DNA Methylation of FKBP5 as Predictor of Overall Survival in Malignant Pleural Mesothelioma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Epigenome-Wide Association Study (EWAS)
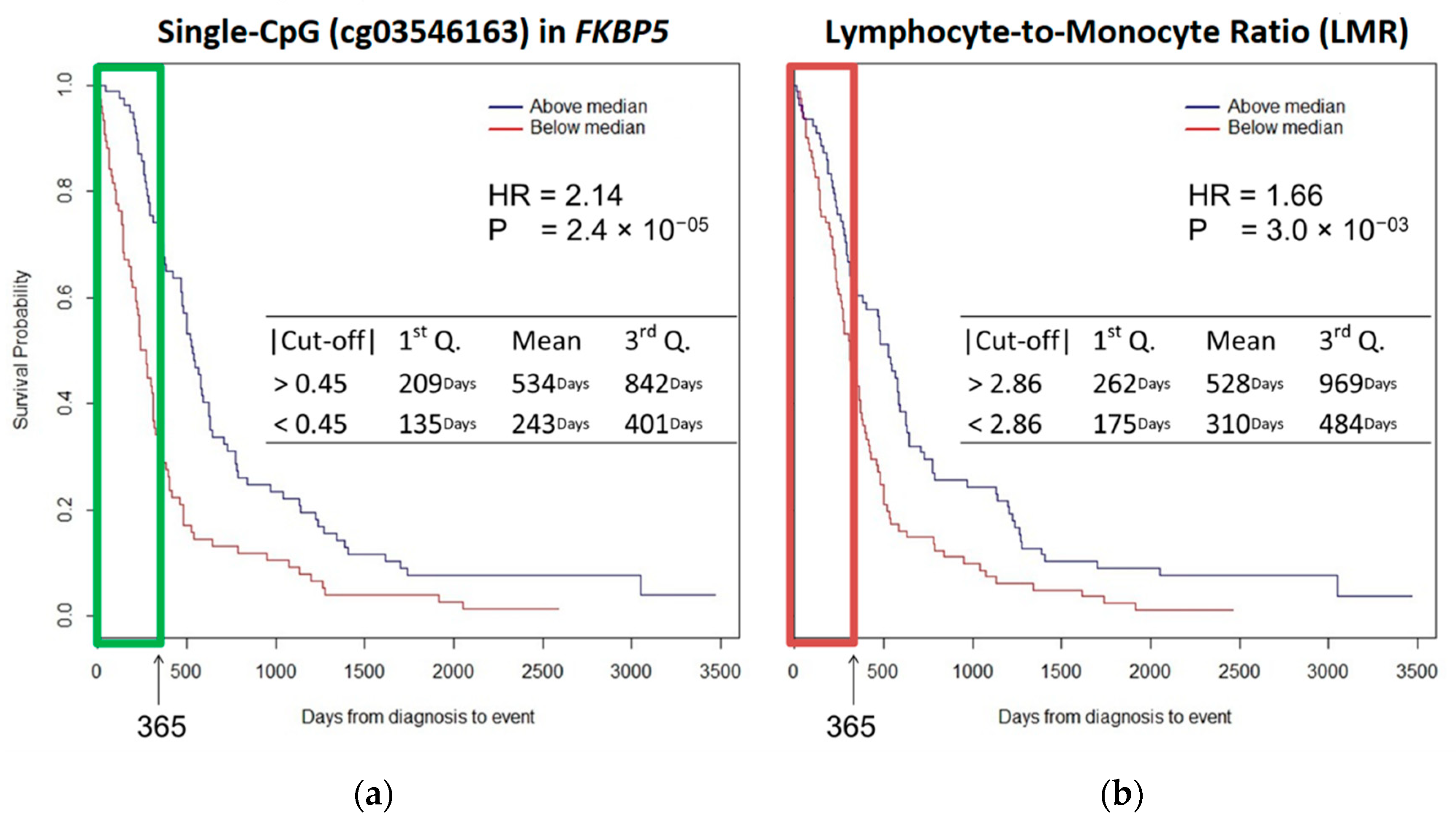
2.2. Survival Analysis
2.3. Validation and Replication
3. Discussion
Limitation of the Study
4. Material and Methods
4.1. Study Population
4.2. Exposure Assessment
4.3. Blood DNAm Analysis
4.4. Beta-Value Extraction
4.5. Batch Effect, Population Stratification, and White Blood Cell Estimations
4.6. Statistical Analyses
Epigenome-Wide Association Study
4.7. Survival Analysis
4.8. Statistical Power
4.9. Validation and Replication
- -
- cg03546163_10FW: aggaagagagTTTTTGTTTAGGATGAATTAGTTTGG;
- -
- cg03546163_T7RV: cagtaatacgactcactatagggagaaggctAAAAACTACAATCTTATCCAATTCCTTT.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sekido, Y. Molecular pathogenesis of malignant mesothelioma. Carcinogenesis 2013, 34, 1413–1419. [Google Scholar] [CrossRef]
- Rossini, M.; Rizzo, P.; Bononi, I.; Clementz, A.; Ferrari, R.; Martini, F.; Tognon, M.G. New Perspectives on Diagnosis and Therapy of Malignant Pleural Mesothelioma. Front. Oncol. 2018, 8, 91. [Google Scholar] [CrossRef] [Green Version]
- Furuya, S.; Chimed-Ochir, O.; Takahashi, K.; David, A.; Takala, J. Global Asbestos Disaster. Int. J. Environ. Res. Public Health 2018, 15, 1000. [Google Scholar] [CrossRef] [Green Version]
- Straif, K.; Benbrahim-Tallaa, L.; Baan, R.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Bouvard, V.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens—Part C: Metals, arsenic, dusts, and fibres. Lancet Oncol. 2009, 10, 453–454. [Google Scholar] [CrossRef]
- Curran, D.; Sahmoud, T.; Therasse, P.; Van Meerbeeck, J.; Postmus, P.E.; Giaccone, G. Prognostic factors in patients with pleural mesothelioma: The European Organization for Research and Treatment of Cancer experience. J. Clin. Oncol. 1998, 16, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Jassem, J.; Ramlau, R.; Santoro, A.; Schuette, W.; Chemaissani, A.; Hong, S.; Blatter, J.; Adachi, S.; Hanauske, A.; Manegold, C. Phase III Trial of Pemetrexed Plus Best Supportive Care Compared with Best Supportive Care in Previously Treated Patients with Advanced Malignant Pleural Mesothelioma. J. Clin. Oncol. 2008, 26, 1698–1704. [Google Scholar] [CrossRef] [PubMed]
