Prospects for Using Expression Patterns of Paramyxovirus Receptors as Biomarkers for Oncolytic Virotherapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Measles Virus as an Oncolytic Agent
- Host: Human
- Origin: Most likely MV originated from a virus of non-human species.
- Genome: MV has a single-stranded, negative-sense, non-segmented RNA genome that is ~16K nucleotides long.
- Virion: MV is an enveloped virus with a lipid membrane.
- Proteins: Nucleoprotein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin (H), large protein (L), and two nonstructural proteins C and V. Protein C is translated from the same mRNA as the P protein but using an alternative start codon in an overlapping ORF. Protein V is translated from an edited P mRNA.
3. Natural MV Receptors
4. MV Retargeting for Binding New Cancer-Associated Proteins
5. SeV as an Oncolytic Agent
- Taxonomy: The virus belongs to the genus Respirovirus within the family Paramyxoviridae [23].
- Host: The virus causes respiratory infections in mice, hamsters, guinea pigs, rats, and other rodents [128].
- Genome: SeV has single-stranded, negative-sense, non-segmented RNA genome that is ~15K nucleotides long [129].
- Virion: SeV is an enveloped virus with a lipid membrane.
- Proteins: Nucleoprotein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN), large protein (L), and nonstructural proteins collectively referred as C-proteins (C’, C, Y1, Y2, V, W) that are translated from an alternative RNA transcript of the P gene [129].
6. SeV receptors
7. Potential Problems of Virus Delivery and Retargeting
7.1. Preexisting Immunity
7.2. Virus Transportation and Tumor Delivery
8. Additional Factors Determining Cell Sensitivity to Viruses
9. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CEA | Carcinoembryonic antigen |
| EGF | Epidermal growth factor |
| EGFR | Epidermal growth factor receptor |
| FOLR1 | Folate receptor 1 |
| HER2/neu | Tyrosine-protein kinase erbB-2 (human epidermal growth factor receptor 2) |
| IFN | Interferon |
| IP | Intraperitoneal delivery |
| IT | Intratumoral delivery |
| IV | Intravenous delivery |
| MV | Measles virus |
| PVRL4 | Poliovirus-receptor-like 4, molecule (nectin-4) |
| SeV | Sendai virus |
| SLAM/SLAMF1/CD150 | Signaling lymphocytic activation molecule 1 |
| SPG | Sialosylparagloboside |
References
- Kaufman, H.L.; Kohlhapp, F.J.; Zloza, A. Oncolytic viruses: A new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015, 14, 642–662. [Google Scholar] [CrossRef]
- Marelli, G.; Howells, A.; Lemoine, N.R.; Wang, Y. Oncolytic Viral Therapy and the Immune System: A Double-Edged Sword Against Cancer. Front. Immunol. 2018, 9, 866. [Google Scholar] [CrossRef]
- Stetson, D.B.; Medzhitov, R. Antiviral defense: Interferons and beyond. J. Exp. Med. 2006, 203, 1837–1841. [Google Scholar] [CrossRef]
- Negishi, H.; Taniguchi, T.; Yanai, H. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb. Perspect. Biol. 2017. [Google Scholar] [CrossRef]
- Katsoulidis, E.; Kaur, S.; Platanias, L.C. Deregulation of interferon signaling in malignant cells. Pharmaceuticals 2010, 3, 406–418. [Google Scholar] [CrossRef]
- Watanabe, M.; Hirano, A.; Stenglein, S.; Nelson, J.; Thomas, G.; Wong, T.C. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J. Virol. 1995, 69, 3206–3210. [Google Scholar] [CrossRef]
- Tashiro, M.; Yokogoshi, Y.; Tobita, K.; Seto, J.T.; Rott, R.; Kido, H. Tryptase Clara, an activating protease for Sendai virus in rat lungs, is involved in pneumopathogenicity. J. Virol. 1992, 66, 7211–7216. [Google Scholar] [CrossRef] [PubMed]
- Kido, H.; Niwa, Y.; Beppu, Y.; Towatari, T. Cellular proteases involved in the pathogenicity of enveloped animal viruses, human immunodeficiency virus, influenza virus A and Sendai virus. Adv. Enzyme Regul. 1996, 36, 325–347. [Google Scholar] [CrossRef]
- Chen, Y.; Shiota, M.; Ohuchi, M.; Towatari, T.; Tashiro, J.; Murakami, M.; Yano, M.; Yang, B.; Kido, H. Mast cell tryptase from pig lungs triggers infection by pneumotropic Sendai and influenza A viruses. Purification and characterization. Eur. J. Biochem. 2000, 267, 3189–3197. [Google Scholar] [CrossRef] [PubMed]
- Le, T.Q.; Kawachi, M.; Yamada, H.; Shiota, M.; Okumura, Y.; Kido, H. Identification of trypsin I as a candidate for influenza A virus and Sendai virus envelope glycoprotein processing protease in rat brain. Biol. Chem. 2006, 387, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Towatari, T.; Ohuchi, M.; Shiota, M.; Akao, M.; Okumura, Y.; Parry, M.A.; Kido, H. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur. J. Biochem. 2001, 268, 2847–2855. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, T.; Gotoh, B.; Suzuki, H.; Asaka, J.; Shimokata, K.; Rott, R.; Nagai, Y. Expression of factor X and its significance for the determination of paramyxovirus tropism in the chick embryo. EMBO J. 1992, 11, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Tahara, M.; Sakai, K.; Yamaguchi, H.; Kanou, K.; Shirato, K.; Kawase, M.; Noda, M.; Kimura, H.; Matsuyama, S.; et al. TMPRSS2 is an activating protease for respiratory parainfluenza viruses. J. Virol. 2013, 87, 11930–11935. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 20 June 2018).
- Matveeva, O.V.; Kochneva, G.V.; Netesov, S.V.; Onikienko, S.B.; Chumakov, P.M. Mechanisms of Oncolysis by Paramyxovirus Sendai. Acta Naturae 2015, 7, 6–16. [Google Scholar] [CrossRef]
- Matveeva, O.V.; Guo, Z.S.; Shabalina, S.A.; Chumakov, P.M. Oncolysis by paramyxoviruses: Multiple mechanisms contribute to therapeutic efficiency. Mol. Ther. Oncolytics 2015, 2, 15011. [Google Scholar] [CrossRef]
- Matveeva, O.V.; Guo, Z.S.; Senin, V.M.; Senina, A.V.; Shabalina, S.A.; Chumakov, P.M. Oncolysis by paramyxoviruses: Preclinical and clinical studies. Mol. Ther. Oncolytics 2015, 2, 15017. [Google Scholar] [CrossRef]
- Jiang, Y.; Qin, Y.; Chen, M. Host-Pathogen Interactions in Measles Virus Replication and Anti-Viral Immunity. Viruses 2016, 8, 308. [Google Scholar] [CrossRef]
- Faisca, P.; Desmecht, D. Sendai virus, the mouse parainfluenza type 1: A longstanding pathogen that remains up-to-date. Res. Vet. Sci. 2007, 82, 115–125. [Google Scholar] [CrossRef]
- Thul, P.J.; Akesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Bjork, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses. Available online: https://talk.ictvonline.org/ (accessed on 25 June 2019).
- Griffin, D.E. Measles Virus. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 1, pp. 1042–1069. [Google Scholar]
- Baldo, A.; Galanis, E.; Tangy, F.; Herman, P. Biosafety considerations for attenuated measles virus vectors used in virotherapy and vaccination. Hum. Vaccin. Immunother. 2016, 12, 1102–1116. [Google Scholar] [CrossRef] [PubMed]
- Msaouel, P.; Opyrchal, M.; Domingo Musibay, E.; Galanis, E. Oncolytic measles virus strains as novel anticancer agents. Expert Opin. Biol. Ther. 2013, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Aref, S.; Bailey, K.; Fielding, A. Measles to the Rescue: A Review of Oncolytic Measles Virus. Viruses 2016, 8, 294. [Google Scholar] [CrossRef]
- Msaouel, P.; Opyrchal, M.; Dispenzieri, A.; Peng, K.W.; Federspiel, M.J.; Russell, S.J.; Galanis, E. Clinical Trials with Oncolytic Measles Virus: Current Status and Future Prospects. Curr. Cancer Drug Targets 2018, 18, 177–187. [Google Scholar] [CrossRef]
- Tatsuo, H.; Ono, N.; Tanaka, K.; Yanagi, Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature 2000, 406, 893–897. [Google Scholar] [CrossRef]
- Tatsuo, H.; Yanagi, Y. The morbillivirus receptor SLAM (CD150). Microbiol. Immunol. 2002, 46, 135–142. [Google Scholar] [CrossRef]
- Muhlebach, M.D.; Mateo, M.; Sinn, P.L.; Prufer, S.; Uhlig, K.M.; Leonard, V.H.; Navaratnarajah, C.K.; Frenzke, M.; Wong, X.X.; Sawatsky, B.; et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 2011, 2, 530–533. [Google Scholar] [CrossRef]
- Noyce, R.S.; Bondre, D.G.; Ha, M.N.; Lin, L.T.; Sisson, G.; Tsao, M.S.; Richardson, C.D. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011, 7, e1002240. [Google Scholar] [CrossRef]
