Interrogation of IDH1 Status in Gliomas by Fourier Transform Infrared Spectroscopy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
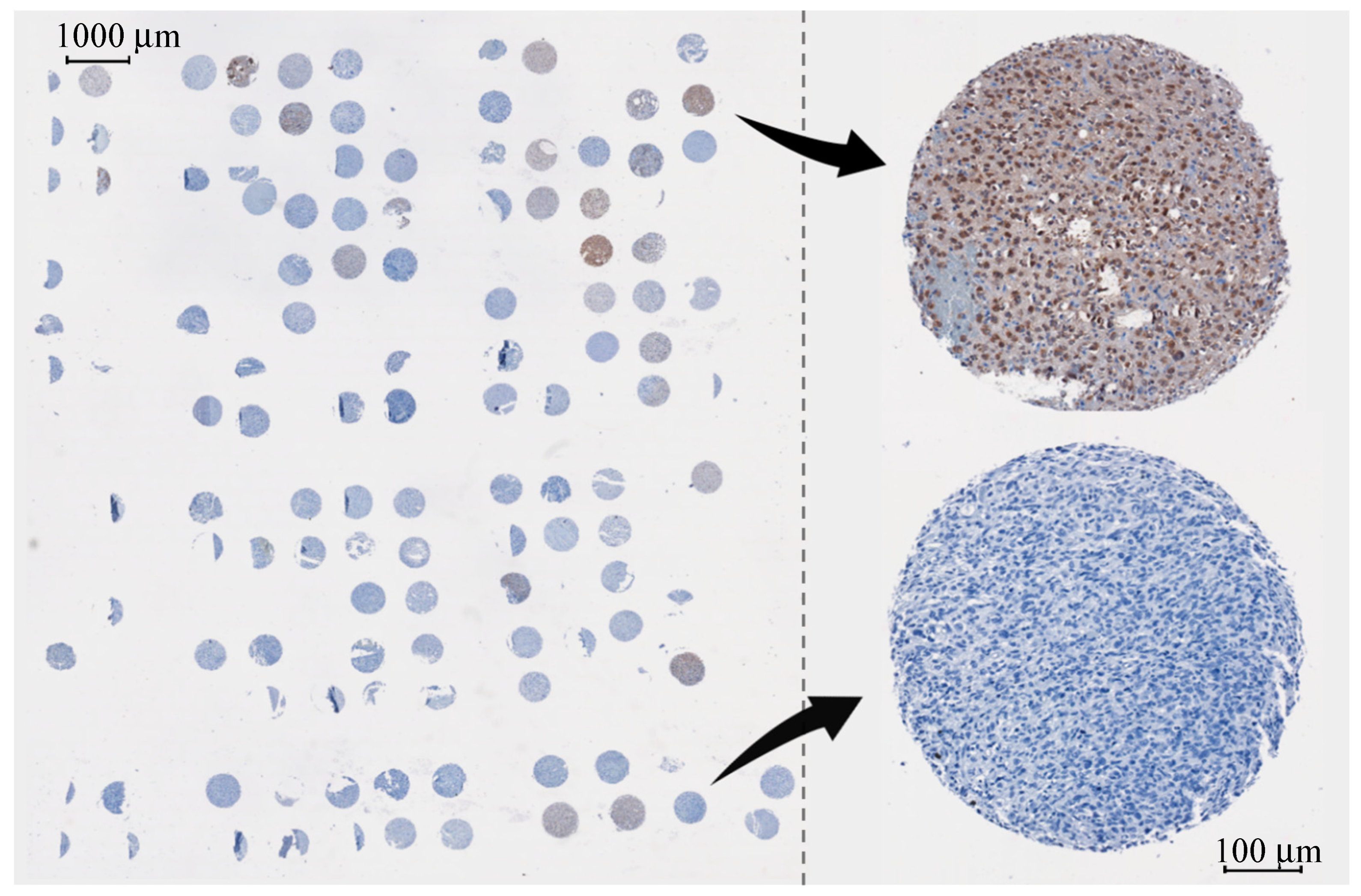
2.1.1. Glioma Tissue
2.1.2. Patient Serum
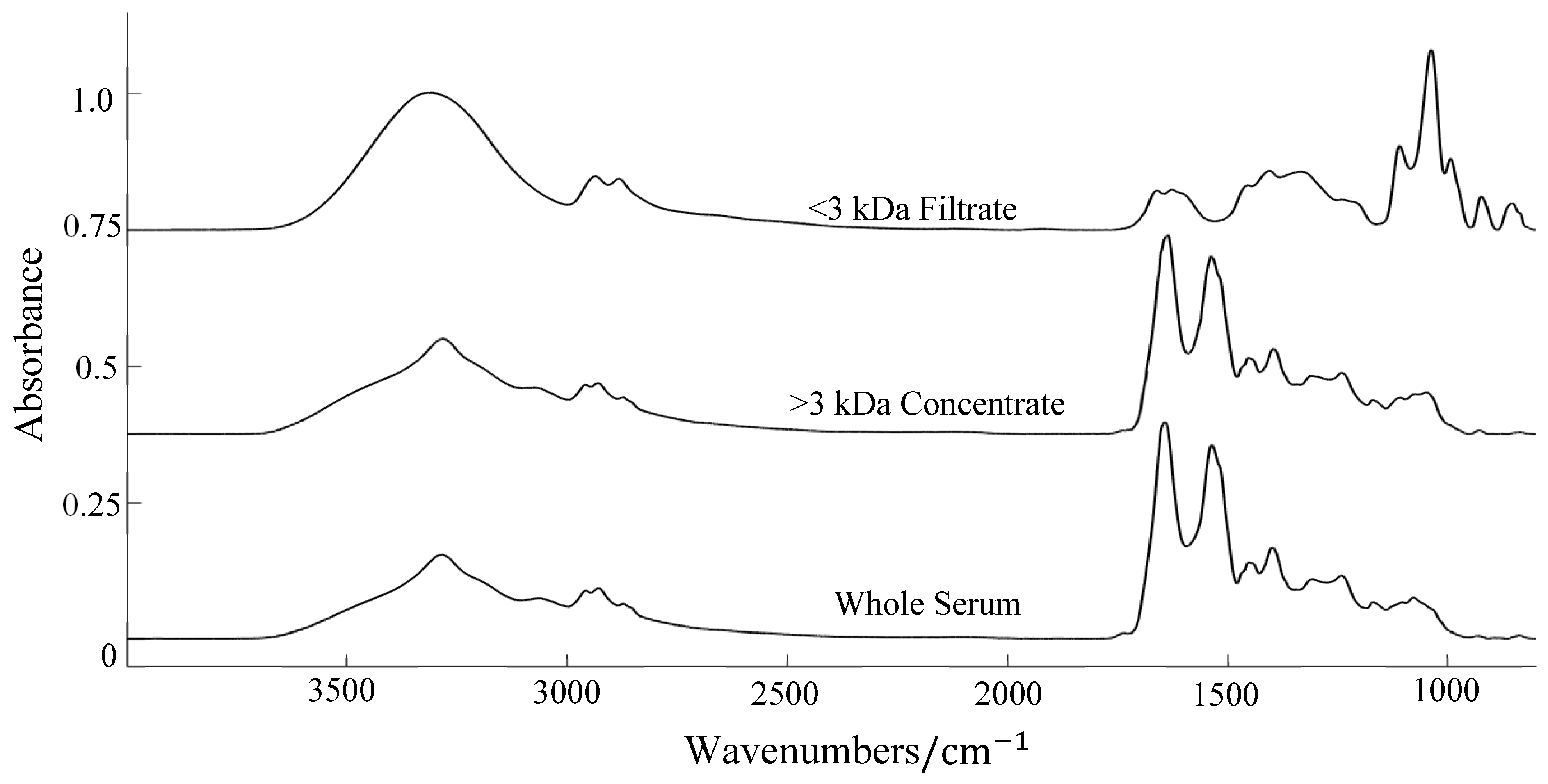
Centrifugal Filtration
2.2. Spectral Collection and Data Analysis
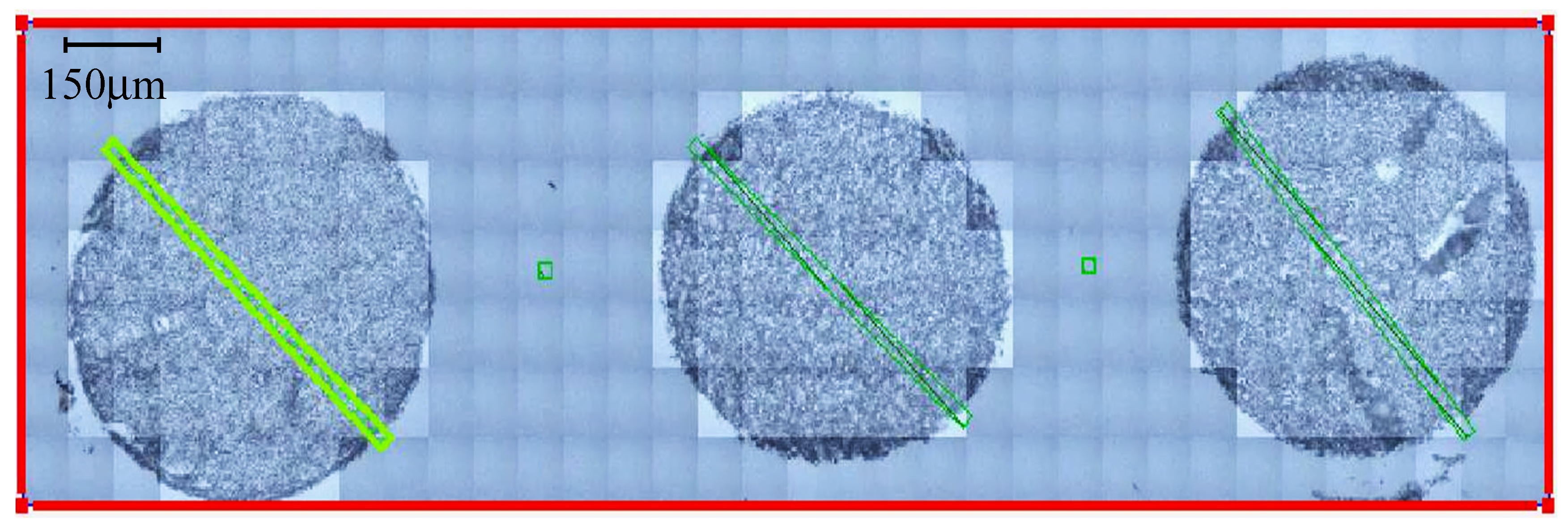
2.2.1. Synchrotron Radiation-Based FTIR Microspectroscopy
2.2.2. ATR-FTIR Spectroscopy
Centrifugal Filtration
3. Results
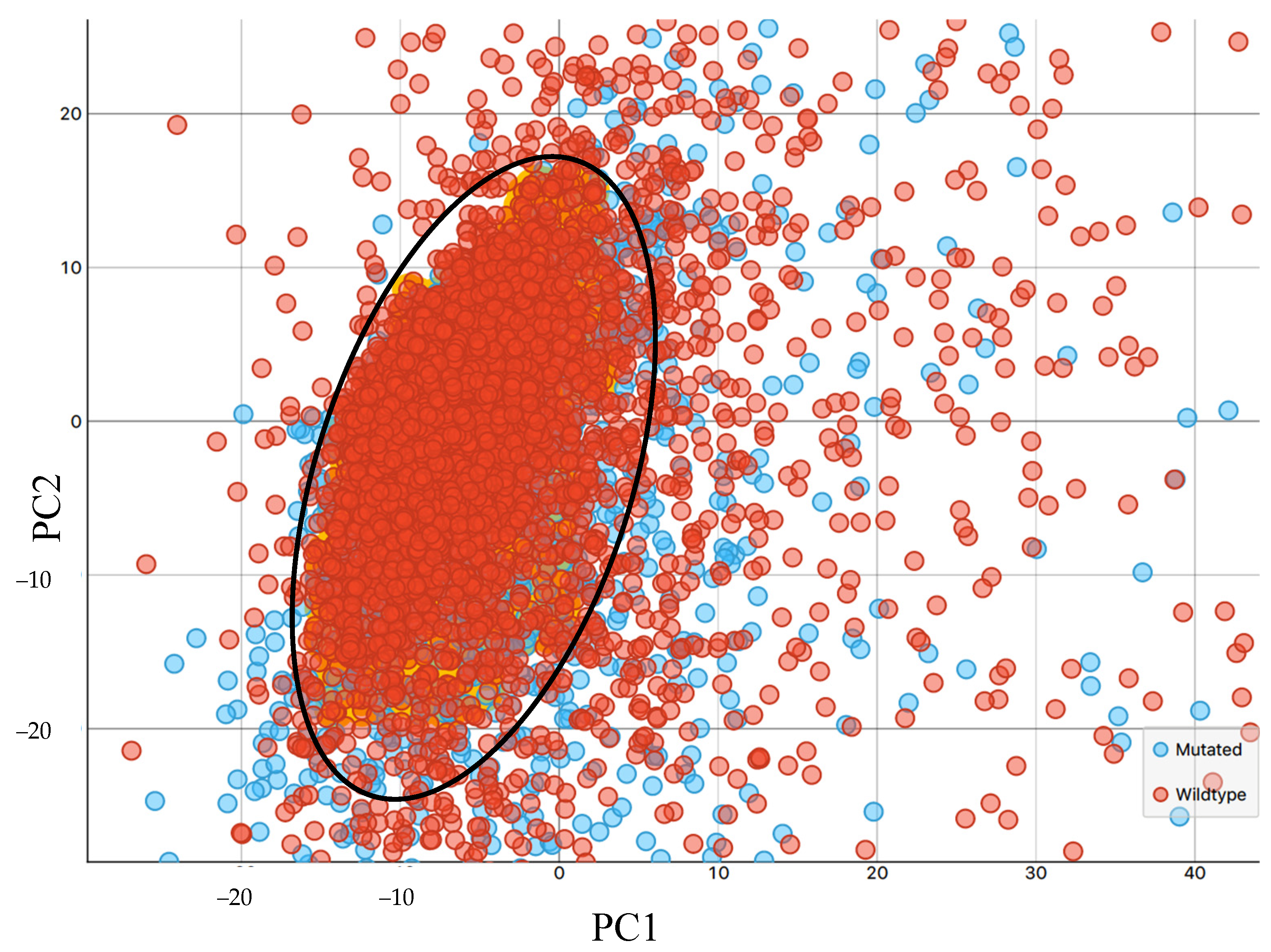
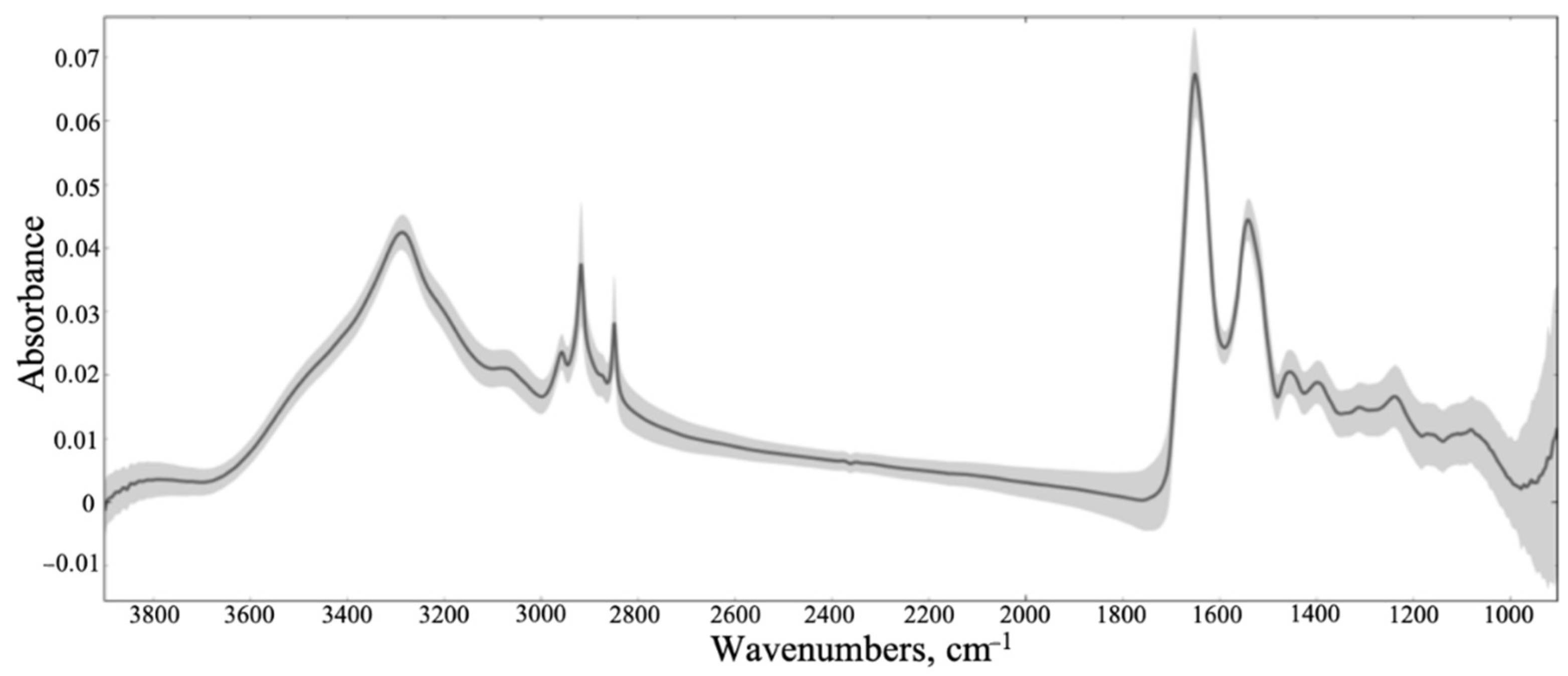
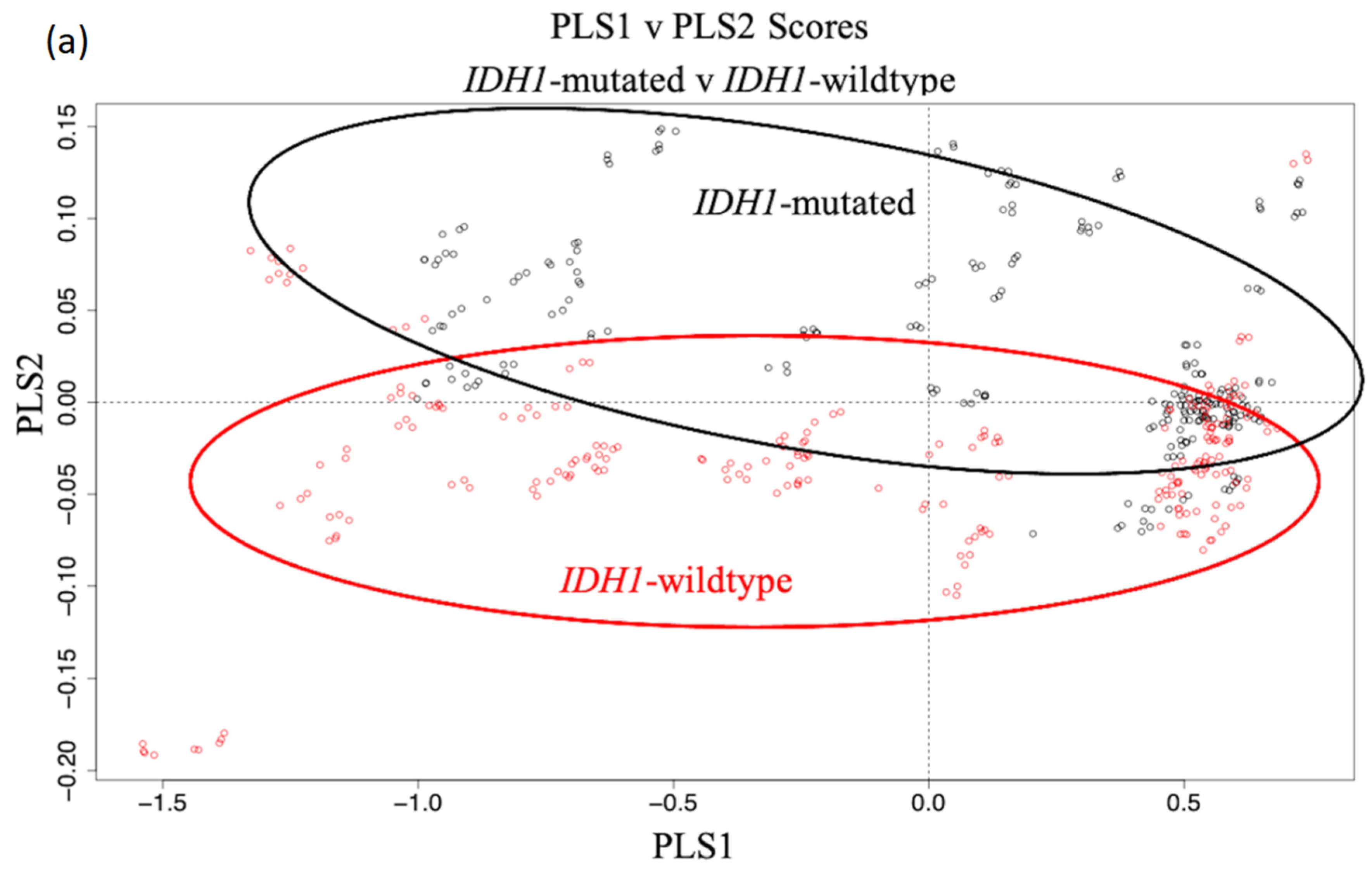
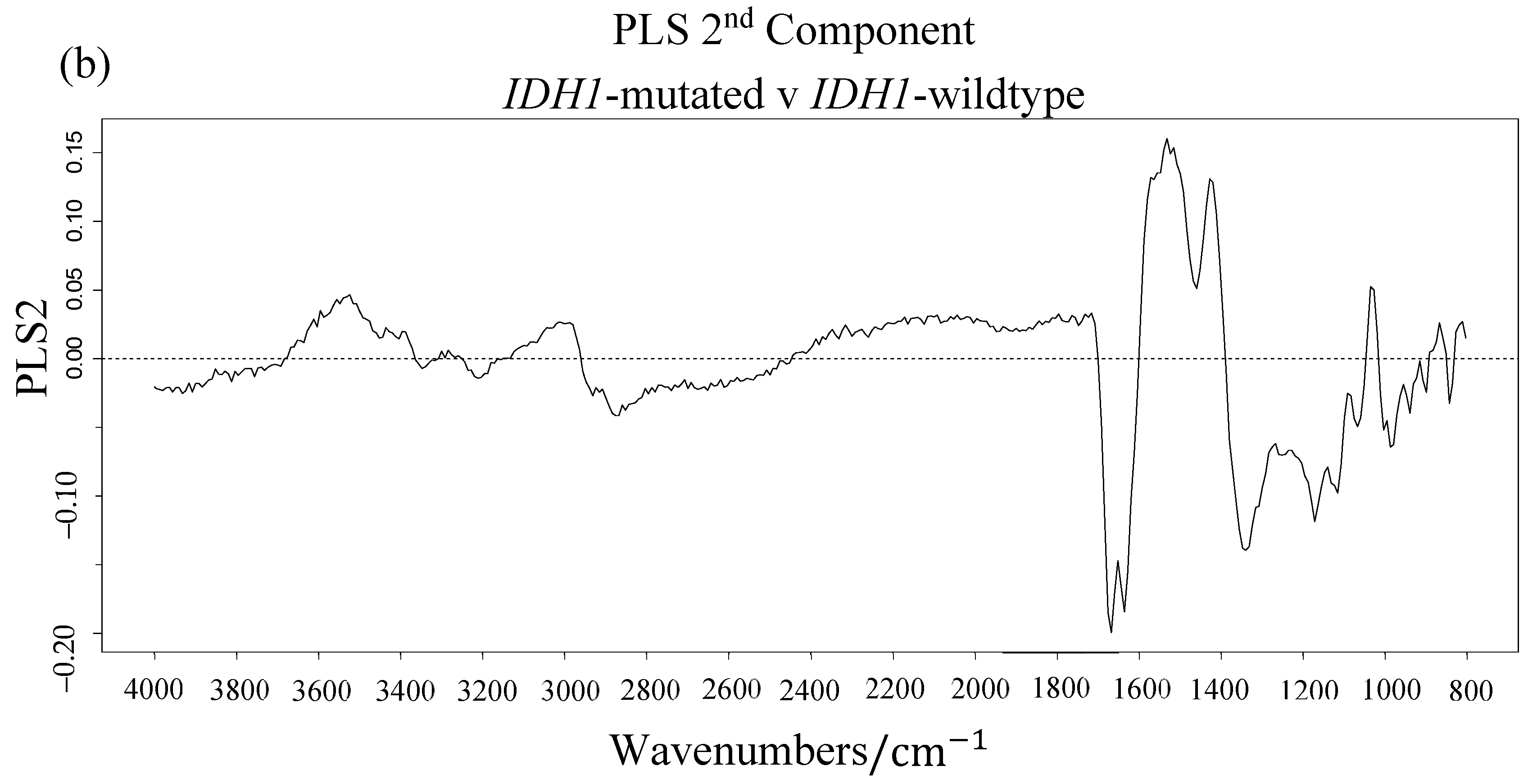
3.1. Synchrotron Microanalysis
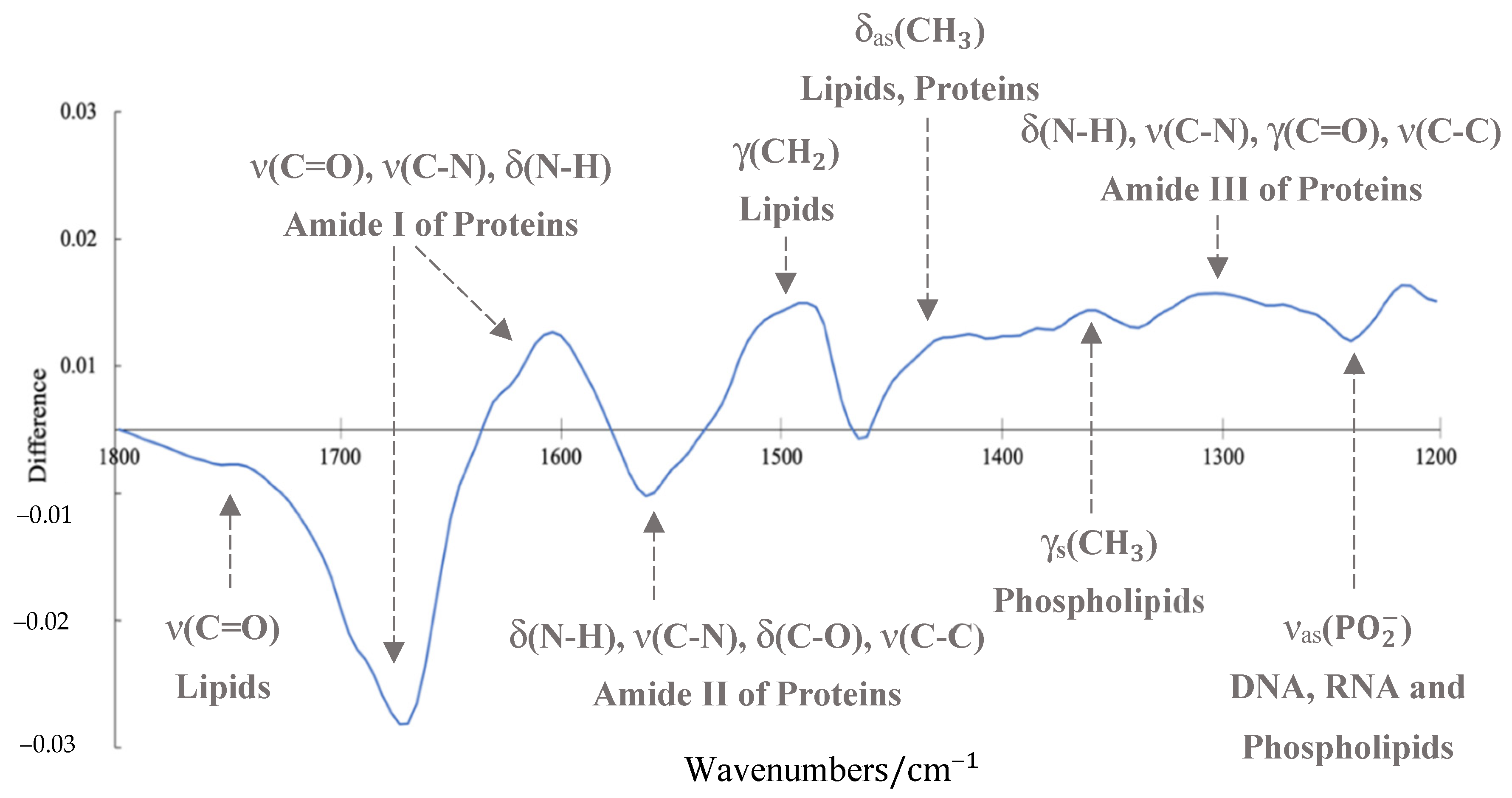
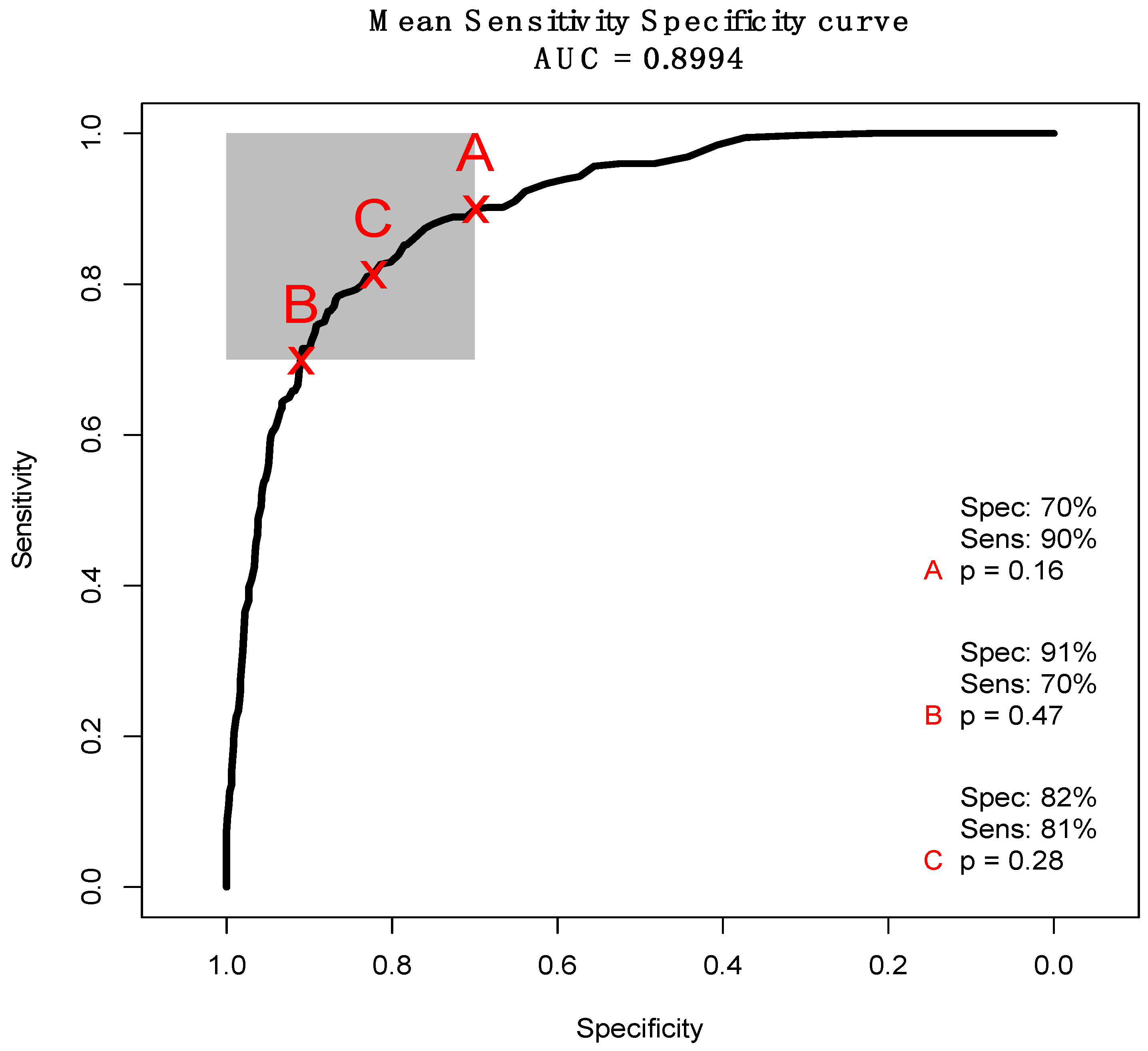
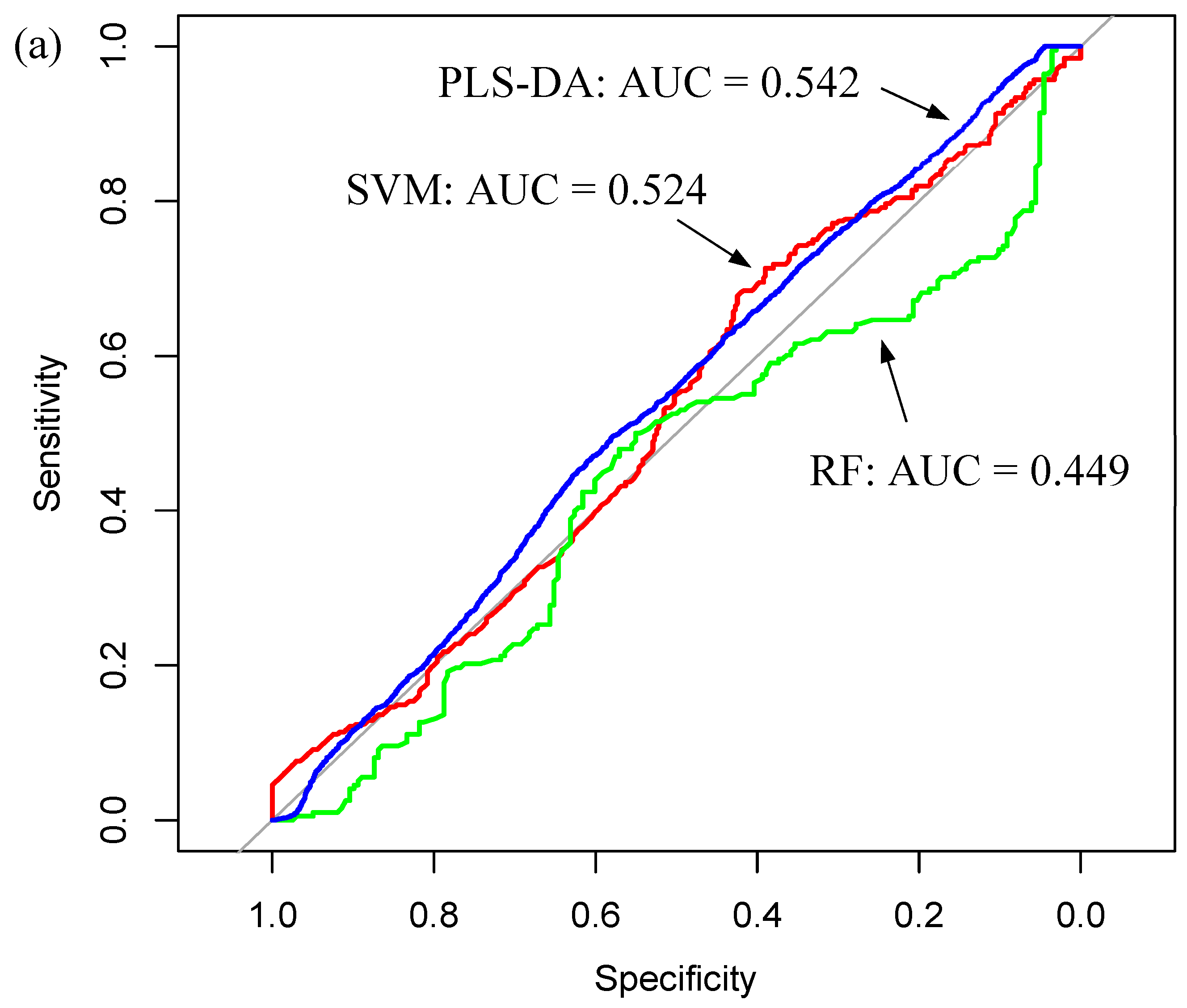
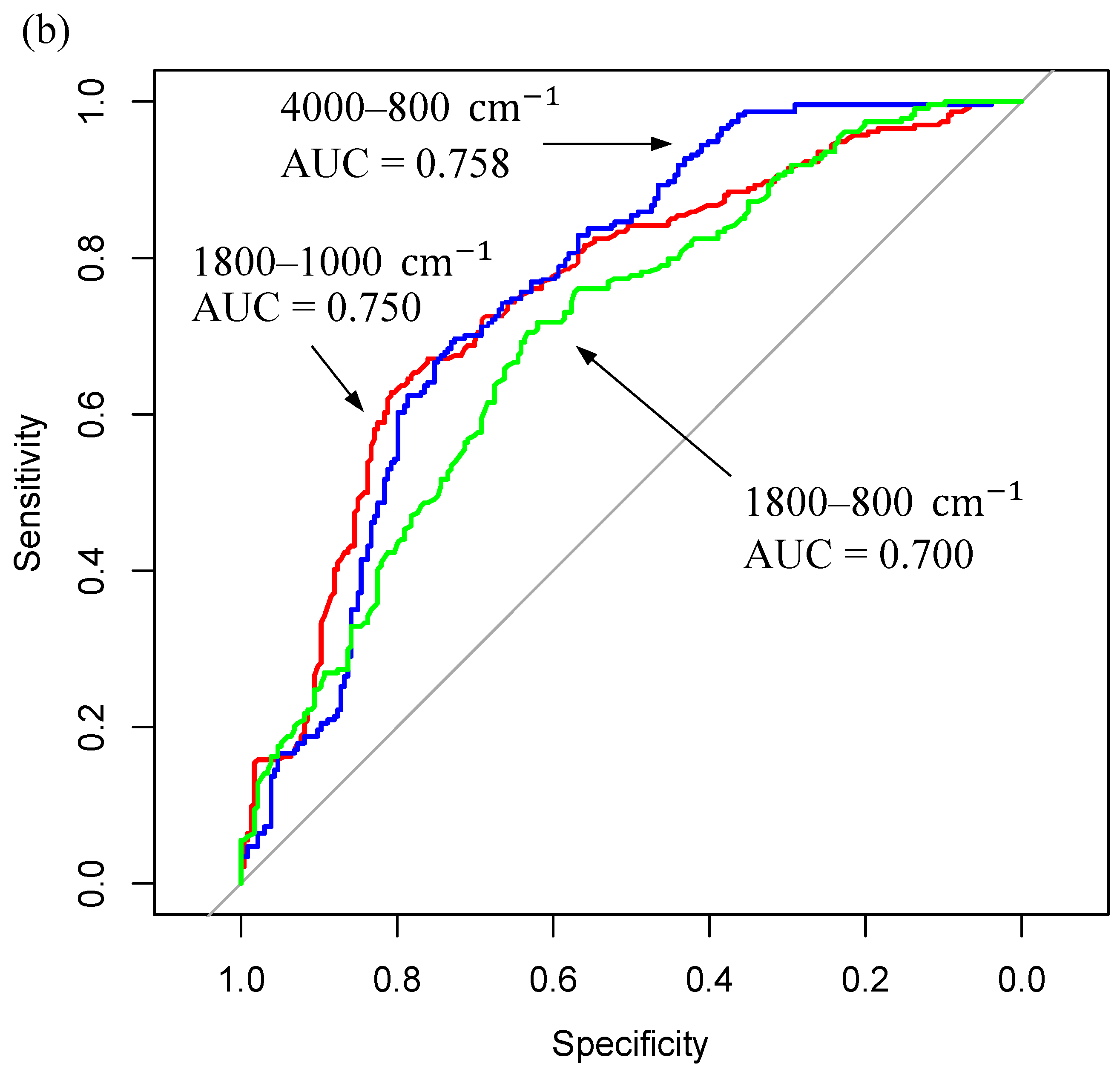
3.2. ATR-FTIR Results
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boots-Sprenger, S.H.; Sijben, A.; Rijntjes, J.; Tops, B.B.; Idema, A.J.; Rivera, A.L.; Bleeker, F.E.; Gijtenbeek, A.M.; Diefes, K.; Heathcock, L.; et al. Significance of complete 1p/19q co-deletion, IDH1 mutation and MGMT promoter methylation in gliomas: Use with caution. Mod. Pathol. 2013, 26, 922. [Google Scholar] [CrossRef] [PubMed]
- Horbinski, C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol. 2013, 125, 621–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, A.L.; Holmen, S.L.; Colman, H. IDH1 and IDH2 Mutations in Gliomas. Curr. Neurol. Neurosci. Rep. 2013, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, C.; Hentschel, B.; Wick, W.; Capper, D.; Felsberg, J.; Simon, M.; Westphal, M.; Schackert, G.; Meyermann, R.; Pietsch, T.; et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: Implications for classification of gliomas. Acta Neuropathol. 2010, 120, 707–718. [Google Scholar] [CrossRef] [Green Version]
