Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
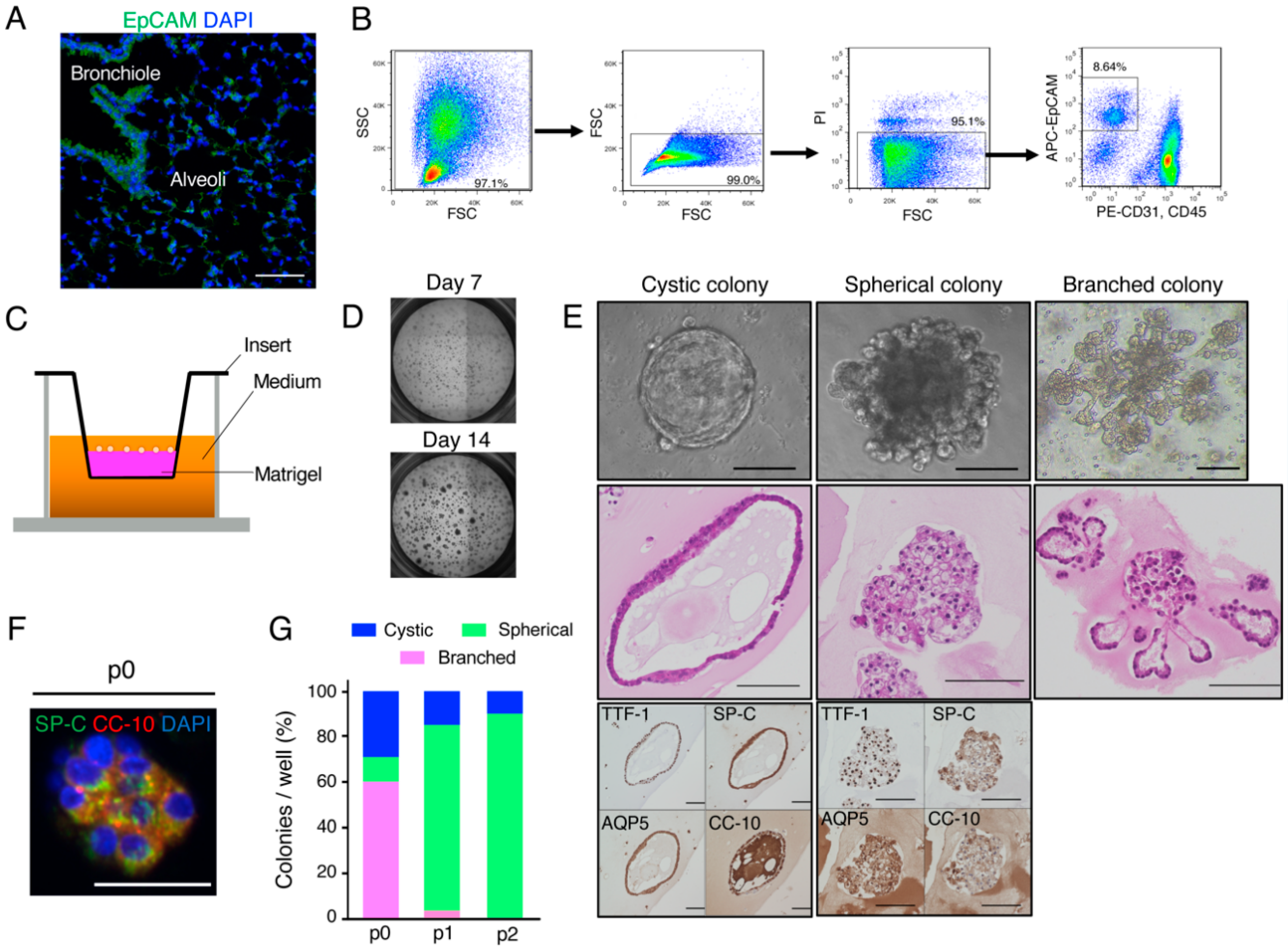
2.1. Organoid Culture of Murine Lung Epithelial Cells
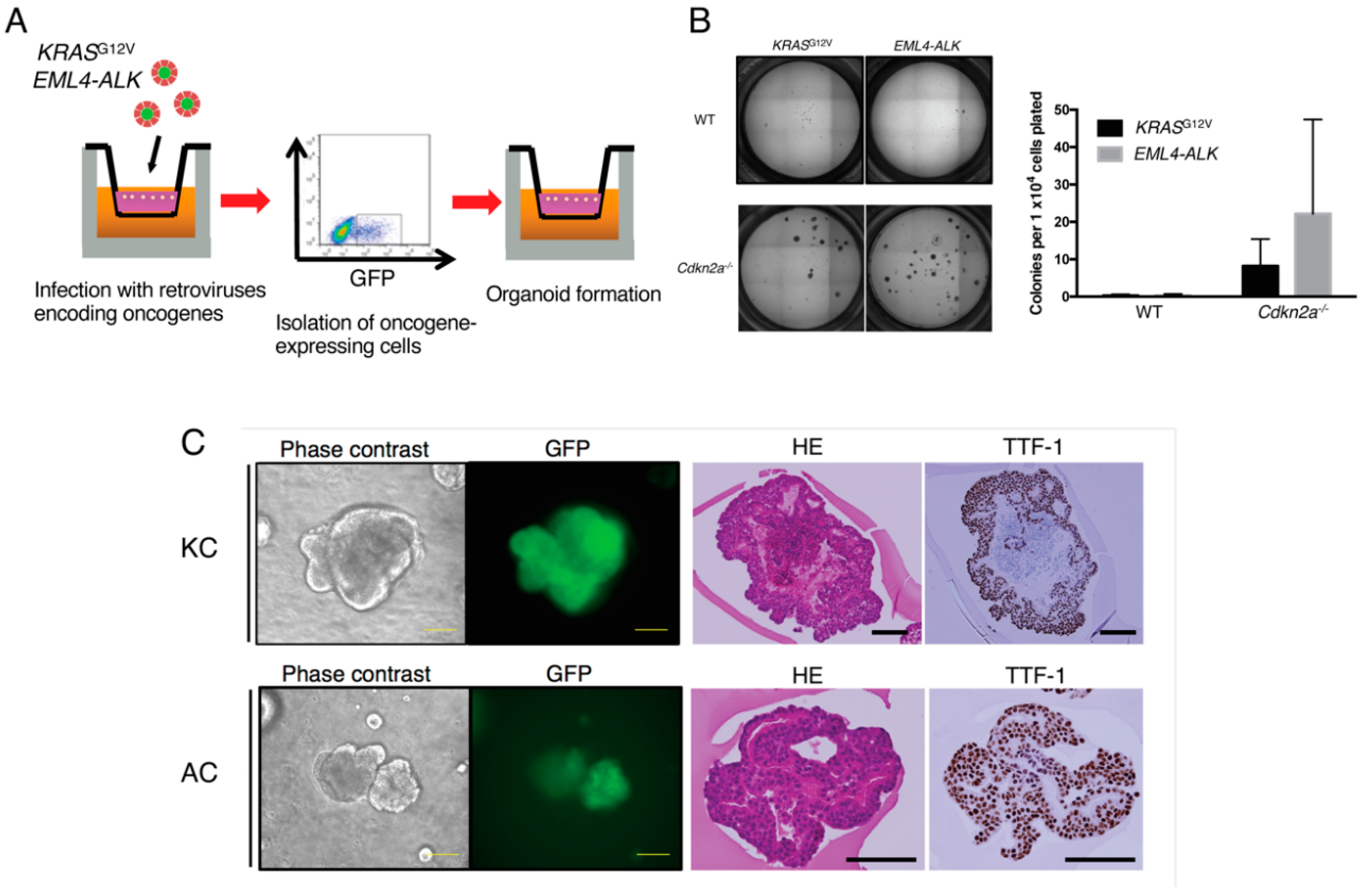
2.2. Expansion of Oncogene-Transformed Lung Epithelial Cells Supported by Cdkn2a Knockout
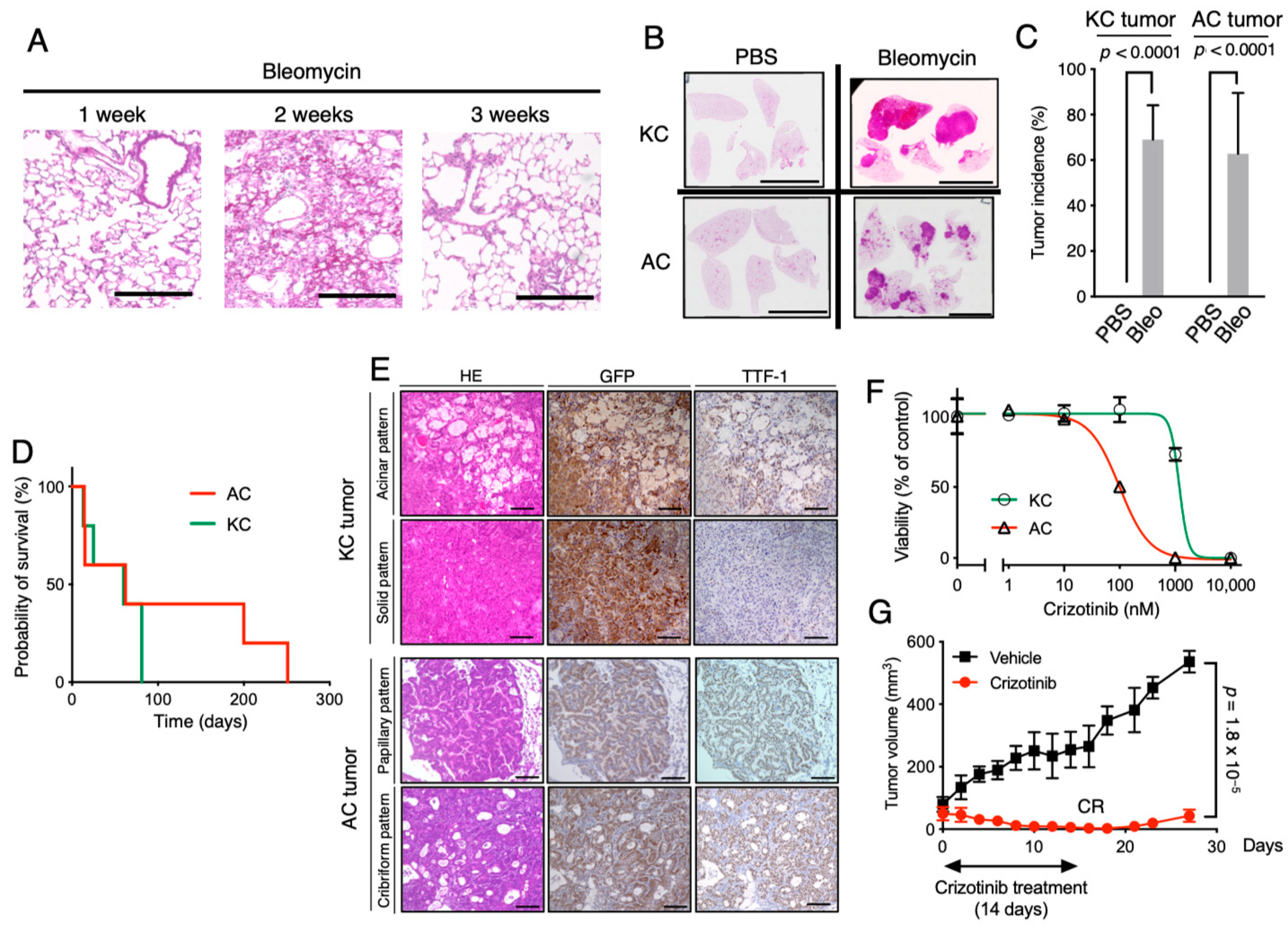
2.3. Establishment of Clinically Relevant Syngeneic Mouse Models of LUAD
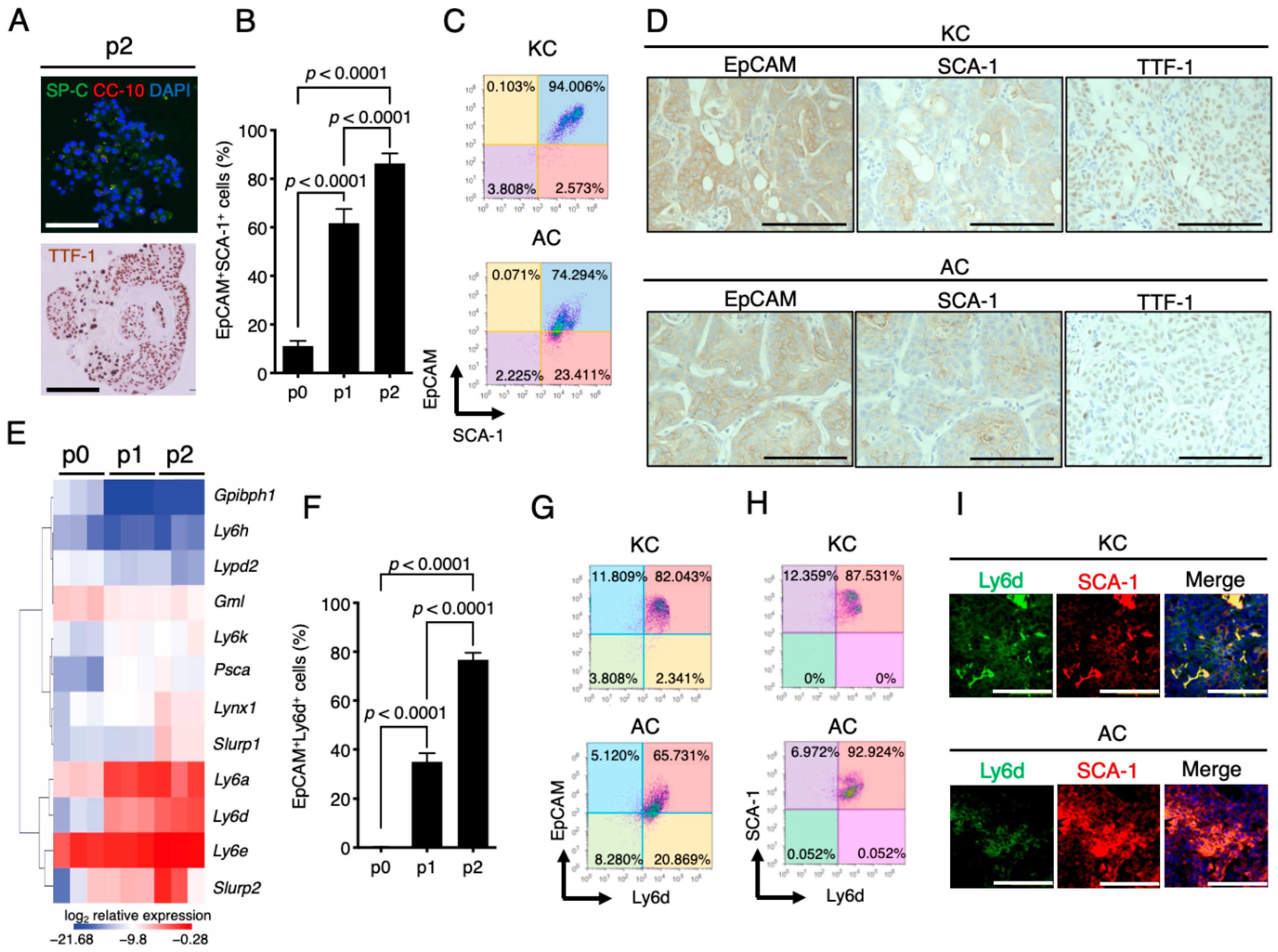
2.4. Stem Cell Properties of KC and AC Cells and Similar Expression Patterns of SCA-1 (Ly6a) and Ly6d in Mouse LUAD-Initiating Cells
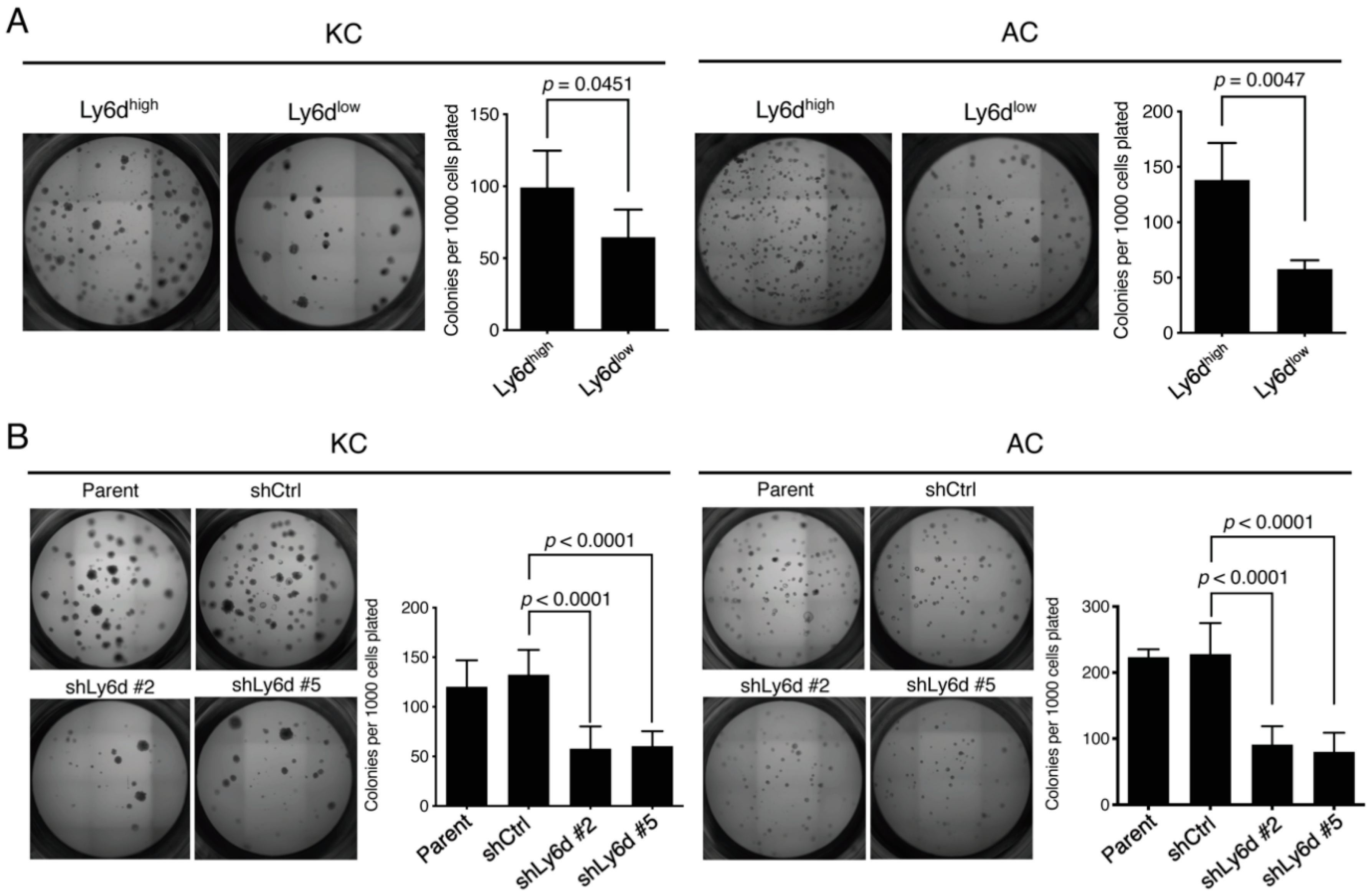
2.5. Ly6d Is Required for Colony Formation by Oncogene-Expressing Lung Epithelial Cells in 3D Culture
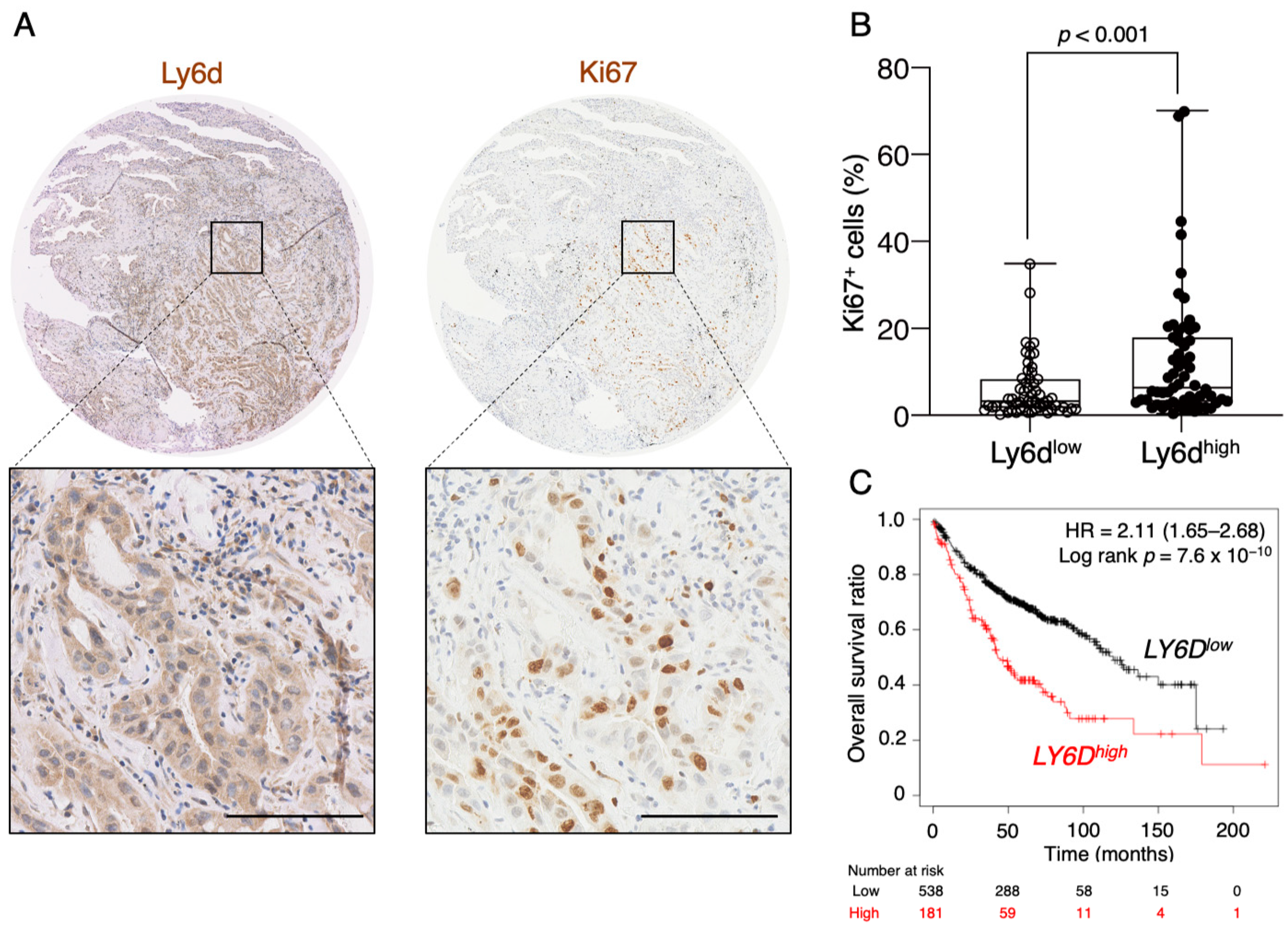
2.6. Expression of Ly6d Is Associated with Poor Prognosis in LUAD Patients
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Isolation of Lung Epithelial Cells from Mice
4.3. Culture of Lung Cells
4.4. Retrovirus-Mediated Oncogene Transduction
4.5. Animal Experiments
4.6. Histological Analysis and Immunostaining
4.7. Flow Cytometry
4.8. Cell Viability Assay
4.9. RNA Interference
4.10. RT-qPCR Analysis
4.11. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Reck, M.; Rabe, K.F. Precision Diagnosis and Treatment for Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 849–861. [Google Scholar] [CrossRef]
- Dela Cruz, C.S.; Tanoue, L.T.; Matthay, R.A. Lung cancer: Epidemiology, etiology, and prevention. Clin. Chest. Med. 2011, 32, 605–644. [Google Scholar] [CrossRef]
- Hammerman, P.S.; Lawrence, M.S.; Voet, D.; Jing, R.; Cibulskis, K.; Sivachenko, A.; Stojanov, P.; McKenna, A.; Lander, E.S.; Gabriel, S.; et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar]
- Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretić, L.; Kong, G.; Leenders, F.; Lu, X.; Fernández-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef]