- Noyce, R.S.; Richardson, C.D. Nectin 4 is the epithelial cell receptor for measles virus. Trends Microbiol. 2012, 20, 429–439. [Google Scholar] [CrossRef]
- Dorig, R.E.; Marcil, A.; Chopra, A.; Richardson, C.D. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 1993, 75, 295–305. [Google Scholar] [CrossRef]
- Naniche, D.; Varior-Krishnan, G.; Cervoni, F.; Wild, T.F.; Rossi, B.; Rabourdin-Combe, C.; Gerlier, D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 1993, 67, 6025–6032. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Morra, M.; Wu, C.; Gullo, C.; Howie, D.; Coyle, T.; Engel, P.; Terhorst, C. CD150 is a member of a family of genes that encode glycoproteins on the surface of hematopoietic cells. Immunogenetics 2001, 53, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Sintes, J.; Romero, X.; Marin, P.; Terhorst, C.; Engel, P. Differential expression of CD150 (SLAM) family receptors by human hematopoietic stem and progenitor cells. Exp. Hematol. 2008, 36, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- De Salort, J.; Sintes, J.; Llinas, L.; Matesanz-Isabel, J.; Engel, P. Expression of SLAM (CD150) cell-surface receptors on human B-cell subsets: From pro-B to plasma cells. Immunol. Lett. 2011, 134, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Maciejczyk, A.; Szelachowska, J.; Szynglarewicz, B.; Szulc, R.; Szulc, A.; Wysocka, T.; Jagoda, E.; Lage, H.; Surowiak, P. CD46 Expression is an unfavorable prognostic factor in breast cancer cases. Appl. Immunohistochem. Mol. Morphol. 2011, 19, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Fabre-Lafay, S.; Monville, F.; Garrido-Urbani, S.; Berruyer-Pouyet, C.; Ginestier, C.; Reymond, N.; Finetti, P.; Sauvan, R.; Adelaide, J.; Geneix, J.; et al. Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer 2007, 7, 73. [Google Scholar] [CrossRef]
- Athanassiadou, A.M.; Patsouris, E.; Tsipis, A.; Gonidi, M.; Athanassiadou, P. The significance of Survivin and Nectin-4 expression in the prognosis of breast carcinoma. Folia Histochemica et Cytobiologica 2011, 49, 26–33. [Google Scholar] [CrossRef]
- Lattanzio, R.; Ghasemi, R.; Brancati, F.; Sorda, R.L.; Tinari, N.; Perracchio, L.; Iacobelli, S.; Mottolese, M.; Natali, P.G.; Piantelli, M. Membranous Nectin-4 expression is a risk factor for distant relapse of T1-T2, N0 luminal-A early breast cancer. Oncogenesis 2014, 3, e118. [Google Scholar] [CrossRef]
- Rajc, J.; Gugic, D.; Frohlich, I.; Marjanovic, K.; Dumencic, B. Prognostic role of Nectin-4 expression in luminal B (HER2 negative) breast cancer. Pathol. Res. Pract. 2017, 213, 1102–1108. [Google Scholar] [CrossRef]
- Erlenhofer, C.; Duprex, W.P.; Rima, B.K.; ter Meulen, V.; Schneider-Schaulies, J. Analysis of receptor (CD46, CD150) usage by measles virus. J. Gen. Virol. 2002, 83, 1431–1436. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Delpeut, S.; Sisson, G.; Black, K.M.; Richardson, C.D. Measles Virus Enters Breast and Colon Cancer Cell Lines through a PVRL4-Mediated Macropinocytosis Pathway. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Papatheodorou, I.; Fonseca, N.A.; Keays, M.; Tang, Y.A.; Barrera, E.; Bazant, W.; Burke, M.; Fullgrabe, A.; Fuentes, A.M.; George, N.; et al. Expression Atlas: Gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018, 46, D246–D251. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.; Opyrchal, M.; Aderca, I.; Schroeder, M.A.; Sarkaria, J.N.; Domingo, E.; Federspiel, M.J.; Galanis, E. Oncolytic measles virus strains have significant antitumor activity against glioma stem cells. Gene Ther. 2012, 20, 444–449. [Google Scholar] [CrossRef]
- Ma, J.; Sheng, Z.; Lv, Y.; Liu, W.; Yao, Q.; Pan, T.; Xu, Z.; Zhang, C.; Xu, G. Expression and clinical significance of Nectin-4 in hepatocellular carcinoma. OncoTargets Ther. 2016, 9, 183–190. [Google Scholar] [CrossRef][Green Version]
- Takano, A.; Ishikawa, N.; Nishino, R.; Masuda, K.; Yasui, W.; Inai, K.; Nishimura, H.; Ito, H.; Nakayama, H.; Miyagi, Y.; et al. Identification of nectin-4 oncoprotein as a diagnostic and therapeutic target for lung cancer. Cancer Res. 2009, 69, 6694–6703. [Google Scholar] [CrossRef]
- Sakuma, T.; Kodama, K.; Hara, T.; Eshita, Y.; Shibata, N.; Matsumoto, M.; Seya, T.; Mori, Y. Levels of complement regulatory molecules in lung cancer: Disappearance of the D17 epitope of CD55 in small-cell carcinoma. Jpn. J. Cancer Res. 1993, 84, 753–759. [Google Scholar] [CrossRef]
- Gordiienko, I.M.; Shlapatska, L.M.; Kovalevska, L.M.; Sidorenko, S.P. Differential expression of CD150/SLAMF1 in normal and malignant B cells on the different stages of maturation. Exp. Oncol. 2016, 38, 101–107. [Google Scholar] [CrossRef]
- Ong, H.T.; Timm, M.M.; Greipp, P.R.; Witzig, T.E.; Dispenzieri, A.; Russell, S.J.; Peng, K.W. Oncolytic measles virus targets high CD46 expression on multiple myeloma cells. Exp. Hematol. 2006, 34, 713–720. [Google Scholar] [CrossRef]
- Sherbenou, D.W.; Aftab, B.T.; Su, Y.; Behrens, C.R.; Wiita, A.; Logan, A.C.; Acosta-Alvear, D.; Hann, B.C.; Walter, P.; Shuman, M.A.; et al. Antibody-drug conjugate targeting CD46 eliminates multiple myeloma cells. J. Clin. Invest. 2016, 126, 4640–4653. [Google Scholar] [CrossRef]
- Surowiak, P.; Materna, V.; Maciejczyk, A.; Kaplenko, I.; Spaczynski, M.; Dietel, M.; Lage, H.; Zabel, M. CD46 expression is indicative of shorter revival-free survival for ovarian cancer patients. Anticancer Res. 2006, 26, 4943–4948. [Google Scholar]
- Derycke, M.S.; Pambuccian, S.E.; Gilks, C.B.; Kalloger, S.E.; Ghidouche, A.; Lopez, M.; Bliss, R.L.; Geller, M.A.; Argenta, P.A.; Harrington, K.M.; et al. Nectin 4 overexpression in ovarian cancer tissues and serum: Potential role as a serum biomarker. Am. J. Clin. Pathol. 2010, 134, 835–845. [Google Scholar] [CrossRef]
- Nabih, E.S.; Abdel Motaleb, F.I.; Salama, F.A. The diagnostic efficacy of nectin 4 expression in ovarian cancer patients. Biomarkers 2014, 19, 498–504. [Google Scholar] [CrossRef]
- Nishiwada, S.; Sho, M.; Yasuda, S.; Shimada, K.; Yamato, I.; Akahori, T.; Kinoshita, S.; Nagai, M.; Konishi, N.; Nakajima, Y. Nectin-4 expression contributes to tumor proliferation, angiogenesis and patient prognosis in human pancreatic cancer. J. Exp. Clin. Cancer Res. 2015, 34, 30. [Google Scholar] [CrossRef]
- Castro, A.G.; Hauser, T.M.; Cocks, B.G.; Abrams, J.; Zurawski, S.; Churakova, T.; Zonin, F.; Robinson, D.; Tangye, S.G.; Aversa, G.; et al. Molecular and functional characterization of mouse signaling lymphocytic activation molecule (SLAM): Differential expression and responsiveness in Th1 and Th2 cells. J. Immunol. 1999, 163, 5860–5870. [Google Scholar]
- Vilar, M.L.; Frutuoso, M.S.; Arruda, S.M.; Lima, D.M.; Bezerra, C.S.; Pompeu, M.M. The role of the SLAM-SAP signaling pathway in the modulation of CD4+ T cell responses. Braz. J. Med. Biol. Res. 2011, 44, 276–282. [Google Scholar] [CrossRef]
- Quiroga, M.F.; Martinez, G.J.; Pasquinelli, V.; Costas, M.A.; Bracco, M.M.; Malbran, A.; Olivares, L.M.; Sieling, P.A.; Garcia, V.E. Activation of signaling lymphocytic activation molecule triggers a signaling cascade that enhances Th1 responses in human intracellular infection. J. Immunol. 2004, 173, 4120–4129. [Google Scholar] [CrossRef]
- Mina, M.J.; Kula, T.; Leng, Y.; Li, M.; de Vries, R.D.; Knip, M.; Siljander, H.; Rewers, M.; Choy, D.F.; Wilson, M.S.; et al. Measles virus infection diminishes preexisting antibodies that offer protection from other pathogens. Science 2019, 366, 599–606. [Google Scholar] [CrossRef]
- Petrova, V.N.; Sawatsky, B.; Han, A.X.; Laksono, B.M.; Walz, L.; Parker, E.; Pieper, K.; Anderson, C.A.; de Vries, R.D.; Lanzavecchia, A.; et al. Incomplete genetic reconstitution of B cell pools contributes to prolonged immunosuppression after measles. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef]
- Sugiyama, T.; Yoneda, M.; Kuraishi, T.; Hattori, S.; Inoue, Y.; Sato, H.; Kai, C. Measles virus selectively blind to signaling lymphocyte activation molecule as a novel oncolytic virus for breast cancer treatment. Gene Ther. 2012, 21, 44. [Google Scholar] [CrossRef]