- Ohgaki, H.; Kleihues, P. The Definition of Primary and Secondary Glioblastoma. Clin. Cancer Res. 2013, 19, 764–772. [Google Scholar] [CrossRef] [Green Version]
- Reifenberger, G.; Wirsching, H.-G.; Knobbe-Thomsen, C.B.; Weller, M. Advances in the molecular genetics of gliomas—Implications for classification and therapy. Nat. Rev. Clin. Oncol. 2017, 14, 434–452. [Google Scholar] [CrossRef]
- Rendina, A.R.; Pietrak, B.; Smallwood, A.; Zhao, H.; Qi, H.; Quinn, C.; Adams, N.D.; Concha, N.; Duraiswami, C.; Thrall, S.H.; et al. Mutant IDH1 Enhances the Production of 2-Hydroxyglutarate Due to Its Kinetic Mechanism. Biochemistry 2013, 52, 4563–4577. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [Green Version]
- Ohba, S.; Hirose, Y. Association between mutant IDHs and tumorigenesis in gliomas. Med. Mol. Morphol. 2018, 51, 194–198. [Google Scholar] [CrossRef]
- Sanson, M.; Marie, Y.; Paris, S.; Idbaih, A.; Laffaire, J.; Ducray, F.; El Hallani, S.; Boisselier, B.; Mokhtari, K.; Hoang-Xuan, K.; et al. Isocitrate Dehydrogenase 1 Codon 132 Mutation Is an Important Prognostic Biomarker in Gliomas. J. Clin. Oncol. 2009, 27, 4150–4154. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Cahill, D.P.; Sloan, A.E.; Nahed, B.V.; Aldape, K.D.; Louis, D.N.; Ryken, T.C.; Kalkanis, S.N.; Olson, J.J. The role of neuropathology in the management of patients with diffuse low grade glioma: A systematic review and evidence-based clinical practice guideline. J. Neurooncol. 2015, 125, 531–549. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Weißert, S.; Balss, J.; Habel, A.; Meyer, J.; Jäger, D.; Ackermann, U.; Tessmer, C.; Korshunov, A.; Zentgraf, H.; et al. Characterization of R132H Mutation-specific IDH1 Antibody Binding in Brain Tumors: IDH1R132H Mutation-specific Antibody. Brain Pathol. 2010, 20, 245–254. [Google Scholar] [CrossRef]