- Vogelzang, N.J.; Rusthoven, J.J.; Symanowski, J.; Denham, C.; Kaukel, E.; Ruffie, P.; Gatzemeier, U.; Boyer, M.; Emri, S.; Manegold, C.; et al. Phase III Study of Pemetrexed in Combination with Cisplatin Versus Cisplatin Alone in Patients with Malignant Pleural Mesothelioma. J. Clin. Oncol. 2003, 21, 2636–2644. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Aerts, J.G.; Popat, S.; Fennell, D.A. Novel insights into mesothelioma biology and implications for therapy. Nat. Rev. Cancer 2017, 17, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Guarrera, S.; Viberti, C.; Cugliari, G.; Allione, A.; Casalone, E.; Betti, M.; Ferrante, D.; Aspesi, A.; Casadio, C.; Grosso, F.; et al. Peripheral Blood DNA Methylation as Potential Biomarker of Malignant Pleural Mesothelioma in Asbestos-Exposed Subjects. J. Thorac. Oncol. 2019, 14, 527–539. [Google Scholar] [CrossRef] [Green Version]
- Matullo, G.; Guarrera, S.; Betti, M.; Fiorito, G.; Ferrante, D.; Voglino, F.; Cadby, G.; Di Gaetano, C.; Rosa, F.; Russo, A.; et al. Genetic Variants Associated with Increased Risk of Malignant Pleural Mesothelioma: A Genome-Wide Association Study. PLoS ONE 2013, 8, e61253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanrikulu, A.C.; Abakay, A.; Komek, H.; Abakay, O. Prognostic value of the lymphocyte-to-monocyte ratio and other inflammatory markers in malignant pleural mesothelioma. Environ. Health Prev. Med. 2016, 21, 304–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, G.; Chmielecki, J.; Goparaju, C.; Heguy, A.; Dolgalev, I.; Carbone, M.; Seepo, S.; Meyerson, M.; Pass, H.I. Whole-Exome Sequencing Reveals Frequent Genetic Alterations in BAP1, NF2, CDKN2A, and CUL1 in Malignant Pleural Mesothelioma. Cancer Res. 2014, 75, 264–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betti, M.; Aspesi, A.; Sculco, M.; Matullo, G.; Magnani, C.; Dianzani, I. Genetic predisposition for malignant mesothelioma: A concise review. Mutat. Res. 2019, 781, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gai, W.; Sun, K. Epigenetic Biomarkers in Cell-Free DNA and Applications in Liquid Biopsy. Genes 2019, 10, 32. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Bononi, A.; Napolitano, A.; Pass, H.I.; Yang, H.; Carbone, M. Latest developments in our understanding of the pathogenesis of mesothelioma and the design of targeted therapies. Expert Rev. Respir. Med. 2015, 9, 633–654. [Google Scholar] [CrossRef] [Green Version]
