicroRNA-21 is a candidate driver gene for 17q23-25 amplification in ovarian clear cell carcinoma. BMC Cancer 2014, 14, 799. [Google Scholar] [CrossRef] [Green Version]
- Lou, Y.; Yang, X.; Wang, F.; Cui, Z.; Huang, Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int. J. Mol. Med. 2010, 26, 521–527. [Google Scholar] [CrossRef] [Green Version]
- Pink, R.C.; Samuel, P.; Massa, D.; Caley, D.P.; Brooks, S.A.; Carter, D.R.F. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecol. Oncol. 2015, 137, 143–151. [Google Scholar] [CrossRef]
- Chan, J.K.; Blansit, K.; Kiet, T.; Sherman, A.; Wong, G.; Earle, C.; Bourguignon, L.Y.W. The inhibition of miR-21 promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol. Oncol. 2014, 132, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Javanmardi, S.; Tamaddon, A.M.; Aghamaali, M.R.; Ghahramani, L.; Abolmaali, S.S. Redox-sensitive, PEG-shielded carboxymethyl PEI nanogels silencing MicroRNA-21, sensitizes resistant ovarian cancer cells to cisplatin. Asian J. Pharm. Sci. 2020, 15, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Echevarría-Vargas, I.M.; Valiyeva, F.; Vivas-Mejía, P.E. Upregulation of miR-21 in cisplatin resistant ovarian cancer via JNK-1/c-Jun pathway. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Li, Y.F.; Ma, H.; Gao, Y.H. Regulation of MYB mediated cisplatin resistance of ovarian cancer cells involves miR-21-wnt signaling axis. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Wang, Q.; Balk, S.; Brown, M.; Lee, G.S.M.; Kantoff, P. The role of microrna-221 and microrna-222 in Androgen- independent prostate cancer cell lines. Cancer Res. 2009, 69, 3356–3363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Wu, J.; Wu, S.H.; Thakur, A.; Bollig, A.; Huang, Y.; Joshua Liao, D. Expression profile of microRNAs in c-Myc induced mouse mammary tumors. Breast Cancer Res. Treat. 2009, 118, 185–196. [Google Scholar] [CrossRef] [Green Version]
- Carrio, M.; Arderiu, G.; Myers, C.; Boudreau, N.J. Homeobox D10 induces phenotypic reversion of breast tumor cells in a three-dimensional culture model. Cancer Res. 2005, 65, 7177–7185. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; Van Der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [Green Version]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef] [PubMed]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Guennewig, B.; Roos, M.; Dogar, A.M.; Gebert, L.F.R.; Zagalak, J.A.; Vongrad, V.; Metzner, K.J.; Hall, J. Synthetic pre-microRNAs reveal dual-strand activity of miR-34a on TNF-α. Rna 2014, 20, 61–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Berg, F.T.; Rossi, J.J.; Arbuthnot, P.; Weinberg, M.S. Design of Effective Primary MicroRNA Mimics With Different Basal Stem Conformations. Mol. Ther. Nucleic Acids 2016, 5, e278. [Google Scholar] [CrossRef] [Green Version]
- Velagapudi, S.P.; Gallo, S.M.; Disney, M.D. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol. 2014, 10, 291–297. [Google Scholar] [CrossRef]
- Gumireddy, K.; Young, D.D.; Xiong, X.; Hogenesch, J.B.; Huang, Q.; Deiters, A. Small-molecule inhibitors of microRNA miR-21 function. Angew. Chemie Int. Ed. 2008, 47, 7482–7484. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.; Zhang, J.; Qian, X.; Han, L.; Zhang, K.; Chen, L.; Liu, J.; Ren, Y.; Yang, M.; Zhang, A.; et al. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013, 73, 5519–5531. [Google Scholar] [CrossRef] [Green Version]
- Melo, S.; Villanueva, A.; Moutinho, C.; Davalos, V.; Spizzo, R.; Ivan, C.; Rossi, S.; Setien, F.; Casanovas, O.; Simo-Riudalbas, L.; et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. USA 2011, 108, 4394–4399. [Google Scholar] [CrossRef] [Green Version]
- Doudna, J.A. The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Jarquin, G. Emerging role of CRISPR/Cas9 technology for MicroRNAs editing in cancer research. Cancer Res. 2017, 77, 6812–6817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| RNA Species | Decoy RNAs | Target miRNA(s) | Ref. |