- Awano, M.; Fujiyuki, T.; Shoji, K.; Amagai, Y.; Murakami, Y.; Furukawa, Y.; Sato, H.; Yoneda, M.; Kai, C. Measles virus selectively blind to signaling lymphocyte activity molecule has oncolytic efficacy against nectin-4-expressing pancreatic cancer cells. Cancer Sci. 2016, 107, 1647–1652. [Google Scholar] [CrossRef]
- Fujiyuki, T.; Yoneda, M.; Amagai, Y.; Obayashi, K.; Ikeda, F.; Shoji, K.; Murakami, Y.; Sato, H.; Kai, C. A measles virus selectively blind to signaling lymphocytic activation molecule shows anti-tumor activity against lung cancer cells. Oncotarget 2015, 6, 24895–24903. [Google Scholar] [CrossRef]
- Leonard, V.H.; Sinn, P.L.; Hodge, G.; Miest, T.; Devaux, P.; Oezguen, N.; Braun, W.; McCray, P.B.J.; McChesney, M.B.; Cattaneo, R. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 2008, 118, 2448–2458. [Google Scholar] [CrossRef]
- Miest, T.S.; Frenzke, M.; Cattaneo, R. Measles virus entry through the signaling lymphocyte activation molecule governs efficacy of mantle cell lymphoma radiovirotherapy. Mol. Ther. 2013, 21, 2019–2031. [Google Scholar] [CrossRef]
- Liszewski, M.K.; Post, T.W.; Atkinson, J.P. Membrane cofactor protein (MCP or CD46): Newest member of the regulators of complement activation gene cluster. Annu. Rev. Immunol. 1991, 9, 431–455. [Google Scholar] [CrossRef]
- Ni Choileain, S.; Astier, A.L. CD46 processing: A means of expression. Immunobiology 2012, 217, 169–175. [Google Scholar] [CrossRef]
- Riley-Vargas, R.C.; Gill, D.B.; Kemper, C.; Liszewski, M.K.; Atkinson, J.P. CD46: Expanding beyond complement regulation. Trends Immunol. 2004, 25, 496–503. [Google Scholar] [CrossRef]
- Buettner, R.; Huang, M.; Gritsko, T.; Karras, J.; Enkemann, S.; Mesa, T.; Nam, S.; Yu, H.; Jove, R. Activated signal transducers and activators of transcription 3 signaling induces CD46 expression and protects human cancer cells from complement-dependent cytotoxicity. Mol. Cancer Res. 2007, 5, 823–832. [Google Scholar] [CrossRef]
- Anderson, B.D.; Nakamura, T.; Russell, S.J.; Peng, K.W. High CD46 receptor density determines preferential killing of tumor cells by oncolytic measles virus. Cancer Res. 2004, 64, 4919–4926. [Google Scholar] [CrossRef]
- Takai, Y.; Miyoshi, J.; Ikeda, W.; Ogita, H. Nectins and nectin-like molecules: Roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell Biol. 2008, 9, 603–615. [Google Scholar] [CrossRef]
- Samanta, D.; Almo, S.C. Nectin family of cell-adhesion molecules: Structural and molecular aspects of function and specificity. Cell. Mol. Life Sci. 2015, 72, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Delpeut, S.; Noyce, R.S.; Richardson, C.D. The tumor-associated marker, PVRL4 (nectin-4), is the epithelial receptor for morbilliviruses. Viruses 2014, 6, 2268–2286. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Pallasch, C.; Elia, A.E.; Braun, C.J.; Westbrook, T.F.; Hemann, M.; Elledge, S.J. A role for PVRL4-driven cell-cell interactions in tumorigenesis. Elife 2013, 2, e00358. [Google Scholar] [CrossRef]
- Suksanpaisan, L.; Russell, S.J.; Peng, K.W. High scFv-receptor affinity does not enhance the antitumor activity of HER2-retargeted measles virus. Cancer Gene Ther. 2014, 21, 256–260. [Google Scholar] [CrossRef][Green Version]
- Hasegawa, K.; Hu, C.; Nakamura, T.; Marks, J.D.; Russell, S.J.; Peng, K.W. Affinity thresholds for membrane fusion triggering by viral glycoproteins. J. Virol. 2007, 81, 13149–13157. [Google Scholar] [CrossRef]
- Schneider, U.; Bullough, F.; Vongpunsawad, S.; Russell, S.J.; Cattaneo, R. Recombinant measles viruses efficiently entering cells through targeted receptors. J. Virol. 2000, 74, 9928–9936. [Google Scholar] [CrossRef]
- Hammond, A.L.; Plemper, R.K.; Zhang, J.; Schneider, U.; Russell, S.J.; Cattaneo, R. Single-chain antibody displayed on a recombinant measles virus confers entry through the tumor-associated carcinoembryonic antigen. J. Virol. 2001, 75, 2087–2096. [Google Scholar] [CrossRef]
- Bucheit, A.D.; Kumar, S.; Grote, D.M.; Lin, Y.; von Messling, V.; Cattaneo, R.B.; Fielding, A.K. An oncolytic measles virus engineered to enter cells through the CD20 antigen. Mol. Ther. 2003, 7, 62–72. [Google Scholar] [CrossRef]
- Peng, K.W.; Donovan, K.A.; Schneider, U.; Cattaneo, R.; Lust, J.A.; Russell, S.J. Oncolytic measles viruses displaying a single-chain antibody against CD38, a myeloma cell marker. Blood 2003, 101, 2557–2562. [Google Scholar] [CrossRef]
- Hallak, L.K.; Merchan, J.R.; Storgard, C.M.; Loftus, J.C.; Russell, S.J. Targeted measles virus vector displaying echistatin infects endothelial cells via alpha(v)beta3 and leads to tumor regression. Cancer Res. 2005, 65, 5292–5300. [Google Scholar] [CrossRef]
- Hammarstrom, S. The carcinoembryonic antigen (CEA) family: Structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999, 9, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Emerson, S.G. Ex vivo expansion of hematopoietic precursors, progenitors, and stem cells: The next generation of cellular therapeutics. Blood 1996, 87, 3082–3088. [Google Scholar] [CrossRef]
- Sanai, N.; Alvarez-Buylla, A.; Berger, M.S. Neural stem cells and the origin of gliomas. N. Engl. J. Med. 2005, 353, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Sun, C.; Feng, F.; Ge, M.; Xia, L. Do relevant markers of cancer stem cells CD133 and Nestin indicate a poor prognosis in glioma patients? A systematic review and meta-analysis. J. Exp. Clin. Cancer Res. 2015, 34, 44. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Qi, X.W.; Yan, G.N.; Zhang, Q.B.; Xu, C.; Bian, X.W. Is CD133 expression a prognostic biomarker of non-small-cell lung cancer? A systematic review and meta-analysis. PLoS ONE 2014, 9, e100168. [Google Scholar] [CrossRef] [PubMed]
- Yin, A.H.; Miraglia, S.; Zanjani, E.D.; Almeida-Porada, G.; Ogawa, M.; Leary, A.G.; Olweus, J.; Kearney, J.; Buck, D.W. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 1997, 90, 5002–5012. [Google Scholar] [CrossRef]
- Corbeil, D.; Roper, K.; Hellwig, A.; Tavian, M.; Miraglia, S.; Watt, S.M.; Simmons, P.J.; Peault, B.; Buck, D.W.; Huttner, W.B. The human AC133 hematopoietic stem cell antigen is also expressed in epithelial cells and targeted to plasma membrane protrusions. J. Biol. Chem. 2000, 275, 5512–5520. [Google Scholar] [CrossRef]
- Hall, P.A.; D’Ardenne, A.J.; Richards, M.A.; Stansfeld, A.G. Lymphoplasmacytoid lymphoma: An immunohistological study. J. Pathol. 1987, 153, 213–223. [Google Scholar] [CrossRef]
- Thomas, D.A.; O’Brien, S.; Jorgensen, J.L.; Cortes, J.; Faderl, S.; Garcia-Manero, G.; Verstovsek, S.; Koller, C.; Pierce, S.; Huh, Y.; et al. Prognostic significance of CD20 expression in adults with de novo precursor B-lineage acute lymphoblastic leukemia. Blood 2009, 113, 6330–6337. [Google Scholar] [CrossRef]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef]
- Tzankov, A.; Krugmann, J.; Fend, F.; Fischhofer, M.; Greil, R.; Dirnhofer, S. Prognostic significance of CD20 expression in classical Hodgkin lymphoma: A clinicopathological study of 119 cases. Clin. Cancer Res. 2003, 9, 1381–1386. [Google Scholar] [PubMed]
- Li, Z.; Xu, Y.; An, G.; Wang, H.; Deng, S.; Zhao, Y.; Qiu, L. The characteristics of 62 cases of CD20-positive multiple myeloma. Zhonghua Xue Ye Xue Za Zhi 2015, 36, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhou, X.J.; Yin, H.L.; Lu, Z.F.; Zhou, H.B. Expression of CD20 in thymomas and its clinical implication. Zhonghua Bing Li Xue Za Zhi 2010, 39, 611–614. [Google Scholar] [PubMed]
- Ibrahim, S.; Keating, M.; Do, K.A.; O’Brien, S.; Huh, Y.O.; Jilani, I.; Lerner, S.; Kantarjian, H.M.; Albitar, M. CD38 expression as an important prognostic factor in B-cell chronic lymphocytic leukemia. Blood 2001, 98, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Burgler, S. Role of CD38 Expression in Diagnosis and Pathogenesis of Chronic Lymphocytic Leukemia and Its Potential as Therapeutic Target. Crit. Rev. Immunol. 2015, 35, 417–432. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.; Li, P.F.; Lu, Y.; Xia, Z.J.; Huang, H.Q.; Zhang, Y.J. CD38 expression predicts poor prognosis and might be a potential therapy target in extranodal NK/T cell lymphoma, nasal type. Ann. Hematol. 2015, 94, 1381–1388. [Google Scholar] [CrossRef]