- Vandermeers, F.; Sriramareddy, S.N.; Costa, C.; Hubaux, R.; Cosse, J.-P.; Willems, L. The role of epigenetics in malignant pleural mesothelioma. Lung Cancer 2013, 81, 311–318. [Google Scholar] [CrossRef]
- Ferrari, L.; Carugno, M.; Mensi, C.; Pesatori, A.C. Circulating Epigenetic Biomarkers in Malignant Pleural Mesothelioma: State of the Art and critical Evaluation. Front. Oncol. 2020, 10, 445. [Google Scholar] [CrossRef] [Green Version]
- Creaney, J.; Yeoman, D.; Demelker, Y.; Segal, A.; Musk, A.; Skates, S.J.; Robinson, B.W. Comparison of Osteopontin, Megakaryocyte Potentiating Factor, and Mesothelin Proteins as Markers in the Serum of Patients with Malignant Mesothelioma. J. Thorac. Oncol. 2008, 3, 851–857. [Google Scholar] [CrossRef] [Green Version]
- Creaney, J.; Dick, I.M.; Meniawy, T.M.; Leong, S.L.; Leon, J.S.; Demelker, Y.; Segal, A.; Musk, A.W. (Bill); Lee, Y.C.G.; Skates, S.J.; et al. Comparison of fibulin-3 and mesothelin as markers in malignant mesothelioma. Thorax 2014, 69, 895–902. [Google Scholar] [CrossRef] [Green Version]
- Tanrikulu, A.C.; Abakay, A.; Kaplan, M.A.; Küçüköner, M.; Palanci, Y.; Evliyaoglu, O.; Sezgi, C.; Sen, H.; Carkanat, A.I.; Kirbas, G. A Clinical, Radiographic and Laboratory Evaluation of Prognostic Factors in 363 Patients with Malignant Pleural Mesothelioma. Respiration 2010, 80, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Costa-Pinheiro, P.; Montezuma, D.; Henrique, R.; Jerónimo, C. Diagnostic and prognostic epigenetic biomarkers in cancer. Epigenomics 2015, 7, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Comba, P.; D’Angelo, M.; Fazzo, L.; Magnani, C.; Marinaccio, A.; Mirabelli, D.; Terracini, B. Mesothelioma in Italy: The Casale Monferrato model to a national epidemiological surveillance system. Annali Istituto Superiore Sanità 2018, 54, 139–148. [Google Scholar]