- Preusser, M.; Capper, D.; Hartmann, C. IDH testing in diagnostic neuropathology: Review and practical guideline article invited by the Euro-CNS research committee. Clin. Neuropathol. 2011, 30, 217–230. [Google Scholar] [CrossRef]
- Preusser, M.; Wöhrer, A.; Stary, S.; Höftberger, R.; Streubel, B.; Hainfellner, J.A. Value and Limitations of Immunohistochemistry and Gene Sequencing for Detection of the IDH1-R132H Mutation in Diffuse Glioma Biopsy Specimens. J. Neuropathol. Exp. Neurol. 2011, 70, 715–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dukor, R. Vibrational spectroscopy in the detection of cancer. In Handbook of Vibrational Spectroscopy; Wiley: Chichester, UK, 2002; Volume 5. [Google Scholar]
- Old, O.J.; Fullwood, L.M.; Scott, R.; Lloyd, G.R.; Almond, L.M.; Shepherd, N.A.; Stone, N.; Barr, H.; Kendall, C. Vibrational spectroscopy for cancer diagnostics. Anal. Methods 2014, 6, 3901. [Google Scholar] [CrossRef]
- Cameron, J.M.; Bruno, C.; Parachalil, D.R.; Baker, M.J.; Bonnier, F.; Butler, H.J.; Byrne, H.J. Vibrational spectroscopic analysis and quantification of proteins in human blood plasma and serum. In Vibrational Spectroscopy in Protein Research; Elsevier: Amsterdam, The Netherlands, 2020; pp. 269–314. ISBN 978-0-12-818610-7. [Google Scholar]
- Walsh, M.J.; Holton, S.E.; Kajdacsy-Balla, A.; Bhargava, R. Attenuated total reflectance Fourier-transform infrared spectroscopic imaging for breast histopathology. Vib. Spectrosc. 2012, 60, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Bird, B.; Bedrossian, K.; Laver, N.; Miljković, M.; Romeo, M.J.; Diem, M. Detection of breast micro-metastases in axillary lymph nodes by infrared micro-spectral imaging. Analyst 2009, 134, 1067. [Google Scholar] [CrossRef] [Green Version]