- San Miguel, J.F.; Garcia-Sanz, R.; Gonzalez, M.; Orfao, A. Immunophenotype and DNA cell content in multiple myeloma. Baillieres Clin. Haematol. 1995, 8, 735–759. [Google Scholar] [CrossRef]
- Mitri, Z.; Constantine, T.; O’Regan, R. The HER2 Receptor in Breast Cancer: Pathophysiology, Clinical Use, and New Advances in Therapy. Chemother. Res. Pract. 2012, 2012, 743193. [Google Scholar] [CrossRef]
- Ruschoff, J.; Hanna, W.; Bilous, M.; Hofmann, M.; Osamura, R.Y.; Penault-Llorca, F.; van de Vijver, M.; Viale, G. HER2 testing in gastric cancer: A practical approach. Mod. Pathol. 2012, 25, 637–650. [Google Scholar] [CrossRef]
- Santin, A.D.; Bellone, S.; Roman, J.J.; McKenney, J.K.; Pecorelli, S. Trastuzumab treatment in patients with advanced or recurrent endometrial carcinoma overexpressing HER2/neu. Int. J. Gynaecol. Obstet. 2008, 102, 128–131. [Google Scholar] [CrossRef]
- Buza, N.; Roque, D.M.; Santin, A.D. HER2/neu in Endometrial Cancer: A Promising Therapeutic Target with Diagnostic Challenges. Arch. Pathol. Lab. Med. 2014, 138, 343–350. [Google Scholar] [CrossRef]
- Teplinsky, E.; Muggia, F. Targeting HER2 in ovarian and uterine cancers: Challenges and future directions. Gynecol. Oncol. 2014, 135, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Siena, S.; Sartore-Bianchi, A.; Marsoni, S.; Hurwitz, H.I.; McCall, S.J.; Penault-Llorca, F.; Srock, S.; Bardelli, A.; Trusolino, L. Targeting the human epidermal growth factor receptor 2 (HER2) oncogene in colorectal cancer. Ann. Oncol. 2018, 29, 1108–1119. [Google Scholar] [CrossRef]
- Tang, T.; Zhang, D.L. Study on extracellular matrix metalloproteinase inducer and human epidermal growth factor receptor-2 protein expression in papillary thyroid carcinoma using a quantum dot-based immunofluorescence technique. Exp. Ther. Med. 2015, 9, 1331–1335. [Google Scholar] [CrossRef][Green Version]
- Kumar, S.; Prajapati, O.; Vaiphei, K.; Parmar, K.M.; Sriharsha, A.S.; Singh, S.K. Human epidermal growth factor receptor 2/neu overexpression in urothelial carcinoma of the bladder and its prognostic significance: Is it worth hype? South Asian J. Cancer 2015, 4, 115–117. [Google Scholar] [CrossRef]
- Hasegawa, K.; Nakamura, T.; Harvey, M.; Ikeda, Y.; Oberg, A.; Figini, M.; Canevari, S.; Hartmann, L.C.; Peng, K.W. The use of a tropism-modified measles virus in folate receptor-targeted virotherapy of ovarian cancer. Clin. Cancer Res. 2006, 12, 6170–6178. [Google Scholar] [CrossRef]
- Chang, S.S. Overview of prostate-specific membrane antigen. Rev. Urol. 2004, 6, S13–S18. [Google Scholar]
- Lal, S.; Raffel, C. Using Cystine Knot Proteins as a Novel Approach to Retarget Oncolytic Measles Virus. Mol. Ther. Oncolytics 2017, 7, 57–66. [Google Scholar] [CrossRef]
- Vongpunsawad, S.; Oezgun, N.; Braun, W.; Cattaneo, R. Selectively receptor-blind measles viruses: Identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J. Virol. 2004, 78, 302–313. [Google Scholar] [CrossRef]
- Hadac, E.M.; Peng, K.W.; Nakamura, T.; Russell, S.J. Reengineering paramyxovirus tropism. Virology 2004, 329, 217–225. [Google Scholar] [CrossRef]
- Nakamura, T.; Peng, K.W.; Harvey, M.; Greiner, S.; Lorimer, I.A.; James, C.D.; Russell, S.J. Rescue and propagation of fully retargeted oncolytic measles viruses. Nat. Biotechnol. 2005, 23, 209–214. [Google Scholar] [CrossRef]
- Bach, P.; Abel, T.; Hoffmann, C.; Gal, Z.; Braun, G.; Voelker, I.; Ball, C.R.; Johnston, I.C.; Lauer, U.M.; Herold-Mende, C.; et al. Specific Elimination of CD133+ Tumor Cells with Targeted Oncolytic Measles Virus. Cancer Res. 2013, 4, 4. [Google Scholar] [CrossRef]
- Kleinlutzum, D.; Hanauer, J.D.S.; Muik, A.; Hanschmann, K.M.; Kays, S.K.; Ayala-Breton, C.; Peng, K.W.; Muhlebach, M.D.; Abel, T.; Buchholz, C.J. Enhancing the Oncolytic Activity of CD133-Targeted Measles Virus: Receptor Extension or Chimerism with Vesicular Stomatitis Virus Are Most Effective. Front. Oncol. 2017, 7, 127. [Google Scholar] [CrossRef]
- Jing, Y.; Bejarano, M.T.; Zaias, J.; Merchan, J.R. In vivo anti-metastatic effects of uPAR retargeted measles virus in syngeneic and xenograft models of mammary cancer. Breast Cancer Res. Treat. 2015, 149, 99–108. [Google Scholar] [CrossRef]
- Jing, Y.; Chavez, V.; Ban, Y.; Acquavella, N.; El-Ashry, D.; Pronin, A.; Chen, X.; Merchan, J.R. Molecular Effects of Stromal-Selective Targeting by uPAR-Retargeted Oncolytic Virus in Breast Cancer. Mol. Cancer Res. 2017, 15, 1410–1420. [Google Scholar] [CrossRef]
- Friedrich, K.; Hanauer, J.R.; Prufer, S.; Munch, R.C.; Volker, I.; Filippis, C.; Jost, C.; Hanschmann, K.M.; Cattaneo, R.; Peng, K.W.; et al. DARPin-targeting of measles virus: Unique bispecificity, effective oncolysis, and enhanced safety. Mol. Ther. 2013, 21, 849–859. [Google Scholar] [CrossRef]
- Hanauer, J.R.; Gottschlich, L.; Riehl, D.; Rusch, T.; Koch, V.; Friedrich, K.; Hutzler, S.; Prufer, S.; Friedel, T.; Hanschmann, K.M.; et al. Enhanced lysis by bispecific oncolytic measles viruses simultaneously using HER2/neu or EpCAM as target receptors. Mol. Ther. Oncolytics 2016, 3, 16003. [Google Scholar] [CrossRef]
- Hanauer, J.R.H.; Koch, V.; Lauer, U.M.; Muhlebach, M.D. High-Affinity DARPin Allows Targeting of MeV to Glioblastoma Multiforme in Combination with Protease Targeting without Loss of Potency. Mol. Ther. Oncolytics 2019, 15, 186–200. [Google Scholar] [CrossRef]
- Liu, C.; Hasegawa, K.; Russell, S.J.; Sadelain, M.; Peng, K.W. Prostate-specific membrane antigen retargeted measles virotherapy for the treatment of prostate cancer. Prostate 2009, 69, 1128–1141. [Google Scholar] [CrossRef]
- Slobod, K.S.; Shenep, J.L.; Lujan-Zilbermann, J.; Allison, K.; Brown, B.; Scroggs, R.A.; Portner, A.; Coleclough, C.; Hurwitz, J.L. Safety and immunogenicity of intranasal murine parainfluenza virus type 1 (Sendai virus) in healthy human adults. Vaccine 2004, 22, 3182–3186. [Google Scholar] [CrossRef]
- Adderson, E.; Branum, K.; Sealy, R.E.; Jones, B.G.; Surman, S.L.; Penkert, R.; Freiden, P.; Slobod, K.S.; Gaur, A.H.; Hayden, R.T.; et al. Safety and immunogenicity of an intranasal Sendai virus-based human parainfluenza virus type 1 vaccine in 3- to 6-year-old children. Clin. Vaccine Immunol. 2015, 22, 298–303. [Google Scholar] [CrossRef]
- Nyombayire, J.; Anzala, O.; Gazzard, B.; Karita, E.; Bergin, P.; Hayes, P.; Kopycinski, J.; Omosa-Manyonyi, G.; Jackson, A.; Bizimana, J.; et al. First-in-Human Evaluation of the Safety and Immunogenicity of an Intranasally Administered Replication-Competent Sendai Virus-Vectored HIV Type 1 Gag Vaccine: Induction of Potent T-Cell or Antibody Responses in Prime-Boost Regimens. J. Infect. Dis. 2017, 215, 95–104. [Google Scholar] [CrossRef]
- Ishii, H.; Matano, T. Development of an AIDS vaccine using Sendai virus vectors. Vaccine 2015, 33, 6061–6065. [Google Scholar] [CrossRef]
- Moriya, C.; Horiba, S.; Kurihara, K.; Kamada, T.; Takahara, Y.; Inoue, M.; Iida, A.; Hara, H.; Shu, T.; Hasegawa, M.; et al. Intranasal Sendai viral vector vaccination is more immunogenic than intramuscular under pre-existing anti-vector antibodies. Vaccine 2011, 29, 8557–8563. [Google Scholar] [CrossRef]
- Institute of Laboratory Animal Resources Committee on Infectious Diseases of Mice and Rats. Infectious Diseases of Mice and Rats; National Academy Press: Washington, WA, USA, 1991; p. xi. 397p. [Google Scholar]
- Nagai, Y.; Takakura, A.; Irie, T.; Yonemitsu, Y.; Gotoh, B. Evolution of Sendai Virus: The Journey from Mouse Pathogen to a State-of-the-Art Tool in Virus Research and Biotechnology. In The Biology of Paramyxoviruses; Caister Academic Press: Maryland, MD, USA, 2011. [Google Scholar]
- Ilyinskaya, G.V.; Mukhina, E.V.; Soboleva, A.V.; Matveeva, O.V.; Chumakov, P.M. Oncolytic Sendai Virus Therapy of Canine Mast Cell Tumors (A Pilot Study). Front. Vet. Sci. 2018, 5, 116. [Google Scholar] [CrossRef]