- Pass, H.I.; Giroux, D.; Kennedy, C.; Ruffini, E.; Cangir, A.K.; Rice, D.; Asamura, H.; Waller, D.; Edwards, J.; Weder, W.; et al. The IASLC Mesothelioma Staging Project: Improving Staging of a Rare Disease Through International Participation. J. Thorac. Oncol. 2016, 11, 2082–2088. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, T.; Fujimoto, N.; Nishi, H.; Miyamoto, Y.; Hara, N.; Asano, M.; Fuchimoto, Y.; Wada, S.; Kitamura, K.; Ozaki, S.; et al. Prognostic significance of the lymphocyte-to-monocyte ratio in patients with malignant pleural mesothelioma. Lung Cancer 2015, 90, 111–117. [Google Scholar] [CrossRef]
- Argentieri, M.A.; Nagarajan, S.; Seddighzadeh, B.; Baccarelli, A.A.; Shields, A.E. Epigenetic Pathways in Human Disease: The Impact of DNA Methylation on Stress-Related Pathogenesis and Current Challenges in Biomarker Development. EBioMedicine 2017, 18, 327–350. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Du, L.; Yi, S.; Wang, Q.; Zhu, Y.; Qiu, Y.; Jiang, Y.; Li, M.; Wang, D.; Wang, Q.; et al. Epigenome-wide association study identifies Behçet’s disease-associated methylation loci in Han Chinese. Rheumatology 2019, 58, 1574–1584. [Google Scholar] [CrossRef]
- Yamaguchi, I.; Nakajima, K.; Shono, K.; Mizobuchi, Y.; Fujihara, T.; Shikata, E.; Yamaguchi, T.; Kitazato, K.; Sampetrean, O.; Saya, H.; et al. Downregulation of PD-L1 via FKBP5 by celecoxib augments antitumor effects of PD-1 blockade in a malignant glioma model. Neuro-Oncology Adv. 2020, 2, vdz058. [Google Scholar] [CrossRef] [Green Version]
- Romano, S.; Simeone, E.; D’Angelillo, A.; D’Arrigo, P.; Russo, M.; Capasso, M.; Lasorsa, V.A.; Zambrano, N.; Ascierto, P.A.; Romano, M.F. FKBP51s signature in peripheral blood mononuclear cells of melanoma patients as a possible predictive factor for immunotherapy. Cancer Immunol. Immunother. 2017, 66, 1143–1151. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Lou, Z.; Wang, L. The role of FKBP5 in cancer aetiology and chemoresistance. Br. J. Cancer 2010, 104, 19–23. [Google Scholar] [CrossRef] [Green Version]