- Bird, B.; Miljković, M.; Remiszewski, S.; Akalin, A.; Kon, M.; Diem, M. Infrared spectral histopathology (SHP): A novel diagnostic tool for the accurate classification of lung cancer. Lab. Investig. 2012, 92, 1358. [Google Scholar] [CrossRef] [Green Version]
- Lasch, P.; Haensch, W.; Naumann, D.; Diem, M. Imaging of colorectal adenocarcinoma using FT-IR microspectroscopy and cluster analysis. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2004, 1688, 176–186. [Google Scholar] [CrossRef] [Green Version]
- Theophilou, G.; Lima, K.M.G.; Martin-Hirsch, P.L.; Stringfellow, H.F.; Martin, F.L. ATR-FTIR spectroscopy coupled with chemometric analysis discriminates normal, borderline and malignant ovarian tissue: Classifying subtypes of human cancer. Analyst 2016, 141, 585–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazi, E.; Baker, M.; Dwyer, J.; Lockyer, N.P.; Gardner, P.; Shanks, J.H.; Reeve, R.S.; Hart, C.A.; Clarke, N.W.; Brown, M.D. A Correlation of FTIR Spectra Derived from Prostate Cancer Biopsies with Gleason Grade and Tumour Stage. Eur. Urol. 2006, 50, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.J.; Gazi, E.; Brown, M.D.; Shanks, J.H.; Gardner, P.; Clarke, N.W. FTIR-based spectroscopic analysis in the identification of clinically aggressive prostate cancer. Br. J. Cancer 2008, 99, 1859–1866. [Google Scholar] [CrossRef] [Green Version]
- Baker, M.J.; Gazi, E.; Brown, M.D.; Shanks, J.H.; Clarke, N.W.; Gardner, P. Investigating FTIR based histopathology for the diagnosis of prostate cancer. J. Biophotonics 2009, 2, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; ur Rehman, D.I. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [Green Version]
- Jermyn, M.; Mok, K.; Mercier, J.; Desroches, J.; Pichette, J.; Saint-Arnaud, K.; Bernstein, L.; Guiot, M.-C.; Petrecca, K.; Leblond, F. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci. Transl. Med. 2015, 7, 274ra19. [Google Scholar] [CrossRef]