- Kinoh, H.; Inoue, M.; Washizawa, K.; Yamamoto, T.; Fujikawa, S.; Tokusumi, Y.; Iida, A.; Nagai, Y.; Hasegawa, M. Generation of a recombinant Sendai virus that is selectively activated and lyses human tumor cells expressing matrix metalloproteinases. Gene Ther. 2004, 11, 1137–1145. [Google Scholar] [CrossRef][Green Version]
- Yonemitsu, Y.; Ueda, Y.; Kinoh, H.; Hasegawa, M. Immunostimulatory virotherapy using recombinant Sendai virus as a new cancer therapeutic regimen. Front. Biosci. 2008, 13, 1892–1898. [Google Scholar] [CrossRef]
- Kinoh, H.; Inoue, M. New cancer therapy using genetically-engineered oncolytic Sendai virus vector. Front. Biosci. 2008, 13, 2327–2334. [Google Scholar] [CrossRef]
- Tatsuta, K.; Tanaka, S.; Tajiri, T.; Shibata, S.; Komaru, A.; Ueda, Y.; Inoue, M.; Hasegawa, M.; Suita, S.; Sueishi, K.; et al. Complete elimination of established neuroblastoma by synergistic action of gamma-irradiation and DCs treated with rSeV expressing interferon-beta gene. Gene Ther. 2009, 16, 240–251. [Google Scholar] [CrossRef]
- Iwadate, Y.; Inoue, M.; Saegusa, T.; Tokusumi, Y.; Kinoh, H.; Hasegawa, M.; Tagawa, M.; Yamaura, A.; Shimada, H. Recombinant Sendai virus vector induces complete remission of established brain tumors through efficient interleukin-2 gene transfer in vaccinated rats. Clin. Cancer Res. 2005, 11, 3821–3827. [Google Scholar] [CrossRef]
- Kurooka, M.; Kaneda, Y. Inactivated Sendai virus particles eradicate tumors by inducing immune responses through blocking regulatory T cells. Cancer Res. 2007, 67, 227–236. [Google Scholar] [CrossRef]
- Kawano, H.; Komaba, S.; Kanamori, T.; Kaneda, Y. A new therapy for highly effective tumor eradication using HVJ-E combined with chemotherapy. BMC Med. 2007, 5, 28. [Google Scholar] [CrossRef]
- Kawano, H.; Komaba, S.; Yamasaki, T.; Maeda, M.; Kimura, Y.; Maeda, A.; Kaneda, Y. New potential therapy for orthotopic bladder carcinoma by combining HVJ envelope with doxorubicin. Cancer Chemother. Pharmacol. 2008, 61, 973–978. [Google Scholar] [CrossRef]
- Fujihara, A.; Kurooka, M.; Miki, T.; Kaneda, Y. Intratumoral injection of inactivated Sendai virus particles elicits strong antitumor activity by enhancing local CXCL10 expression and systemic NK cell activation. Cancer Immunol. Immunother. 2008, 57, 73–84. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Miyamoto, Y.; Inoue, T.; Kaneda, Y. Efficient eradication of hormone-resistant human prostate cancers by inactivated Sendai virus particle. Int. J. Cancer 2009, 124, 2478–2487. [Google Scholar] [CrossRef]
- Lallemand, C.; Blanchard, B.; Palmieri, M.; Lebon, P.; May, E.; Tovey, M.G. Single-stranded RNA viruses inactivate the transcriptional activity of p53 but induce NOXA-dependent apoptosis via post-translational modifications of IRF-1, IRF-3 and CREB. Oncogene 2007, 26, 328–338. [Google Scholar] [CrossRef]
- Shah, N.R.; Sunderland, A.; Grdzelishvili, V.Z. Cell type mediated resistance of vesicular stomatitis virus and Sendai virus to ribavirin. PLoS ONE 2010, 5, e11265. [Google Scholar] [CrossRef]
- Takeuchi, K.; Komatsu, T.; Kitagawa, Y.; Sada, K.; Gotoh, B. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J. Virol. 2008, 82, 10102–10110. [Google Scholar] [CrossRef]
- Zainutdinov, S.S.; Grazhdantseva, A.A.; Kochetkov, D.V.; Chumakov, P.M.; Netesov, S.V.; Matveeva, O.V.; Kochneva, G.V. Change in Oncolytic Activity of Sendai Virus during Adaptation to Cell Cultures. Mol. Gen. Microbiol. Virol. 2017, 32, 212–217. [Google Scholar] [CrossRef]
- Bitzer, M.; Lauer, U.; Baumann, C.; Spiegel, M.; Gregor, M.; Neubert, W.J. Sendai virus efficiently infects cells via the asialoglycoprotein receptor and requires the presence of cleaved F0 precursor proteins for this alternative route of cell entry. J. Virol. 1997, 71, 5481–5486. [Google Scholar] [CrossRef]
- Keskinen, P.; Nyqvist, M.; Sareneva, T.; Pirhonen, J.; Melen, K.; Julkunen, I. Impaired antiviral response in human hepatoma cells. Virology 1999, 263, 364–375. [Google Scholar] [CrossRef]
- Sumpter, R.J.; Loo, Y.M.; Foy, E.; Li, K.; Yoneyama, M.; Fujita, T.; Lemon, S.M.; Gale, M.J. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005, 79, 2689–2699. [Google Scholar] [CrossRef]
- Buggele, W.A.; Horvath, C.M. MicroRNA profiling of Sendai virus-infected A549 cells identifies miR-203 as an interferon-inducible regulator of IFIT1/ISG56. J. Virol. 2013, 87, 9260–9270. [Google Scholar] [CrossRef]
- Bedsaul, J.R.; Zaritsky, L.A.; Zoon, K.C. Type I Interferon-Mediated Induction of Antiviral Genes and Proteins Fails to Protect Cells from the Cytopathic Effects of Sendai Virus Infection. J. Interferon Cytokine Res. 2016, 36, 652–665. [Google Scholar] [CrossRef]
- Genoyer, E.; Kulej, K.; Hung, C.T.; Thibault, P.A.; Azarm, K.; Takimoto, T.; Garcia, B.A.; Lee, B.; Lakdawala, S.; Weitzman, M.D.; et al. The Viral Polymerase Complex Mediates the Interaction of Viral Ribonucleoprotein Complexes with Recycling Endosomes during Sendai Virus Assembly. mBio 2020, 11. [Google Scholar] [CrossRef]
- Mandhana, R.; Horvath, C.M. Sendai Virus Infection Induces Expression of Novel RNAs in Human Cells. Sci. Rep. 2018, 8, 16815. [Google Scholar] [CrossRef]
- Belova, A.A.; Sosnovtseva, A.O.; Lipatova, A.V.; Njushko, K.M.; Volchenko, N.N.; Belyakov, M.M.; Sudalenko, O.V.; Krasheninnikov, A.A.; Shegai, P.V.; Sadritdinova, A.F.; et al. Biomarkers of prostate cancer sensitivity to the Sendai virus. Mol. Biol. 2017, 51, 94–103. [Google Scholar] [CrossRef]
- Wheelock, E.F.; Dingle, J.H. Observations on the Repeated Administration of Viruses to a Patient with Acute Leukemia. A Preliminary Report. N. Engl. J. Med. 1964, 271, 645–651. [Google Scholar] [CrossRef]
- Senina, A.; Matveeva, O.; Senin, V. Method for Cancer Immunotherapy and Pharmaceutical Compositions Based on Oncolytic Non-Pathogenic Sendai Virus. US Patent 9526779, 27 December 2016. [Google Scholar]
- Suzuki, Y.; Suzuki, T.; Matsumoto, M. Isolation and characterization of receptor sialoglycoprotein for hemagglutinating virus of Japan (Sendai virus) from bovine erythrocyte membrane. J. Biochem. 1983, 93, 1621–1633. [Google Scholar] [CrossRef]
- Markwell, M.A.; Portner, A.; Schwartz, A.L. An alternative route of infection for viruses: Entry by means of the asialoglycoprotein receptor of a Sendai virus mutant lacking its attachment protein. Proc. Natl. Acad. Sci. USA 1985, 82, 978–982. [Google Scholar] [CrossRef]
- Wybenga, L.E.; Epand, R.F.; Nir, S.; Chu, J.W.; Sharom, F.J.; Flanagan, T.D.; Epand, R.M. Glycophorin as a receptor for Sendai virus. Biochemistry 1996, 35, 9513–9518. [Google Scholar] [CrossRef]
- Muthing, J. Influenza A and Sendai viruses preferentially bind to fucosylated gangliosides with linear poly-N-acetyllactosaminyl chains from human granulocytes. Carbohydr. Res. 1996, 290, 217–224. [Google Scholar] [CrossRef]
- Villar, E.; Barroso, I.M. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: A minireview. Glycoconj. J. 2006, 23, 5–17. [Google Scholar] [CrossRef]
- Macher, B.A.; Beckstead, J.H. Distribution of VIM-2 and SSEA-1 glycoconjugate epitopes among human leukocytes and leukemia cells. Leuk. Res. 1990, 14, 119–130. [Google Scholar] [CrossRef]
- Holmgren, J.; Svennerholm, L.; Elwing, H.; Fredman, P.; Strannegard, O. Sendai virus receptor: Proposed recognition structure based on binding to plastic-adsorbed gangliosides. Proc. Natl. Acad. Sci. USA 1980, 77, 1947–1950. [Google Scholar] [CrossRef]
- Markwell, M.A.; Svennerholm, L.; Paulson, J.C. Specific gangliosides function as host cell receptors for Sendai virus. Proc. Natl. Acad. Sci. USA 1981, 78, 5406–5410. [Google Scholar] [CrossRef]
- Suzuki, Y.; Suzuki, T.; Matsunaga, M.; Matsumoto, M. Gangliosides as paramyxovirus receptor. Structural requirement of sialo-oligosaccharides in receptors for hemagglutinating virus of Japan (Sendai virus) and Newcastle disease virus. J. Biochem. 1985, 97, 1189–1199. [Google Scholar] [CrossRef]