- Staibano, S.; Mascolo, M.; Ilardi, G.; Siano, M.; De Rosa, G. Immunohistochemical analysis of FKBP51 in human cancers. Curr. Opin. Pharmacol. 2011, 11, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Zannas, A.S.; Jia, M.; Hafner, K.; Baumert, J.; Wiechmann, T.; Pape, J.C.; Arloth, J.; Ködel, M.; Martinelli, S.; Roitman, M.; et al. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-kappaB-driven inflammation and cardiovascular risk. Proc. Natl. Acad. Sci. USA 2019, 116, 11370–11379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, M.F.; Avellino, R.; Petrella, A.; Bisogni, R.; Romano, S.; Venuta, S. Rapamycin inhibits doxorubicin-induced NF-kappaB/Rel nuclear activity and enhances the apoptosis of melanoma cells. Eur. J. Cancer 2004, 40, 2829–2836. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.-K.; Huang, S.-L.; Chang, P.-Y.; Lu, H.-P.; Chao, C.C.-K. Transcriptomic profiling of taxol-resistant ovarian cancer cells identifies FKBP5 and the androgen receptor as critical markers of chemotherapeutic response. Oncotarget 2014, 5, 11939–11956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, S.; D’Angelillo, A.; Romano, M.F. Pleiotropic roles in cancer biology for multifaceted proteins FKBPs. Biochim. Biophys. Acta 2015, 1850, 2061–2068. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, D.; Bertolotti, M.; Todesco, A.; Mirabelli, D.; Terracini, B.; Magnani, C. Cancer Mortality and Incidence of Mesothelioma in a Cohort of Wives of Asbestos Workers in Casale Monferrato, Italy. Environ. Health Perspect. 2007, 115, 1401–1405. [Google Scholar] [CrossRef]
- Dianzani, I.; Gibello, L.; Biava, A.; Mirabelli, D.; Terracini, B.; Magnani, C. Polymorphisms in DNA repair genes as risk factors for asbestos-related malignant mesothelioma in a general population study. Mutat. Res. 2006, 599, 124–134. [Google Scholar] [CrossRef]
- Betti, M.; Ferrante, D.; Padoan, M.; Guarrera, S.; Giordano, M.; Aspesi, A.; Mirabelli, D.; Casadio, C.; Ardissone, F.; Ruffini, E.; et al. XRCC1 and ERCC1 variants modify malignant mesothelioma risk: A case—Control study. Mutat. Res. Mol. Mech. Mutagen. 2011, 708, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Betti, M.; Casalone, E.; Ferrante, D.; Aspesi, A.; Morleo, G.; Biasi, A.; Sculco, M.; Mancuso, G.; Guarrera, S.; Righi, L.; et al. Germline mutations in DNA repair genes predispose asbestos-exposed patients to malignant pleural mesothelioma. Cancer Lett. 2017, 405, 38–45. [Google Scholar] [CrossRef]