- Desroches, J.; Jermyn, M.; Pinto, M.; Picot, F.; Tremblay, M.-A.; Obaid, S.; Marple, E.; Urmey, K.; Trudel, D.; Soulez, G.; et al. A new method using Raman spectroscopy for in vivo targeted brain cancer tissue biopsy. Sci. Rep. 2018, 8, 1792. [Google Scholar] [CrossRef]
- Hollon, T.; Lewis, S.; Freudiger, C.W.; Sunney Xie, X.; Orringer, D.A. Improving the accuracy of brain tumor surgery via Raman-based technology. Neurosurg. Focus 2016, 40, E9. [Google Scholar] [CrossRef]
- Broadbent, B.; Tseng, J.; Kast, R.; Noh, T.; Brusatori, M.; Kalkanis, S.N.; Auner, G.W. Shining light on neurosurgery diagnostics using Raman spectroscopy. J. Neurooncol. 2016, 130, 1–9. [Google Scholar] [CrossRef]
- Beleites, C.; Steiner, G.; Sowa, M.G.; Baumgartner, R.; Sobottka, S.; Schackert, G.; Salzer, R. Classification of human gliomas by infrared imaging spectroscopy and chemometric image processing. Vib. Spectrosc. 2005, 38, 143–149. [Google Scholar] [CrossRef]
- Krafft, C.; Thümmler, K.; Sobottka, S.B.; Schackert, G.; Salzer, R. Classification of malignant gliomas by infrared spectroscopy and linear discriminant analysis. Biopolymers 2006, 82, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Krafft, C.; Shapoval, L.; Sobottka, S.B.; Schackert, G.; Salzer, R. Identification of Primary Tumors of Brain Metastases by Infrared Spectroscopic Imaging and Linear Discriminant Analysis. Technol. Cancer Res. Treat. 2006, 5, 291–298. [Google Scholar] [CrossRef] [Green Version]
- Gajjar, K.; Heppenstall, L.D.; Pang, W.; Ashton, K.M.; Trevisan, J.; Patel, I.I.; Llabjani, V.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Dawson, T.; et al. Diagnostic segregation of human brain tumours using Fourier-transform infrared and/or Raman spectroscopy coupled with discriminant analysis. Anal. Methods 2013, 5, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Sala, A.; Anderson, D.J.; Brennan, P.M.; Butler, H.J.; Cameron, J.M.; Jenkinson, M.D.; Rinaldi, C.; Theakstone, A.G.; Baker, M.J. Biofluid Diagnostics by FTIR Spectroscopy: A Platform Technology for Cancer Detection. Cancer Lett. 2020, S0304383520300835. [Google Scholar] [CrossRef]