- Paez, J.G.; Janne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef]
- Kwak, E.L.; Bang, Y.J.; Camidge, D.R.; Shaw, A.T.; Solomon, B.; Maki, R.G.; Ou, S.H.; Dezube, B.J.; Janne, P.A.; Costa, D.B.; et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010, 363, 1693–1703. [Google Scholar] [CrossRef]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nature Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Altorki, N.K.; Markowitz, G.J.; Gao, D.; Port, J.L.; Saxena, A.; Stiles, B.; McGraw, T.; Mittal, V. The lung microenvironment: An important regulator of tumour growth and metastasis. Nat. Rev. Cancer 2019, 19, 9–31. [Google Scholar] [CrossRef] [PubMed]
- Kellar, A.; Egan, C.; Morris, D. Preclinical Murine Models for Lung Cancer: Clinical Trial Applications. Biomed. Res. Int. 2015, 2015, 621324. [Google Scholar] [CrossRef]
- Kwon, M.-c.; Berns, A. Mouse models for lung cancer. Mol. Oncol. 2013, 7, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Guerin, M.V.; Finisguerra, V.; Van den Eynde, B.J.; Bercovici, N.; Trautmann, A. Preclinical murine tumor models: A structural and functional perspective. Elife 2020, 9, e50740. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ishikawa, T.; Sugihara, E.; Kuninaka, S.; Miyamoto, T.; Mabuchi, Y.; Matsuzaki, Y.; Tsunoda, T.; Miya, F.; Morioka, H.; et al. c-MYC overexpression with loss of Ink4a/Arf transforms bone marrow stromal cells into osteosarcoma accompanied by loss of adipogenesis. Oncogene 2010, 29, 5687–5699. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, E.; Shimizu, T.; Kojima, K.; Onishi, N.; Kai, K.; Ishizawa, J.; Nagata, K.; Hashimoto, N.; Honda, H.; Kanno, M.; et al. Ink4a and Arf are crucial factors in the determination of the cell of origin and the therapeutic sensitivity of Myc-induced mouse lymphoid tumor. Oncogene 2012, 31, 2849–2861. [Google Scholar] [CrossRef][Green Version]