- Ferrante, D.; Mirabelli, D.; Tunesi, S.; Terracini, B.; Magnani, C. Pleural mesothelioma and occupational and non-occupational asbestos exposure: A case—Control study with quantitative risk assessment. Occup. Environ. Med. 2016, 73, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Bibikova, M.; Barnes, B.; Tsan, C.; Ho, V.; Klotzle, B.; Le, J.M.; Delano, D.; Zhang, L.; Schroth, G.P.; Gunderson, K.L.; et al. High density DNA methylation array with single CpG site resolution. Genomics 2011, 98, 288–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibikova, M.; Lin, Z.; Zhou, L.; Chudin, E.; Garcia, E.W.; Wu, B.; Doucet, D.; Thomas, N.J.; Wang, Y.; Vollmer, E.; et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006, 16, 383–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, P.; Zhang, X.; Huang, C.-C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar]
- Lehne, B.; Drong, A.W.; Loh, M.; Zhang, W.; Scott, W.R.; Tan, S.-T.; Afzal, U.; Scott, J.; Jarvelin, M.-R.; Elliott, P.; et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015, 16, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campanella, G.; Polidoro, S.; Di Gaetano, C.; Fiorito, G.; Guarrera, S.; Krogh, V.; Palli, D.; Panico, S.; Sacerdote, C.; Tumino, R.; et al. Epigenetic signatures of internal migration in Italy. Int. J. Epidemiol. 2014, 44, 1442–1449. [Google Scholar] [CrossRef] [Green Version]
- Di Gaetano, C.; Voglino, F.; Guarrera, S.; Fiorito, G.; Rosa, F.; Di Blasio, A.M.; Manzini, P.; Dianzani, I.; Betti, M.; Cusi, D.; et al. An Overview of the Genetic Structure within the Italian Population from Genome-Wide Data. PLoS ONE 2012, 7, e43759. [Google Scholar] [CrossRef]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef] [Green Version]
- Ehrich, M.; Nelson, M.R.; Stanssens, P.; Zabeau, M.; Liloglou, T.; Xinarianos, G.; Cantor, C.R.; Field, J.K.; Boom, D.V.D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. USA 2005, 102, 15785–15790. [Google Scholar] [CrossRef] [Green Version]
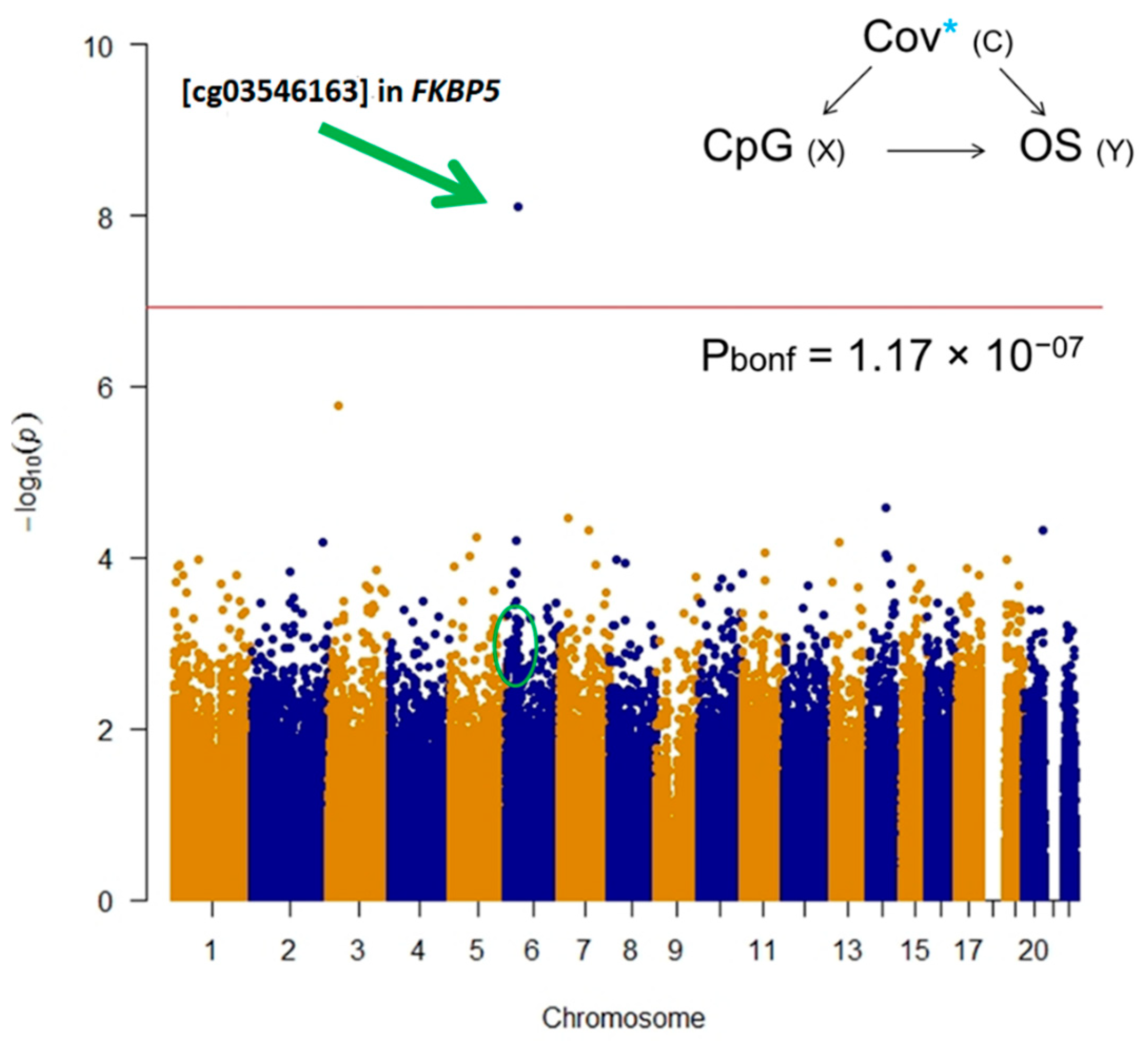
| TargetID | CHR | UCSC RefGene Group | Enhancer | Probe Start | Probe End | Closest TSS | Distance Closest TSS | Closest TSS Gene Name | Effect Size | SE | p value | Bonferroni | Significance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cg03546163 | 6 | 5′UTR;5′UTR;5’UTR;5′UTR | NA | 35654313 | 35654363 | 35656691 | 2329 | FKBP5 | 0.12 | 0.02 | 7.71E-09 | 0.003280418 | *§ |