- Yu, R.K.; Tsai, Y.T.; Ariga, T.; Yanagisawa, M. Structures, biosynthesis, and functions of gangliosides—An overview. J. Oleo Sci. 2011, 60, 537–544. [Google Scholar] [CrossRef]
- Suzuki, T.; Portner, A.; Scroggs, R.A.; Uchikawa, M.; Koyama, N.; Matsuo, K.; Suzuki, Y.; Takimoto, T. Receptor specificities of human respiroviruses. J. Virol. 2001, 75, 4604–4613. [Google Scholar] [CrossRef]
- Umeda, M.; Nojima, S.; Inoue, K. Activity of human erythrocyte gangliosides as a receptor to HVJ. Virology 1984, 133, 172–182. [Google Scholar] [CrossRef]
- Liang, J.X.; Liang, Y.; Gao, W. Clinicopathological and prognostic significance of sialyl Lewis X overexpression in patients with cancer: A meta-analysis. OncoTargets Ther. 2016, 9, 3113–3125. [Google Scholar] [CrossRef][Green Version]
- Ogawa, J.; Sano, A.; Inoue, H.; Koide, S. Expression of Lewis-related antigen and prognosis in stage I non-small cell lung cancer. Ann. Thorac. Surg. 1995, 59, 412–415. [Google Scholar] [CrossRef]
- Yu, C.J.; Shih, J.Y.; Lee, Y.C.; Shun, C.T.; Yuan, A.; Yang, P.C. Sialyl Lewis antigens: Association with MUC5AC protein and correlation with post-operative recurrence of non-small cell lung cancer. Lung Cancer 2005, 47, 59–67. [Google Scholar] [CrossRef]
- Sterner, E.; Flanagan, N.; Gildersleeve, J.C. Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database. ACS Chem. Biol. 2016, 11, 1773–1783. [Google Scholar] [CrossRef]
- Fukuoka, K.; Narita, N.; Saijo, N. Increased expression of sialyl Lewis(x) antigen is associated with distant metastasis in lung cancer patients: Immunohistochemical study on bronchofiberscopic biopsy specimens. Lung Cancer 1998, 20, 109–116. [Google Scholar] [CrossRef]
- Nakamori, S.; Kameyama, M.; Imaoka, S.; Furukawa, H.; Ishikawa, O.; Sasaki, Y.; Izumi, Y.; Irimura, T. Involvement of carbohydrate antigen sialyl Lewis(x) in colorectal cancer metastasis. Dis. Colon Rectum 1997, 40, 420–431. [Google Scholar] [CrossRef]
- Nakagoe, T.; Fukushima, K.; Tanaka, K.; Sawai, T.; Tsuji, T.; Jibiki, M.; Nanashima, A.; Yamaguchi, H.; Yasutake, T.; Ayabe, H.; et al. Evaluation of sialyl Lewis(a), sialyl Lewis(x), and sialyl Tn antigens expression levels as predictors of recurrence after curative surgery in node-negative colorectal cancer patients. J. Exp. Clin. Cancer Res. 2002, 21, 107–113. [Google Scholar]
- Yamadera, M.; Shinto, E.; Tsuda, H.; Kajiwara, Y.; Naito, Y.; Hase, K.; Yamamoto, J.; Ueno, H. Sialyl Lewis(x) expression at the invasive front as a predictive marker of liver recurrence in stage II colorectal cancer. Oncol. Lett. 2018, 15, 221–228. [Google Scholar] [CrossRef]
- Nakagoe, T.; Fukushima, K.; Sawai, T.; Tsuji, T.; Jibiki, M.; Nanashima, A.; Tanaka, K.; Yamaguchi, H.; Yasutake, T.; Ayabe, H.; et al. Increased expression of sialyl Lewis(x) antigen as a prognostic factor in patients with stage 0, I, and II gastric cancer. Cancer Lett. 2002, 175, 213–221. [Google Scholar] [CrossRef]
- Trinchera, M.; Aronica, A.; Dall’Olio, F. Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis x in Gastrointestinal Cancers. Biology 2017, 6, 16. [Google Scholar] [CrossRef]
- Nakagoe, T.; Fukushima, K.; Itoyanagi, N.; Ikuta, Y.; Oka, T.; Nagayasu, T.; Ayabe, H.; Hara, S.; Ishikawa, H.; Minami, H. Expression of ABH/Lewis-related antigens as prognostic factors in patients with breast cancer. J. Cancer Res. Clin. Oncol. 2002, 128, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, U.; Mylonas, I.; Shabani, N.; Kunert-Keil, C.; Schindlbeck, C.; Gerber, B.; Friese, K. Expression of sialyl Lewis X, Sialyl Lewis A, E-cadherin and cathepsin-D in human breast cancer: Immunohistochemical analysis in mammary carcinoma in situ, invasive carcinomas and their lymph node metastasis. Anticancer Res. 2005, 25, 1615–1622. [Google Scholar]
- Carrascal, M.A.; Silva, M.; Ferreira, J.A.; Azevedo, R.; Ferreira, D.; Silva, A.M.N.; Ligeiro, D.; Santos, L.L.; Sackstein, R.; Videira, P.A. A functional glycoproteomics approach identifies CD13 as a novel E-selectin ligand in breast cancer. Biochimica et Biophysica Acta 2018. [Google Scholar] [CrossRef] [PubMed]
- Dimitroff, C.J.; Lechpammer, M.; Long-Woodward, D.; Kutok, J.L. Rolling of human bone-metastatic prostate tumor cells on human bone marrow endothelium under shear flow is mediated by E-selectin. Cancer Res. 2004, 64, 5261–5269. [Google Scholar] [CrossRef]
- Munkley, J. Glycosylation is a global target for androgen control in prostate cancer cells. Endocr. Relat. Cancer 2017, 24, R49–R64. [Google Scholar] [CrossRef]
- Idikio, H.A. Sialyl-Lewis-X, Gleason grade and stage in non-metastatic human prostate cancer. Glycoconj. J. 1997, 14, 875–877. [Google Scholar] [CrossRef] [PubMed]
- Takada, A.; Ohmori, K.; Yoneda, T.; Tsuyuoka, K.; Hasegawa, A.; Kiso, M.; Kannagi, R. Contribution of carbohydrate antigens sialyl Lewis A and Sialyl Lewis X to adhesion of human cancer cells to vascular endothelium. Cancer Res. 1993, 53, 354–361. [Google Scholar] [PubMed]
- Majdic, O.; Bettelheim, P.; Stockinger, H.; Aberer, W.; Liszka, K.; Lutz, D.; Knapp, W. M2, a novel myelomonocytic cell surface antigen and its distribution on leukemic cells. Int. J. Cancer 1984, 33, 617–623. [Google Scholar] [CrossRef]
- Noguchi, M.; Sato, N.; Sugimori, H.; Mori, K.; Oshimi, K. A minor E-selectin ligand, CD65, is critical for extravascular infiltration of acute myeloid leukemia cells. Leuk. Res. 2001, 25, 847–853. [Google Scholar] [CrossRef]
- Liang, Y.J.; Ding, Y.; Levery, S.B.; Lobaton, M.; Handa, K.; Hakomori, S.I. Differential expression profiles of glycosphingolipids in human breast cancer stem cells vs. cancer non-stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 4968–4973. [Google Scholar] [CrossRef]
- Hatano, K.; Miyamoto, Y.; Nonomura, N.; Kaneda, Y. Expression of gangliosides, GD1a, and sialyl paragloboside is regulated by NF-κB-dependent transcriptional control of α2,3-sialyltransferase I, II, and VI in human castration-resistant prostate cancer cells. Int. J. Cancer 2011, 129, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, H.; Aoyagi, M.; Kasama, T.; Handa, S.; Hirakawa, K.; Taki, T. GT1b in human metastatic brain tumors: GT1b as a brain metastasis-associated ganglioside. Biochimica et Biophysica Acta 1999, 1437, 93–99. [Google Scholar] [CrossRef]
- Merritt, W.D.; Sztein, M.B.; Taylor, B.; Reaman, G.H. Immunoreactivity of leukemic lymphoblasts of T-cell and B-cell precursor origin with monoclonal anti-GD3 and anti-GM3 antibodies. Leukemia 1991, 5, 1087–1091. [Google Scholar] [PubMed]
- Westrick, M.A.; Lee, W.M.; Macher, B.A. Isolation and characterization of gangliosides from chronic myelogenous leukemia cells. Cancer Res. 1983, 43, 5890–5894. [Google Scholar]
- Hara, H.; Hara, H.; Hironaka, T.; Inoue, M.; Iida, A.; Shu, T.; Hasegawa, M.; Nagai, Y.; Falsey, A.R.; Kamali, A.; et al. Prevalence of specific neutralizing antibodies against Sendai virus in populations from different geographic areas: Implications for AIDS vaccine development using Sendai virus vectors. Hum. Vaccin. 2011, 7, 639–645. [Google Scholar] [CrossRef]
- Makela, M.J.; Marusyk, R.G.; Norrby, E.; Tyrrell, D.L.; Salmi, A.A. Antibodies to measles virus surface polypeptides in an immunized student population before and after booster vaccination. Vaccine 1989, 7, 541–545. [Google Scholar] [CrossRef]
- Roy, D.G.; Bell, J.C. Cell carriers for oncolytic viruses: Current challenges and future directions. Oncolytic Virother. 2013, 2, 47–56. [Google Scholar] [CrossRef]
- Ong, H.T.; Federspiel, M.J.; Guo, C.M.; Ooi, L.L.; Russell, S.J.; Peng, K.W.; Hui, K.M. Systemically delivered measles virus-infected mesenchymal stem cells can evade host immunity to inhibit liver cancer growth. J. Hepatol. 2013, 16, 00451–00457. [Google Scholar] [CrossRef]
- Grosjean, I.; Caux, C.; Bella, C.; Berger, I.; Wild, F.; Banchereau, J.; Kaiserlian, D. Measles virus infects human dendritic cells and blocks their allostimulatory properties for CD4+ T cells. J. Exp. Med. 1997, 186, 801–812. [Google Scholar] [CrossRef]