- Sampetrean, O.; Saga, I.; Nakanishi, M.; Sugihara, E.; Fukaya, R.; Onishi, N.; Osuka, S.; Akahata, M.; Kai, K.; Sugimoto, H.; et al. Invasion Precedes Tumor Mass Formation in a Malignant Brain Tumor Model of Genetically Modified Neural Stem Cells. Neoplasia 2011, 13, 784-IN3. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Shimizu, T.; Naoe, H.; Ueki, A.; Ishizawa, J.; Chiyoda, T.; Onishi, N.; Sugihara, E.; Nagano, O.; Banno, K.; et al. Establishment of a choriocarcinoma model from immortalized normal extravillous trophoblast cells transduced with HRASV12. Am. J. Pathol. 2011, 179, 1471–1482. [Google Scholar] [CrossRef]
- Takamoto, Y.; Arima, Y.; Saya, H. Development of mouse models of malignant phyllodes tumors by transplantation of syngeneic mammary gland cells expressing mutant H-Ras. Genes Cells 2016, 21, 1244–1252. [Google Scholar] [CrossRef][Green Version]
- Motohara, T.; Masuko, S.; Ishimoto, T.; Yae, T.; Onishi, N.; Muraguchi, T.; Hirao, A.; Matsuzaki, Y.; Tashiro, H.; Katabuchi, H.; et al. Transient depletion of p53 followed by transduction of c-Myc and K-Ras converts ovarian stem-like cells into tumor-initiating cells. Carcinogenesis 2011, 32, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, A.; Semba, T.; Sato, R.; Nobusue, H.; Sugihara, E.; Takaishi, H.; Kanai, T.; Saya, H.; Arima, Y. Oncogenic KRAS-expressing organoids with biliary epithelial stem cell properties give rise to biliary tract cancer in mice. Cancer Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.F.B.; Jackson, E.L.; Woolfenden, A.E.; Lawrence, S.; Babar, I.; Vogel, S.; Crowley, D.; Bronson, R.T.; Jacks, T. Identification of Bronchioalveolar Stem Cells in Normal Lung and Lung Cancer. Cell 2005, 121, 823–835. [Google Scholar] [CrossRef] [PubMed]