|---|---|---|---|
| Pseudogenes | PTENP1 | miR-20a, miR-19b, miR-21, miR-26a, and miR-214 | [48] |
| TUSC2P | miR-17, miR-93, miR-299-3p, miR-520a, miR-608, and miR-661 | [49] | |
| LincRNAs | LincRNA-p21 | let-7 | [50] |
| Linc-RoR | miR-145-5p, miR-181a-5p, and miR-99b-3p | [51] | |
| Linc00673 mutant | miR-1231 | [52] | |
| T-UCRs | Uc.160 | miR-155, miR-24 | [53] |
| Uc.346A | miR-155 | ||
| Uc.348 | miR-29b | ||
| Uc.339 | miR-339-3p, miR-663b-3p, and miR-95-5p | [54] | |
| CircRNAs | ciRS-7 | miR-7 | [55] |
| Sry | miR-138 | ||
| CDR1as | miR-7 | [56] |
| Annotation | Mature Products | Previous ID | Clustered miRNAs | Type of Cancer | Targets | Biological Implications | Ref. |
|---|---|---|---|---|---|---|---|
| Oncogenic pre-miRNAs | miR-17-5p/3p | miR-17/miR-17 * | miR-17~92 | Prostate cancer | TIMP3 | Growth↑, invasion↑ | [72] |
| Hepatocellular carcinoma | PTEN, GalNT7, VIM | Proliferation↑, migration↑ | [73] | ||||
| miR-10b-5p/3p | miR-10b/miR-10b * | _ | Esophageal squamous cell carcinoma | DAZAP1, TSGA10 | Oncogenic autophagy↑, growth↑, metastasis↑ | [74,75] | |
| miR-221-5p/3p | miR-221 */miR-221 | miR-222/221 | Prostate cancer | SOCS1, HECTD2, RAB1A | Proliferation↑, migration↑, androgen independent growth↑ | [76,77] | |
| Tumor-suppressive pre-miRNAs | miR-126-5p/3p | miR-126 */miR-126 | _ | Breast cancer | CXCL12 | Metastasis↓ | [78] |
| miR-144-5p/3p | miR-144 */miR-144 | miR-4732~451a | Lung squamous cell carcinoma | NCS1 | Proliferation↓, invasion↓, migration↓ | [79] | |
| miR-524-5p/3p | miR-524 */miR-524 | miR-520b~520d | Glioblastoma | TEAD1, SMAD2, HES1 | Migration↓, proliferation↓ | [80] | |
| miR-491-5p/3p | miR-491/- | _ | Glioblastoma | BCL2L1, EGFR, CDK6, IGFBP2 | Proliferation↓, invasion↓, glioma stem cell propagation↓ | [81] | |
| Ambivalent pre-miRNAs | miR-31-5p/3p | miR-31/miR-31 * | _ | Oral squamous cell carcinoma | FIH, RHOA | Viability↑, migration↑↓, growth↓ | [82,83] |
| miR-9-5p/3p | miR-9/miR-9 * | _ | Breast cancer | CDH1, ITGB1 | Angiogenesis↑, motility↑, invasion↑, drug resistance↓ | [84,85] | |
| miR-10b-5p/3p | miR-10b/miR-10b * | _ | Breast cancer | HOXD10, BUB1, PLK1, CCNA2 | Metastasis↑, cell cycle regulation↓, proliferation↓ | [86,87] | |
| miR-221-5p/3p | miR-221*/miR-221 | miR-222/221 | Colorectal cancer | MBD2, QKI | Migration↓, motility↓, stemness↑ | [88,89] |
| Intervention/Treatment (Company) | Target miRNA | Oligonucleotide Format | Condition(s) or Disease(s) | Stage | Clinical Trials.gov Identifier: | Ref. |
|---|---|---|---|---|---|---|
| Mature miRNA-based strategies | ||||||
| Miravirsen (Santaris Pharma A/S) | miR-122 | Locked Nucleic Acid-modified DNA phosphorothioate antisense oligonucleotide | Hepatitis C | Phase 2a | NCT01200420 | [112] |
| MRX34 (Mirna Therapeutics, Inc.) | miR-34a | Liposomal mimic | Primary liver cancer Lymphoma Melanoma Multiple myeloma Renal cell carcinoma Lung cancer | Phase 1 | NCT01829971 | [113] |
| TargomiRs (EnGeneIC Limited) | miR-16 | Minicells (EnGeneIC Dream Vectors) loaded with mimic microRNA | Malignant pleural mesothelioma Nonsmall cell lung cancer | Phase 1 | NCT02369198 | [114] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Huang, K.; Yi, K.; Huang, Y.; Tian, X.; Kang, C. Premature MicroRNA-Based Therapeutic: A “One-Two Punch” against Cancers. Cancers 2020, 12, 3831. https://doi.org/10.3390/cancers12123831
Chen L, Huang K, Yi K, Huang Y, Tian X, Kang C. Premature MicroRNA-Based Therapeutic: A “One-Two Punch” against Cancers. Cancers. 2020; 12(12):3831. https://doi.org/10.3390/cancers12123831
Chicago/Turabian StyleChen, Luyue, Kai Huang, Kaikai Yi, Yanlin Huang, Xinhua Tian, and Chunsheng Kang. 2020. "Premature MicroRNA-Based Therapeutic: A “One-Two Punch” against Cancers" Cancers 12, no. 12: 3831. https://doi.org/10.3390/cancers12123831