- Cameron, J.M.; Butler, H.J.; Anderson, D.J.; Christie, L.; Confield, L.; Spalding, K.E.; Finlayson, D.; Murray, S.; Panni, Z.; Rinaldi, C.; et al. Exploring pre-analytical factors for the optimisation of serum diagnostics: Progressing the clinical utility of ATR-FTIR spectroscopy. Vib. Spectrosc. 2020, 109, 103092. [Google Scholar] [CrossRef]
- Surowka, A.D.; Adamek, D.; Szczerbowska-Boruchowska, M. The combination of artificial neural networks and synchrotron radiation-based infrared micro-spectroscopy for a study on the protein composition of human glial tumors. Analyst 2015, 140, 2428–2438. [Google Scholar] [CrossRef]
- Uckermann, O.; Juratli, T.A.; Galli, R.; Conde, M.; Wiedemuth, R.; Krex, D.; Geiger, K.; Temme, A.; Schackert, G.; Koch, E.; et al. Optical Analysis of Glioma: Fourier-Transform Infrared Spectroscopy Reveals the IDH1 Mutation Status. Clin. Cancer Res. 2018, 24, 2530–2538. [Google Scholar] [CrossRef] [Green Version]
- Theophilou, G.; Morais, C.L.M.; Halliwell, D.E.; Lima, K.M.G.; Drury, J.; Martin-Hirsch, P.L.; Stringfellow, H.F.; Hapangama, D.K.; Martin, F.L. Synchrotron- and focal plane array-based Fourier-transform infrared spectroscopy differentiates the basalis and functionalis epithelial endometrial regions and identifies putative stem cell regions of human endometrial glands. Anal. Bioanal. Chem. 2018, 410, 4541–4554. [Google Scholar] [CrossRef] [Green Version]
- Marcelli, A.; Cinque, G. Chapter 3. Infrared Synchrotron Radiation Beamlines: High Brilliance Tools for IR Spectromicroscopy. In RSC Analytical Spectroscopy Series; Moss, D., Ed.; Royal Society of Chemistry: Cambridge, UK, 2010; pp. 67–104. ISBN 978-0-85404-154-1. [Google Scholar]