- McQualter, J.L.; Yuen, K.; Williams, B.; Bertoncello, I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc. Natl. Acad. Sci. USA 2010, 107, 1414–1419. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Bhang, D.H.; Beede, A.; Huang, T.L.; Stripp, B.R.; Bloch, K.D.; Wagers, A.J.; Tseng, Y.H.; Ryeom, S.; Kim, C.F. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 2014, 156, 440–455. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Lin, A.W.; McCurrach, M.E.; Beach, D.; Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88, 593–602. [Google Scholar] [CrossRef]
- Sharpless, N.E.; Bardeesy, N.; Lee, K.H.; Carrasco, D.; Castrillon, D.H.; Aguirre, A.J.; Wu, E.A.; Horner, J.W.; DePinho, R.A. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 2001, 413, 86–91. [Google Scholar] [CrossRef]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Peeper, D.S. The essence of senescence. Genes Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef]
- Seike, M.; Gemma, A.; Hosoya, Y.; Hemmi, S.; Taniguchi, Y.; Fukuda, Y.; Yamanaka, N.; Kudoh, S. Increase in the frequency of p16INK4 gene inactivation by hypermethylation in lung cancer during the process of metastasis and its relation to the status of p53. Clin. Cancer Res. 2000, 6, 4307–4313. [Google Scholar]
- Pyfferoen, L.; Brabants, E.; Everaert, C.; De Cabooter, N.; Heyns, K.; Deswarte, K.; Vanheerswynghels, M.; De Prijck, S.; Waegemans, G.; Dullaers, M.; et al. The transcriptome of lung tumor-infiltrating dendritic cells reveals a tumor-supporting phenotype and a microRNA signature with negative impact on clinical outcome. Oncoimmunology 2017, 6, e1253655. [Google Scholar] [CrossRef]
- Moll, H.P.; Mohrherr, J.; Breitenecker, K.; Haber, M.; Voronin, V.; Casanova, E. Orthotopic Transplantation of Syngeneic Lung Adenocarcinoma Cells to Study PD-L1 Expression. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [PubMed]