- de Swart, R.L.; Ludlow, M.; de Witte, L.; Yanagi, Y.; van Amerongen, G.; McQuaid, S.; Yuksel, S.; Geijtenbeek, T.B.; Duprex, W.P.; Osterhaus, A.D. Predominant infection of CD150+ lymphocytes and dendritic cells during measles virus infection of macaques. PLoS Pathog. 2007, 3, e178. [Google Scholar] [CrossRef]
- Derakhshani, S.; Kurz, A.; Japtok, L.; Schumacher, F.; Pilgram, L.; Steinke, M.; Kleuser, B.; Sauer, M.; Schneider-Schaulies, S.; Avota, E. Measles Virus Infection Fosters Dendritic Cell Motility in a 3D Environment to Enhance Transmission to Target Cells in the Respiratory Epithelium. Front. Immunol. 2019, 10, 1294. [Google Scholar] [CrossRef] [PubMed]
- Kiener, R.; Fleischmann, M.; Wiegand, M.A.; Lemmermann, N.A.W.; Schwegler, C.; Kaufmann, C.; Renzaho, A.; Thomas, S.; Felder, E.; Niller, H.H.; et al. Efficient Delivery of Human Cytomegalovirus T Cell Antigens by Attenuated Sendai Virus Vectors. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Okano, S.; Yonemitsu, Y.; Onimaru, M.; Sata, S.; Nagata-Takeshita, H.; Inoue, M.; Zhu, T.; Hasegawa, M.; Moroi, Y.; et al. Induction of efficient antitumor immunity using dendritic cells activated by recombinant Sendai virus and its modulation by exogenous IFN-β gene. J. Immunol. 2006, 177, 3564–3576. [Google Scholar] [CrossRef] [PubMed]
- Ilett, E.J.; Prestwich, R.J.; Kottke, T.; Errington, F.; Thompson, J.M.; Harrington, K.J.; Pandha, H.S.; Coffey, M.; Selby, P.J.; Vile, R.G.; et al. Dendritic cells and T cells deliver oncolytic reovirus for tumour killing despite pre-existing anti-viral immunity. Gene Ther. 2009, 16, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Ilett, E.J.; Barcena, M.; Errington-Mais, F.; Griffin, S.; Harrington, K.J.; Pandha, H.S.; Coffey, M.; Selby, P.J.; Limpens, R.W.; Mommaas, M.; et al. Internalization of oncolytic reovirus by human dendritic cell carriers protects the virus from neutralization. Clin. Cancer Res. 2011, 17, 2767–2776. [Google Scholar] [CrossRef] [PubMed]
- Aref, S.; Castleton, A.Z.; Bailey, K.; Burt, R.; Dey, A.; Leongamornlert, D.; Mitchell, R.J.; Okasha, D.; Fielding, A.K. Type 1 Interferon Responses Underlie Tumor-Selective Replication of Oncolytic Measles Virus. Mol. Ther. 2020, 28, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Allagui, F.; Achard, C.; Panterne, C.; Combredet, C.; Labarriere, N.; Dreno, B.; Elgaaied, A.B.; Pouliquen, D.; Tangy, F.; Fonteneau, J.F.; et al. Modulation of the Type I Interferon Response Defines the Sensitivity of Human Melanoma Cells to Oncolytic Measles Virus. Curr. Gene Ther. 2017, 16, 419–428. [Google Scholar] [CrossRef]
- Tanabe, L.M.; List, K. The role of type II transmembrane serine protease-mediated signaling in cancer. FEBS J. 2017, 284, 1421–1436. [Google Scholar] [CrossRef]
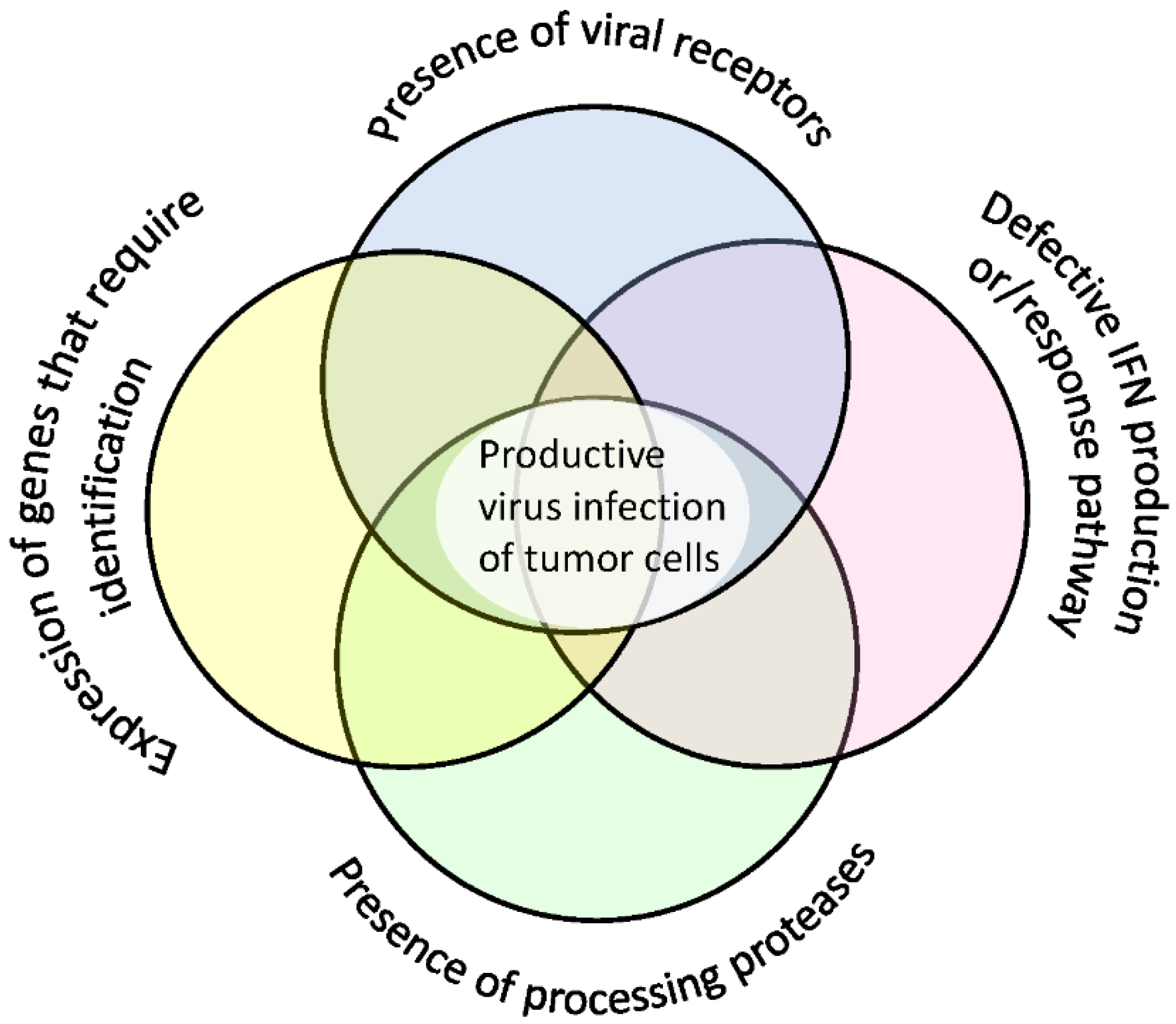

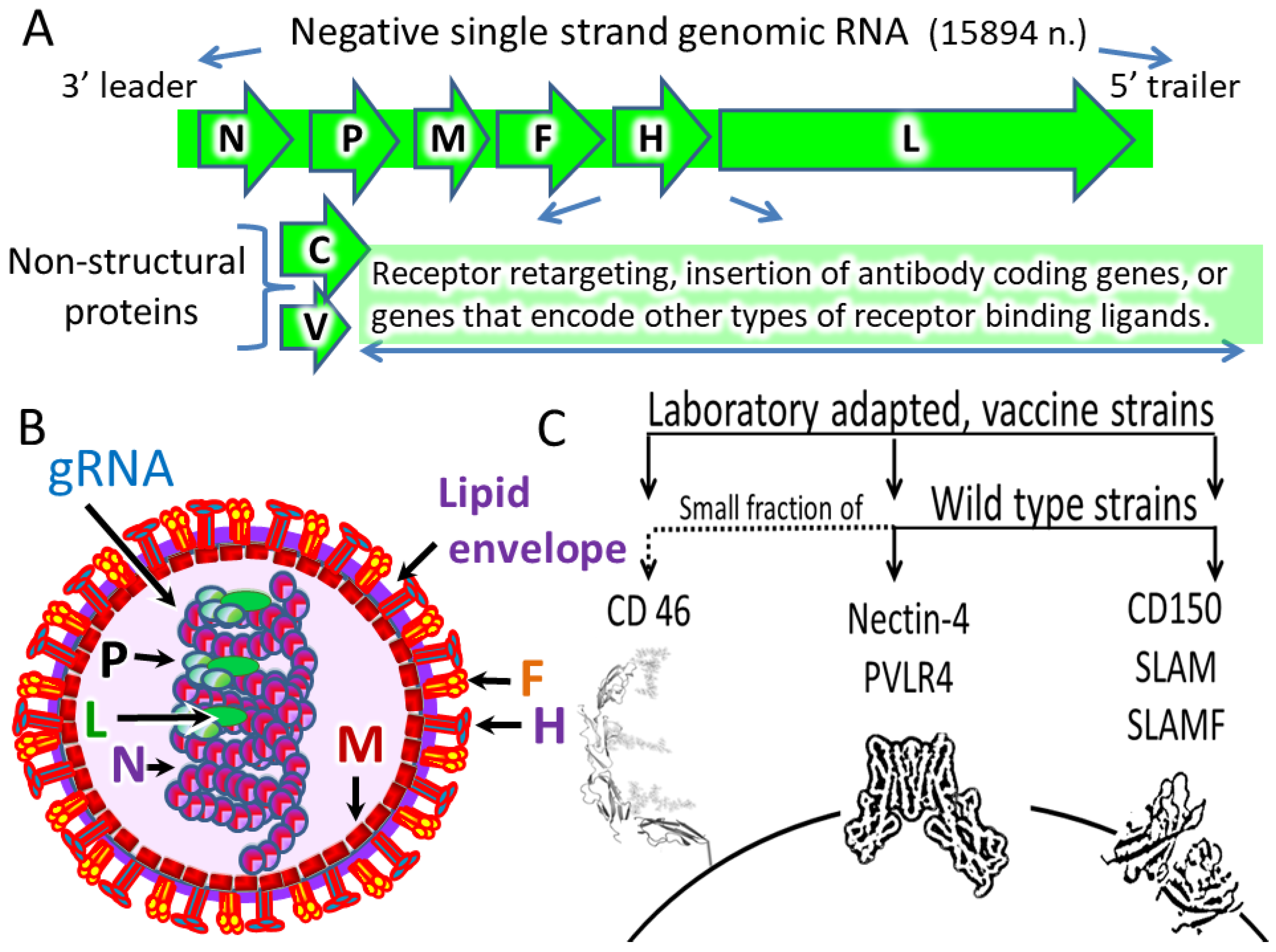
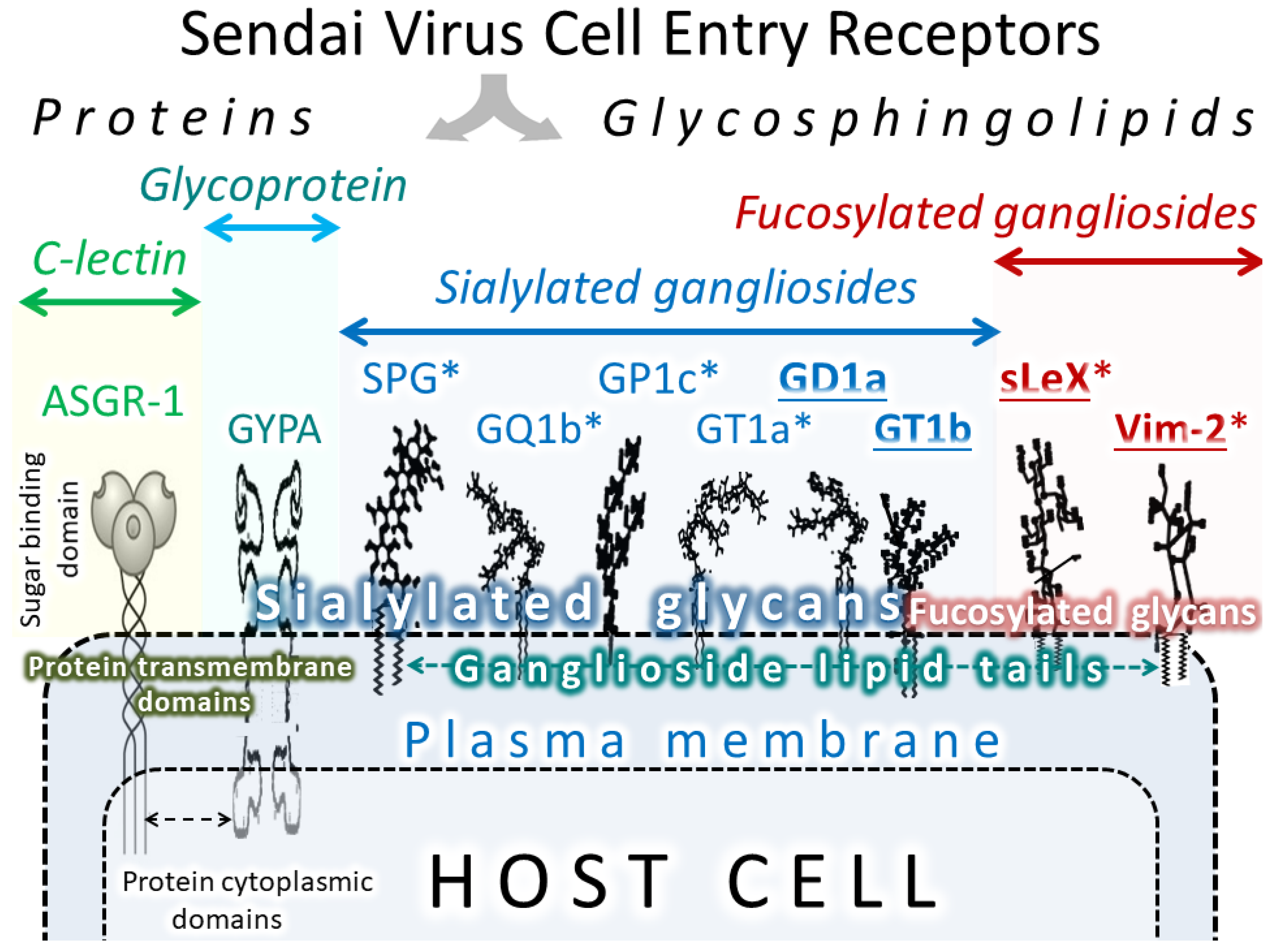
| Receptor | High Expression in Normal Cells | Expression Evaluation |
|---|---|---|
| CD150/SLAM | Hematopoietic stem and progenitor cells including T, B, natural killer, and dendritic cells [36,37,38] | Multicolor flow-cytometry |
| Spleen red pulp cells and thymus cortical and medullary cells [14,15,16] | Immunohistochemical tissue staining AB: HPA069319,CAB002438 | |
| Nectin-4/PVLR 4 | Glandular cells of breast, stomach colon, gall bladder and others [14,15,16] | Immunohistochemical tissue staining AB: HPA016903,CAB010401 |
| CD46/membrane cofactor protein | Glandular cells of breast, stomach, colon and others [14,15,16] | Immunohistochemical tissue staining AB: HPA010775 |
| Malignancy | CD150/SLAM (Ref/Evidence) | CD46/Membrane Cofactor Protein (Ref/Evidence) | Nectin-4 (Ref/Evidence) |
|---|---|---|---|
| Breast cancer | [39]/IS, [14,15,16]/IS, TCGA dataset | [33,40,41,42,43,44,45]/IS, FC, PCR | |
| Cervical cancer | [14,15,16]/IS, TCGA dataset | ||
| Colorectal cancer | [14,15,16]/IS, TCGA dataset, [46]/oligo-array | [14,15,16,33,45,46]/IS, TCGA dataset | |
| Endometrial cancer | [14,15,16]/IS, TCGA dataset | ||
| Glioma | [47] IS, FC | ||
| Liver cancer | [14,15,16]/IS, TCGA dataset | [48]/PCR, IS | |
| Lung cancer | [33,49]/IS, ELISA | ||
| Non-small cell lung cancer | [50] IS, FC | [46]/Oligo-array | |
| Lymphoma | [51]/PCR, IS, WB, FC | [52] IS, FC | |
| Melanoma | [14,15,16]/IS, TCGA | ||
| Multiple myeloma | [53]/IS, [46]/Oligo-array | ||
| Ovarian cancer | [54]/IS, WB | [55,56]/PCR, IS, WB, FC | |
| Pancreatic cancer | [14,15,16]/IS, TCGA dataset | [57]/IS | |
| Prostate cancer | |||
| Stomach cancer | |||
| Thyroid cancer | [14,15,16]/IS, TCGA dataset | ||
| Urothelial cancer | [14,15,16]/IS, TCGA dataset |
| Blinding to | Effect | Cell Type or Malignancy | Model and Type of Virus Delivery to Animals | Reference |
|---|---|---|---|---|
| CD150/SLAM, no natural CD46 | Viability of CD150-positive lymphoid cells unaffected; reduced infection of CD46-positive primary normal human cells; tumor stabilized or regressed | Breast carcinoma | Xenografts; IT virus delivery | [63] |
| Tumor stabilized; animal survival prolonged | Pancreatic carcinomas | Xenografts; IT virus delivery | [64] | |
| Lung carcinoma | [65] |
| Blinding to | Introducing Property to Bind | Effect | Cell Type or Malignancy | Model and Type of Virus Delivery to Animals | Reference |
|---|---|---|---|---|---|
| Via fusion of viral H protein with epidermal growth factor (EGF) or insulin-like growth factor 1 (IGF1) receptor binding domains | |||||
| None | EGF or IGF1 receptors | Infection of EGF or IGF1 receptor positive and CD46 negative cells | EGF or IGF1 receptor positive cells | Cell culture | [79] |
| Via fusion of viral H protein with a single-chain variable fragment (scFv) | |||||
| Carcino-embryonic antigen (CEA) | Infection of CEA positive cells | CEA-positive cells | Cell culture | [80] | |
| CD20 | Delayed growth | Fibrosarcoma | Xenografts; IP virus delivery | [81] | |
| CD38 | Malignant cells less tumorigenic, animal survival prolonged | Multiple myeloma | Xenografts; construct premixed with tumor cells before implantation | [82] | |
| Via fusion of viral H protein with echistatin, which is a 49-residue peptide from family of disintegrins | |||||
| Integrin alpha(v)beta3 | Tumor regressed or stabilized | Multiple myeloma | Xenografts; IT virus delivery | [83] | |
| Receptor Name | Alternative Name | Expression | |
|---|---|---|---|
| In Malignancies | In Normal Cells and Tissues | ||
| CEA glyco-proteins | Carcinoembryonic antigen-related cell adhesion molecules | High or moderate in gastric, colorectal, lung ovarian, breast, and cervical cancers [84] | Different subfamily members expressed to different degrees in hematopoietic cells, glandular cells of colon, etc. [14,15,16] |
| CD133 | Prominin-1 (PROM1) | High in leukemias [85], gliomas [86,87], colorectal, prostate, endometrial, pancreatic and thyroid cancers [14,15,16], and non-small cell lung cancers [88]. Levels in gliomas [87] and non-small cell lung cancers [88] are negatively correlated with patient survival [87] | High in glandular cells of gall bladder, endometrium, cervix and uterus [14,15,16]. Also expressed on the surfaces of hematopoietic stem cells [89], epithelial progenitor cells [90], and neural and glial stem cells [86] |
| CD20 | MS4A1, B1, Bp35, CVID5, LEU-16, MS4A2, S7, membrane spanning 4-domains A1 | Frequently high in B-cell lymphomas [91], B-cell leukemias [92], and melanoma stem cells [93]. Less frequent in Hodgkin’s lymphoma [94], myeloma [95], and thymoma [96] | Low and moderate expression in white and red pulp in spleen, hematopoietic cells of bone marrow, lymphoid tissues of appendix. and other tissues [14,15,16] |
| CD38 | Cyclic ADP ribose hydrolase | High in chronic lymphocytic leukemia [97,98] in NK/T-cell lymphomas [99], and in multiple myeloma [100] | High in large spectrum of immune cells as well as glandular cells of prostate and seminal vesicles [14,15,16] |
| Epidermal growth factor receptor 1 (EGFR1) | ErbB 1, HER1 | Particular high in gliomas, high in renal, urothelial, lung, liver, and many other cancers [14,15,16] | Low levels in a number of normal tissues but high levels in trophoblastic cells of placenta [14,15,16] |