- Miller, L.M.; Smith, R.J. Synchrotrons versus globars, point-detectors versus focal plane arrays: Selecting the best source and detector for specific infrared microspectroscopy and imaging applications. Vib. Spectrosc. 2005, 38, 237–240. [Google Scholar] [CrossRef]
- Szopa, W.; Burley, T.A.; Kramer-Marek, G.; Kaspera, W. Diagnostic and Therapeutic Biomarkers in Glioblastoma: Current Status and Future Perspectives. BioMed Res. Int. 2017, 2017, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shankar, G.M.; Francis, J.M.; Rinne, M.L.; Ramkissoon, S.H.; Huang, F.W.; Venteicher, A.S.; Akama-Garren, E.H.; Kang, Y.J.; Lelic, N.; Kim, J.C.; et al. Rapid Intraoperative Molecular Characterization of Glioma. JAMA Oncol. 2015, 1, 662. [Google Scholar] [CrossRef] [PubMed]
- Diamond Light Source MIRIAM B22 Beamline. Available online: https://www.diamond.ac.uk/B22 (accessed on 10 March 2020).
- Cinque, G.; Frogley, M.; Wehbe, K.; Filik, J.; Pijanka, J. Multimode InfraRed Imaging and Microspectroscopy (MIRIAM) Beamline at Diamond. Synchrotron Radiat. News 2011, 24, 24–33. [Google Scholar] [CrossRef]
- Bassan, P.; Sachdeva, A.; Kohler, A.; Hughes, C.; Henderson, A.; Boyle, J.; Shanks, J.H.; Brown, M.; Clarke, N.W.; Gardner, P. FTIR microscopy of biological cells and tissue: Data analysis using resonant Mie scattering (RMieS) EMSC algorithm. Analyst 2012, 137, 1370. [Google Scholar] [CrossRef] [PubMed]
- Toplak, M.; Birarda, G.; Read, S.; Sandt, C.; Rosendahl, S.M.; Vaccari, L.; Demšar, J.; Borondics, F. Infrared Orange: Connecting Hyperspectral Data with Machine Learning. Synchrotron Radiat. News 2017, 30, 40–45. [Google Scholar] [CrossRef]
- Raschka, S. Linear Discriminant Analysis-Bit by Bit. Available online: https://sebastianraschka.com/Articles/2014_python_lda.html (accessed on 10 March 2020).