- Semba, T.; Sugihara, E.; Kamoshita, N.; Ueno, S.; Fukuda, K.; Yoshino, M.; Takao, K.; Yoshikawa, K.; Izuhara, K.; Arima, Y.; et al. Periostin antisense oligonucleotide suppresses bleomycin-induced formation of a lung premetastatic niche for melanoma. Cancer Sci. 2018, 109, 1447–1454. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.M.; Lawson, W.E.; Oury, T.D.; Sisson, T.H.; Raghavendran, K.; Hogaboam, C.M. Animal models of fibrotic lung disease. Am. J. Respir. Cell Mol. Biol. 2013, 49, 167–179. [Google Scholar] [CrossRef]
- Solomon, B.J.; Mok, T.; Kim, D.W.; Wu, Y.L.; Nakagawa, K.; Mekhail, T.; Felip, E.; Cappuzzo, F.; Paolini, J.; Usari, T.; et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 2014, 371, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.B.; Kobayashi, S.; Pandya, S.S.; Yeo, W.L.; Shen, Z.; Tan, W.; Wilner, K.D. CSF concentration of the anaplastic lymphoma kinase inhibitor crizotinib. J. Clin. Oncol. 2011, 29, e443–e445. [Google Scholar] [CrossRef]
- Hrustanovic, G.; Olivas, V.; Pazarentzos, E.; Tulpule, A.; Asthana, S.; Blakely, C.M.; Okimoto, R.A.; Lin, L.; Neel, D.S.; Sabnis, A.; et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat. Med. 2015, 21, 1038–1047. [Google Scholar] [CrossRef]
- Leeman, K.T.; Pessina, P.; Lee, J.H.; Kim, C.F. Mesenchymal Stem Cells Increase Alveolar Differentiation in Lung Progenitor Organoid Cultures. Sci. Rep. 2019, 9, 6479. [Google Scholar] [CrossRef]
- Grange, C.; Lanzardo, S.; Cavallo, F.; Camussi, G.; Bussolati, B. Sca-1 identifies the tumor-initiating cells in mammary tumors of BALB-neuT transgenic mice. Neoplasia 2008, 10, 1433–1443. [Google Scholar] [CrossRef] [PubMed]
- Curtis, S.J.; Sinkevicius, K.W.; Li, D.; Lau, A.N.; Roach, R.R.; Zamponi, R.; Woolfenden, A.E.; Kirsch, D.G.; Wong, K.K.; Kim, C.F. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell 2010, 7, 127–133. [Google Scholar] [CrossRef]