| cg00052684 | 6 | 5′UTR | TRUE | 35694195 | 35694245 | 35696396 | 2152 | FKBP5 | 0.04 | 0.02 | 0.014589031 | 1 | * |
| cg00130530 | 6 | 5′UTR;TSS1500;TSS1500;TSS1500 | NA | 35657152 | 35657202 | 35656718 | −483 | FKBP5 | 0.03 | 0.01 | 0.001490825 | 1 | * |
| cg19226017 | 6 | TSS1500;Body | NA | 35697185 | 35697235 | 35696396 | −788 | FKBP5 | 0.03 | 0.01 | 0.021639194 | 1 | * |
| cg08915438 | 6 | TSS1500;Body | NA | 35697709 | 35697759 | 35696396 | −1362 | FKBP5 | 0.02 | 0.01 | 0.050779639 | 1 | * |
| cg14642437 | 6 | 5′UTR;5′UTR;5′UTR;5’UTR | NA | 35652471 | 35652521 | 35656691 | 4171 | FKBP5 | 0.02 | 0.01 | 0.030718193 | 1 | * |
| cg25114611 | 6 | TSS1500;Body | NA | 35696820 | 35696870 | 35696396 | −473 | FKBP5 | 0.02 | 0.01 | 0.080435168 | 1 | |
| cg16052510 | 6 | Body;Body;Body;Body | TRUE | 35603093 | 35603143 | 35656691 | 53549 | FKBP5 | 0.01 | 0.01 | 0.201783727 | 1 | |
| cg03591753 | 6 | 5′UTR | NA | 35659141 | 35659191 | 35656718 | −2422 | FKBP5 | 0.01 | 0.01 | 0.071287867 | 1 | |
| cg23416081 | 6 | 5′UTR | TRUE | 35693573 | 35693623 | 35696396 | 2824 | FKBP5 | 0.01 | 0.01 | 0.300181524 | 1 | |
| cg19014730 | 6 | 5′UTR;5′UTR;5′UTR;5′UTR | TRUE | 35635985 | 35636035 | 35656691 | 20707 | FKBP5 | 0.01 | 0.01 | 0.510924063 | 1 | |
| cg20813374 | 6 | 5′UTR;TSS1500;TSS1500;TSS1500 | NA | 35657130 | 35657180 | 35656718 | −461 | FKBP5 | 0.01 | 0.01 | 0.538622493 | 1 | |
| cg07061368 | 6 | 5′UTR;5′UTR;5′UTR;5′UTR | TRUE | 35631736 | 35631786 | 35656691 | 24956 | FKBP5 | 0.00 | 0.01 | 0.440719926 | 1 | |
| cg08636224 | 6 | 5′UTR;TSS1500;TSS1500;TSS1500 | NA | 35657871 | 35657921 | 35656718 | −1202 | FKBP5 | 0.00 | 0.00 | 0.18248273 | 1 | |
| cg01294490 | 6 | TSS200;TSS200;5′UTR;TSS1500 | NA | 35656906 | 35656956 | 35656718 | −187 | FKBP5 | 0.00 | 0.01 | 0.421300242 | 1 | |
| cg07485685 | 6 | 5′UTR;Body | NA | 35696060 | 35696110 | 35696396 | 336 | FKBP5 | 0.00 | 0.00 | 0.847941933 | 1 | |
| cg14284211 | 6 | Body;Body;Body;Body | TRUE | 35570224 | 35570274 | 35656691 | 86468 | FKBP5 | 0.00 | 0.01 | 0.974344781 | 1 | |
| cg17030679 | 6 | 5′UTR;Body;1stExon | NA | 35696300 | 35696350 | 35696396 | 97 | FKBP5 | 0.00 | 0.00 | 0.955719442 | 1 | |