- Cameron, J.M.; Butler, H.J.; Smith, B.R.; Hegarty, M.G.; Jenkinson, M.D.; Syed, K.; Brennan, P.M.; Ashton, K.; Dawson, T.; Palmer, D.S.; et al. Developing infrared spectroscopic detection for stratifying brain tumour patients: Glioblastoma multiforme vs. lymphoma. Analyst 2019, 144, 6736–6750. [Google Scholar] [CrossRef]
- Butler, H.J.; Brennan, P.M.; Cameron, J.M.; Finlayson, D.; Hegarty, M.G.; Jenkinson, M.D.; Palmer, D.S.; Smith, B.R.; Baker, M.J. Development of high-throughput ATR-FTIR technology for rapid triage of brain cancer. Nat. Commun. 2019, 10, 4501. [Google Scholar] [CrossRef] [Green Version]
- Cameron, J.M.; Butler, H.J.; Palmer, D.S.; Baker, M.J. Biofluid spectroscopic disease diagnostics: A review on the processes and spectral impact of drying. J. Biophotonics 2018, 11, e201700299. [Google Scholar] [CrossRef] [Green Version]
- Lovergne, L.; Clemens, G.; Untereiner, V.; Lukaszweski, R.A.; Sockalingum, G.D.; Baker, M.J. Investigating optimum sample preparation for infrared spectroscopic serum diagnostics. Anal. Methods 2015, 7, 7140–7149. [Google Scholar] [CrossRef] [Green Version]
- Lovergne, L.; Bouzy, P.; Untereiner, V.; Garnotel, R.; Baker, M.J.; Thiéfin, G.; Sockalingum, G.D. Biofluid infrared spectro-diagnostics: Pre-analytical considerations for clinical applications. Faraday Discuss 2016, 187, 521–537. [Google Scholar] [CrossRef] [Green Version]
- Cameron, J.M.; Rinaldi, C.; Butler, H.J.; Hegarty, M.G.; Brennan, P.M.; Jenkinson, M.D.; Syed, K.; Ashton, K.M.; Dawson, T.P.; Palmer, D.S.; et al. Stratifying Brain Tumour Histological Sub-Types: The Application of ATR-FTIR Serum Spectroscopy in Secondary Care. Cancers 2020, 12, 1710. [Google Scholar] [CrossRef] [PubMed]
- Bassan, P.; Byrne, H.J.; Bonnier, F.; Lee, J.; Dumas, P.; Gardner, P. Resonant Mie scattering in infrared spectroscopy of biological materials—Understanding the ‘dispersion artefact’. Analyst 2009, 134, 1586. [Google Scholar] [CrossRef] [PubMed]
- Viera, A.J.; Garrett, J.M. Understanding Interobserver Agreement: The Kappa Statistic. Fam. Med. 2005, 37, 360–363. [Google Scholar]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Mandrekar, J.N. Receiver Operating Characteristic Curve in Diagnostic Test Assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Smith, B.R.; Ashton, K.M.; Brodbelt, A.; Dawson, T.; Jenkinson, M.D.; Hunt, N.T.; Palmer, D.S.; Baker, M.J. Combining random forest and 2D correlation analysis to identify serum spectral signatures for neuro-oncology. Analyst 2016, 141, 3668–3678. [Google Scholar] [CrossRef] [Green Version]
- Dumas, P.; Miller, L.M.; Tobin, M.J. Challenges in Biology and Medicine with Synchrotron Infrared Light. Acta Phys. Pol. A 2009, 115, 446–454. [Google Scholar] [CrossRef]
- Carr, G.L. Resolution limits for infrared microspectroscopy explored with synchrotron radiation. Rev. Sci. Instrum. 2001, 72, 1613. [Google Scholar] [CrossRef]
- Kaur, G.; Dufour, J.M. Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2012, 2, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Kong, J.; Yu, S. Fourier Transform Infrared Spectroscopic Analysis of Protein Secondary Structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta BBA Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petricoin, E.F.; Belluco, C.; Araujo, R.P.; Liotta, L.A. The blood peptidome: A higher dimension of information content for cancer biomarker discovery. Nat. Rev. Cancer 2006, 6, 961–967. [Google Scholar] [CrossRef]
- Bonnier, F.; Baker, M.J.; Byrne, H.J. Vibrational spectroscopic analysis of body fluids: Avoiding molecular contamination using centrifugal filtration. Anal. Methods 2014, 6, 5155. [Google Scholar] [CrossRef] [Green Version]
- Bonnier, F.; Petitjean, F.; Baker, M.J.; Byrne, H.J. Improved protocols for vibrational spectroscopic analysis of body fluids: Improved protocols for vibrational spectroscopic analysis of body fluids. J. Biophotonics 2014, 7, 167–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Glioma Entity | WHO Grade | IDH1 Mutation | Additional Associated Alterations |
|---|---|---|---|
| Pilocytic astrocytoma | I | Extremely rare | BRAF, KRAS, NF1, FGFR1 |
| Diffuse astrocytoma | II | Common | IDH2, TP53, ATRX, LOH 17p |
| Anaplastic astrocytoma | III | Common | IDH2, TP53, ATRX, LOH 17p |
| Oligodendroglioma | II | Majority of cases | IDH2, 1p/19q co-deletion |
| Anaplastic oligodendroglioma | III | Majority of cases | IDH2, 1p/19q co-deletion |
| Glioblastoma (primary) | IV | Rare | TERT, PTEN, TP53, MGMT hypermethylation, EGFR, 7+/10− |
| Glioblastoma (secondary) | IV | Extremely Common | IDH2, TP53, ATRX, LOH 17p |
| Parameter | Variations | |||
|---|---|---|---|---|
| Normalisation (n) | None (0) | Min-max (1) | Vector (2) | Amide I (3) |
| Derivative (l) | None (0) | First (1) | Second (2) | - |
| Binning (b) | 1 | 2 | 4 | 8 |
| Smoothing with Savitzky–Golay filter (s) | None (0) | 2 | 3 | 4 |
| Spectral cut (p) | None (0) | 1800–1000 cm−1 | 1800–1200 cm−1 | - |
| Statistic | Mean | Standard Deviation |
|---|---|---|
| Sensitivity (%) | 82.4 | 16.8 |
| Specificity (%) | 83.4 | 8.2 |
| Balanced Accuracy (%) | 82.9 | 9.6 |
| Sample Fraction | Model | Sensitivity (%) | Specificity (%) | Balanced Accuracy (%) | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| Whole Serum | RF | 50.3 | 15.2 | 45.4 | 15.1 | 47.9 | 8.6 |
| PLS-DA | 69.3 | 13.8 | 35.3 | 14.7 | 52.3 | 7.4 | |
| SVM | 75.9 | 17.5 | 28.0 | 14.6 | 51.9 | 7.7 | |
| Sample Fraction | Model | Sensitivity (%) | Specificity (%) | Balanced Accuracy (%) | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| <3kDa Filtered Serum (4000–800 cm−1) | RF | 68.4 | 16.2 | 67.5 | 15.9 | 68.0 | 11.1 |
| PLS-DA | 75.5 | 12.3 | 62.6 | 15.5 | 69.1 | 9.0 | |
| SVM | 68.4 | 16.5 | 64.2 | 16.0 | 66.4 | 10.2 | |
| <3kDa Filtered Serum (1800–800 cm−1) | RF | 70.6 | 17.8 | 66.4 | 14.5 | 68.5 | 11.2 |
| PLS-DA | 65.0 | 14.6 | 64.6 | 16.5 | 64.8 | 8.7 | |
| SVM | 63.2 | 16.3 | 63.8 | 16.9 | 63.5 | 9.6 | |
| <3kDa Filtered Serum (1800–1000 cm−1) | RF | 66.6 | 15.4 | 68.1 | 14.1 | 67.4 | 9.9 |
| PLS-DA | 65.9 | 14.6 | 56.2 | 15.5 | 61.1 | 9.1 | |
| SVM | 68.1 | 15.6 | 56.8 | 15.6 | 62.5 | 10.1 | |
| ∑ Gini | Vibrational Modes | |
|---|---|---|
| 1124.5 | 12.31 | C-O stretch |
| 1172.5 | 11.22 | C-O, C-OH stretch |
| 1164.5 | 9.07 | C-C, C-O and C-OH stretch |
| 1180.5 | 6.43 | twisting |
| 1116.5 | 5.39 | RNA; C-OH stretch |
| 1028.5 | 5.01 | Carbohydrate; C-O stretch |
| 1188.5 | 4.46 | DNA; Symmetric stretch |
| 1740.5 | 4.19 | Lipids; C = O stretch |
| 1020.5 | 3.60 | Glycogen; C-O stretch |
| 1132.5 | 3.49 | C-O and C-C stretch |
| 1588.5 | 2.77 | Amide I; C = O and C-N stretch, N-H bending |
| 1548.5 | 2.73 | Amide II; N-H bending, C-N stretching |
| 1444.5 | 2.57 | Lipids; bending |
| 1468.5 | 2.52 | Lipids/Proteins; bending |
| 1612.5 | 2.45 | Amide I; C = O and C-N stretch, N-H bending |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cameron, J.M.; Conn, J.J.A.; Rinaldi, C.; Sala, A.; Brennan, P.M.; Jenkinson, M.D.; Caldwell, H.; Cinque, G.; Syed, K.; Butler, H.J.; et al. Interrogation of IDH1 Status in Gliomas by Fourier Transform Infrared Spectroscopy. Cancers 2020, 12, 3682. https://doi.org/10.3390/cancers12123682
Cameron JM, Conn JJA, Rinaldi C, Sala A, Brennan PM, Jenkinson MD, Caldwell H, Cinque G, Syed K, Butler HJ, et al. Interrogation of IDH1 Status in Gliomas by Fourier Transform Infrared Spectroscopy. Cancers. 2020; 12(12):3682. https://doi.org/10.3390/cancers12123682
Chicago/Turabian StyleCameron, James M., Justin J. A. Conn, Christopher Rinaldi, Alexandra Sala, Paul M. Brennan, Michael D. Jenkinson, Helen Caldwell, Gianfelice Cinque, Khaja Syed, Holly J. Butler, and et al. 2020. "Interrogation of IDH1 Status in Gliomas by Fourier Transform Infrared Spectroscopy" Cancers 12, no. 12: 3682. https://doi.org/10.3390/cancers12123682
APA StyleCameron, J. M., Conn, J. J. A., Rinaldi, C., Sala, A., Brennan, P. M., Jenkinson, M. D., Caldwell, H., Cinque, G., Syed, K., Butler, H. J., Hegarty, M. G., Palmer, D. S., & Baker, M. J. (2020). Interrogation of IDH1 Status in Gliomas by Fourier Transform Infrared Spectroscopy. Cancers, 12(12), 3682. https://doi.org/10.3390/cancers12123682