- Park, J.W.; Park, J.M.; Park, D.M.; Kim, D.Y.; Kim, H.K. Stem Cells Antigen-1 Enriches for a Cancer Stem Cell-Like Subpopulation in Mouse Gastric Cancer. Stem Cells 2016, 34, 1177–1187. [Google Scholar] [CrossRef][Green Version]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Loughner, C.L.; Bruford, E.A.; McAndrews, M.S.; Delp, E.E.; Swamynathan, S.; Swamynathan, S.K. Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum. Genom. 2016, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, G. Emerging Role of Lymphocyte Antigen-6 Family of Genes in Cancer and Immune Cells. Front. Immunol. 2019, 10, 819. [Google Scholar] [CrossRef] [PubMed]
- Inlay, M.A.; Bhattacharya, D.; Sahoo, D.; Serwold, T.; Seita, J.; Karsunky, H.; Plevritis, S.K.; Dill, D.L.; Weissman, I.L. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009, 23, 2376–2381. [Google Scholar] [CrossRef]
- Rodrigues, P.F.; Alberti-Servera, L.; Eremin, A.; Grajales-Reyes, G.E.; Ivanek, R.; Tussiwand, R. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 2018, 19, 711–722. [Google Scholar] [CrossRef]
- Barros-Silva, J.D.; Linn, D.E.; Steiner, I.; Guo, G.; Ali, A.; Pakula, H.; Ashton, G.; Peset, I.; Brown, M.; Clarke, N.W.; et al. Single-Cell Analysis Identifies LY6D as a Marker Linking Castration-Resistant Prostate Luminal Cells to Prostate Progenitors and Cancer. Cell Rep. 2018, 25, 3504–3518 e3506. [Google Scholar] [CrossRef]
- Martin, B.; Paesmans, M.; Mascaux, C.; Berghmans, T.; Lothaire, P.; Meert, A.P.; Lafitte, J.J.; Sculier, J.P. Ki-67 expression and patients survival in lung cancer: Systematic review of the literature with meta-analysis. Br. J. Cancer 2004, 91, 2018–2025. [Google Scholar] [CrossRef]
- Kretzschmar, K.; Clevers, H. Organoids: Modeling Development and the Stem Cell Niche in a Dish. Dev. Cell 2016, 38, 590–600. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef]
- Reichert, M.; Takano, S.; Heeg, S.; Bakir, B.; Botta, G.P.; Rustgi, A.K. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nat. Protoc. 2013, 8, 1354–1365. [Google Scholar] [CrossRef] [PubMed]
- Huch, M.; Dorrell, C.; Boj, S.F.; van Es, J.H.; Li, V.S.; van de Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013, 494, 247–250. [Google Scholar] [CrossRef]