| Epidermal growth factor receptor 2 (EGFR2) | Receptor tyrosine-protein kinase, ErbB-2, HER2/neu, ERBB2, CD340 | Frequently highly overexpressed in malignancies including breast [101], stomach [102], endometrial [103,104], ovarian, uterine [105] colorectal [106], thyroid [107], urothelial [108] | Medium levels in glandular cells of appendix, breast, and cervix, myocytes, respiratory epithelium, and urothelial cells [14,15,16] |
| Insulin-like growth factor receptor (IGF1R) | IGF-1 receptor | High in lymphomas, thyroid, liver, pancreatic, and many other cancers [14,15,16] | Low level of expression in bone marrow hematopoietic cells, respiratory cells, and glandular cells of gallbladder [14,15,16] |
| Folate receptor 1 (FOLR1) | Folate receptor alpha, Glutamate carboxypeptidase II (GCPII), and folate hydrolase 1 | High in ovarian cancers [14,15,16]; particularly strong and frequent expression of mRNA observed in non-mucinous ovarian cancers [109] | Medium levels in brain, lung, and salivary gland tissues [14,15,16] |
| Prostate specific membrane antigen, (PSMA) | High expression in malignant prostate cells [110] | High expression in prostate tissues [110] | |
| Urokinase receptor | UPA, UPAR, CD87, PLAUR | Infrequently expressed in malignant cells [14,15,16] | High in bone marrow, lymphoid tissues, neutrophils, and respiratory epithelial cells of the nasopharynx and bronchus |
| Blinding to | Introducing Property to Bind | Effect | Cell Type or Malignancy | Model; Route of Virus Delivery to Animals | Reference |
|---|---|---|---|---|---|
| CD150/SLAM and CD46 | Via fusion of viral H protein with scFv | ||||
| CD38 or EGFR | Tumor stabilized; animal survival prolonged | CD38 or EGFR positive cancers | Xenografts; IT or IV | [113,114] | |
| Folate receptor 1 (FOLR1) | Biodistribution more specific towards malignant tissues; tumor stabilized; animal survival prolonged | Ovarian cancer | Xenografts; IV | [109] | |
| Prostate specific membrane antigen, (PSMA) | Tumor stabilized; animal survival prolonged | Prostate cancer | Xenografts; IT | [122] | |
| HER2 protein | Malignant cells infected in vitro, tumor regressed, animal survival prolonged | Ovarian cancer | Xenografts; IP | [77,78] | |
| CD133, Prominin1 (PROM1) | Tumor formation inhibited; animal survival prolonged | Glioblastoma, lung metastases of colon cancer and hepatocellular carcinoma | Xenografts; IT or IV | [115,116] | |
| Urokinase receptor | Delayed development of lung metastases, animal survival prolonged | Breast cancer | Syngeneic and xenografts; IV | [117,118] | |
| Via fusion of viral H protein with cystine knot proteins | |||||
| Integrins | Malignant cells killed in vitro; cytopathic effects produced in vivo | Glioblastoma, medullo-blastoma, melanoma | Glioblastoma xenografts; IV | [111] | |
| Via fusion of viral H protein with designed ankyrin repeat proteins (DARPin) | |||||
| Bispecific binding to HER2/neu, and/or EpCAM | Animal survival significantly prolonged, tumor burden reduced | Ovarian cancer | Xenografts; IT | [119,120] | |
| EGFR | Malignant cells killed in vitro | Glioblastoma multiforme | Cell lines | [121] | |
| Cell Line | Type of Malignancy | Reference |
|---|---|---|
| Human origin | ||
| MCF7 | Breast carcinoma | [141] |
| HeLa | Cervical carcinoma | [142] |
| CaCo2 | Colon carcinoma | [13] |
| U118 | Glioblastoma | [143] |
| U87MG | Most likely, human glioma | [144] |
| Hep G2 | Hepatic carcinoma | [142,145,146] |
| Huh7 | [146,147] | |
| A549 | Lung carcinoma | [142,148,149,150] |
| Calu-3 | [13] | |
| U937 | Histiocytic lymphoma | [149] |
| Namalwa | Burkitt’s lymphoma | [149,151] |
| PC-3 | Prostate carcinoma derived from metastatic site in bone | [152] |
| DU145 | Prostate carcinoma derived from metastatic site in brain | |
| Murine origin | ||
| 4T1 | Mammary gland metastatic adenocarcinoma | [142] |
| Sub-Type of Molecule | Receptor | Affinity to SeV | Ref. | Function in Normal Human Cells | Expression in Normal Human Cells |
|---|---|---|---|---|---|
| Glycoproteins | |||||
| Human asialoglyco-protein receptor 1 | ASGR1 | High | [145,156] | Removes the target glycoproteins from circulation in the liver | Hepatocytes [14,15,16] |
| Bovine glycoprotein 2 | Glycoprotein2/GP2 | High | [155] | - | |
| Human sialo-glycoprotein | Glycophorin A/GYPA/CD235a | High | [157] | Defines the antigenic determinants for some blood groups | Bone marrow, immune cells, [14,15,16] |
| Fucosylated glycans | |||||
| Tetra-saccharide | Sialyl Lewis-x antigen (sLeX/CD15s) | High | [158] | Serves as a blood group antigen and participates in cell-cell recognition process. | Bone marrow, erythrocytes [14,15,16] |
| Ceramide-dodeca-saccharide | VIM-2 antigen (CD65s) | Unknown | Granulocytes and monocytes [158] | ||
| Sialylated gangliosides | |||||
| Ganglio-series | GD1a, GT1b, and GQ1b, | Not reported | [159] | Cell-cell recognition, adhesion, and signal transduction | Granulocytes, normal myeloid cells [160] |
| GT1a, GP1c | High | [161,162,163] | - | Many cell types, but mainly the cells of the nervous system [164] | |
| GD1a, GT1b | Moderate | [161,162,163] | - | ||
| GQ1b | Very high | ||||
| GM3 | Low | [165] | Cell–cell recognition | Blood cells, liver | |
| Neolacto-series | Sialosylparagloboside (SPG, NeuAcα2-3PG) | Very high | [163,166] | - | Common for non-neural cells |
| NeuAcα2-3I NeuAcα2-3i | [165] | - | |||
| NeuGca2-3I NeuAca2-6PG NeuAca2-6I | Moderate | - | |||
| Receptor | Malignancy/Effect of Receptor Expression | Ref. | Monoclonal AB Availability |
|---|---|---|---|
| Human asialoglyco-protein receptor 1 | High expression in liver cancer and occasionally moderate expression in gliomas, renal, pancreatic, colorectal, and ovarian cancers | [14,15,16] | Two variants [14,15,16] |
| Sialyl-Lewisx Antigen (sLeX/CD15) | Non-small cell lung cancer/enhances post-operative recurrence | [168,169] | Many variants [170] |
| Lung cancer, distant metastases | [171] | ||
| Colorectal cancer/promotes liver metastases, decreases time of disease-free survival | [172,173,174] | ||
| Gastric cancers/decreases patient survival time | [175,176] | ||
| Breast cancer/decreases patient survival time | [177,178,179] | ||
| Prostate tumor/promotes bone metastases | [180,181,182] | ||
| Cell lines of variable origin/high expression enhances adhesion of malignant cells to vascular endothelium | [183] | ||
| Variable cancers/high expression related to lymphatic invasion, venous invasion, T stage, N stage, M stage, tumor stage, recurrence, and overall patient survival | Review [167] | ||
| VIM-2 antigen (CD65s) | Acute myeloblastic leukemias | [160,184,185] | One variant [170] |
| GD1a | Breast cancer stem cells | [186] | Many variants [170] |
| Castration-resistant prostate cancer cells | [187] | ||
| GT1b | Brain metastases from colon, renal, lung, esophagus, pancreas, and mammary carcinomas | [188] | Three variants [170] |
| SPG | Castration-resistant prostate cancer cells | [187] | One variant [189] |
| Lymphoid leukemia cells | [189,190] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matveeva, O.V.; Shabalina, S.A. Prospects for Using Expression Patterns of Paramyxovirus Receptors as Biomarkers for Oncolytic Virotherapy. Cancers 2020, 12, 3659. https://doi.org/10.3390/cancers12123659
Matveeva OV, Shabalina SA. Prospects for Using Expression Patterns of Paramyxovirus Receptors as Biomarkers for Oncolytic Virotherapy. Cancers. 2020; 12(12):3659. https://doi.org/10.3390/cancers12123659
Chicago/Turabian StyleMatveeva, Olga V., and Svetlana A. Shabalina. 2020. "Prospects for Using Expression Patterns of Paramyxovirus Receptors as Biomarkers for Oncolytic Virotherapy" Cancers 12, no. 12: 3659. https://doi.org/10.3390/cancers12123659
APA StyleMatveeva, O. V., & Shabalina, S. A. (2020). Prospects for Using Expression Patterns of Paramyxovirus Receptors as Biomarkers for Oncolytic Virotherapy. Cancers, 12(12), 3659. https://doi.org/10.3390/cancers12123659