- Katsuda, T.; Kawamata, M.; Hagiwara, K.; Takahashi, R.U.; Yamamoto, Y.; Camargo, F.D.; Ochiya, T. Conversion of Terminally Committed Hepatocytes to Culturable Bipotent Progenitor Cells with Regenerative Capacity. Cell Stem Cell 2017, 20, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Salwig, I.; Spitznagel, B.; Vazquez-Armendariz, A.I.; Khalooghi, K.; Guenther, S.; Herold, S.; Szibor, M.; Braun, T. Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. EMBO J. 2019, 38, e102099. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef]
- Meads, M.B.; Gatenby, R.A.; Dalton, W.S. Environment-mediated drug resistance: A major contributor to minimal residual disease. Nat. Rev. Cancer 2009, 9, 665–674. [Google Scholar] [CrossRef]
- Borovski, T.; De Sousa, E.; Melo, F.; Vermeulen, L.; Medema, J.P. Cancer stem cell niche: The place to be. Cancer Res. 2011, 71, 634–639. [Google Scholar] [CrossRef]
- Su, S.; Chen, J.; Yao, H.; Liu, J.; Yu, S.; Lao, L.; Wang, M.; Luo, M.; Xing, Y.; Chen, F.; et al. CD10(+)GPR77(+) Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018, 172, 841–856 e816. [Google Scholar] [CrossRef]
- Ansari, A.M.; Ahmed, A.K.; Matsangos, A.E.; Lay, F.; Born, L.J.; Marti, G.; Harmon, J.W.; Sun, Z. Cellular GFP Toxicity and Immunogenicity: Potential Confounders in in Vivo Cell Tracking Experiments. Stem Cell Rev. Rep. 2016, 12, 553–559. [Google Scholar] [CrossRef]
- Birjandi, S.Z.; Palchevskiy, V.; Xue, Y.Y.; Nunez, S.; Kern, R.; Weigt, S.S.; Lynch, J.P., 3rd; Chatila, T.A.; Belperio, J.A. CD4(+)CD25(hi)Foxp3(+) Cells Exacerbate Bleomycin-Induced Pulmonary Fibrosis. Am. J. Pathol. 2016, 186, 2008–2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ayaub, E.A.; Wang, B.; Puchulu-Campanella, E.; Li, Y.H.; Hettiarachchi, S.U.; Lindeman, S.D.; Luo, Q.; Rout, S.; Srinivasarao, M.; et al. Reprogramming of profibrotic macrophages for treatment of bleomycin-induced pulmonary fibrosis. EMBO Mol. Med. 2020, 12, e12034. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-J.; Ho, C.-C.; Chang, Y.-L.; Chen, H.-Y.; Lin, C.-A.; Ling, T.-Y.; Yu, S.-L.; Yuan, S.-S.; Chen, Y.-J.L.; Lin, C.-Y.; et al. Cancer-associated fibroblasts regulate the plasticity of lung cancer stemness via paracrine signalling. Nature Commun. 2014, 5, 3472. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodriguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csoszi, T.; Fulop, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Koshino, Y.; Shibata, F.; Oki, T.; Nakajima, H.; Nosaka, T.; Kumagai, H. Retrovirus-mediated gene transfer and expression cloning: Powerful tools in functional genomics. Exp. Hematol. 2003, 31, 1007–1014. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Nagy, A.; Lanczky, A.; Menyhart, O.; Gyorffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semba, T.; Sato, R.; Kasuga, A.; Suina, K.; Shibata, T.; Kohno, T.; Suzuki, M.; Saya, H.; Arima, Y. Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d. Cancers 2020, 12, 3805. https://doi.org/10.3390/cancers12123805
Semba T, Sato R, Kasuga A, Suina K, Shibata T, Kohno T, Suzuki M, Saya H, Arima Y. Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d. Cancers. 2020; 12(12):3805. https://doi.org/10.3390/cancers12123805
Chicago/Turabian StyleSemba, Takashi, Ryo Sato, Akiyoshi Kasuga, Kentaro Suina, Tatsuhiro Shibata, Takashi Kohno, Makoto Suzuki, Hideyuki Saya, and Yoshimi Arima. 2020. "Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d" Cancers 12, no. 12: 3805. https://doi.org/10.3390/cancers12123805
APA StyleSemba, T., Sato, R., Kasuga, A., Suina, K., Shibata, T., Kohno, T., Suzuki, M., Saya, H., & Arima, Y. (2020). Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d. Cancers, 12(12), 3805. https://doi.org/10.3390/cancers12123805