| cg00862770 | 6 | 5′UTR;5′UTR;5′UTR;5′UTR | NA | 35655764 | 35655814 | 35656691 | 928 | FKBP5 | 0.00 | 0.00 | 0.939904147 | 1 | |
| cg00140191 | 6 | 5′UTR;5′UTR;5′UTR;5′UTR | NA | 35656193 | 35656243 | 35656691 | 450 | FKBP5 | 0.00 | 0.00 | 0.882388191 | 1 | |
| cg00610228 | 6 | 5′UTR;Body | NA | 35695934 | 35695984 | 35696396 | 463 | FKBP5 | 0.00 | 0.00 | 0.87376216 | 1 | |
| cg07633853 | 6 | Body;Body;Body;Body | TRUE | 35569421 | 35569471 | 35656691 | 87221 | FKBP5 | 0.00 | 0.01 | 0.965427693 | 1 | |
| cg10300814 | 6 | Body;Body;Body;Body | TRUE | 35565066 | 35565116 | 35480646 | −84469 | TULP1 | 0.00 | 0.00 | 0.620677997 | 1 | |
| cg16012111 | 6 | TSS200;TSS200;TSS200;5′UTR | NA | 35656758 | 35656808 | 35656718 | −39 | FKBP5 | 0.00 | 0.00 | 0.519047184 | 1 | |
| cg06937024 | 6 | 5′UTR;Body | NA | 35695440 | 35695490 | 35696396 | 908 | FKBP5 | 0.00 | 0.00 | 0.135004544 | 1 | |
| cg08586216 | 6 | 5′UTR;5′UTR;5′UTR;5′UTR | TRUE | 35612301 | 35612351 | 35656691 | 44341 | FKBP5 | 0.00 | 0.00 | 0.105631333 | 1 | |
| cg17085721 | 6 | 5′UTR;5′UTR;5′UTR;5′UTR | TRUE | 35645291 | 35645341 | 35656691 | 11351 | FKBP5 | 0.00 | 0.00 | 0.211582562 | 1 | |
| cg02665568 | 6 | Body;Body;Body | NA | 35544468 | 35544518 | 35480646 | −63821 | TULP1 | −0.01 | 0.01 | 0.294757699 | 1 | |
| cg15929276 | 6 | 5′UTR | TRUE | 35687456 | 35687506 | 35696396 | 8940 | FKBP5 | −0.01 | 0.01 | 0.455969031 | 1 | |
| cg06087101 | 6 | Body;3′UTR;Body;Body | NA | 35551882 | 35551932 | 35480646 | −71285 | TULP1 | −0.02 | 0.02 | 0.203783874 | 1 |
| Categorical Variable | Level | Low OS (n = 79) | High OS (n = 80) | ||
| N | % | N | % | ||
| Centre | Casale | 50 | 63.3 | 46 | 57.5 |
| Torino | 29 | 36.7 | 34 | 42.5 | |
| Gender | Males | 59 | 74.7 | 50 | 62.5 |
| Females | 20 | 25.3 | 30 | 37.5 | |
| Smoke | Current | 20 | 26.3 | 8 | 10.3 |
| Former | 24 | 31.6 | 29 | 37.2 | |
| Never | 32 | 42.1 | 41 | 52.6 | |
| Histotype | Epithelioid | 44 | 55.7 | 61 | 76.3 |
| Sarcomatoid | 14 | 17.7 | 2 | 2.5 | |
| Biphasic | 17 | 21.5 | 11 | 13.8 | |
| Undefined | 2 | 2.5 | 1 | 1.3 | |
| Not known | 2 | 2.5 | 5 | 6.3 | |
| Continuous Variable | Level | Low OS | High OS | ||
| Mean | SD | Mean | SD | ||
| Overall Survival (days) | 198.7 | 101.6 | 957.8 | 698.7 | |
| Age (years) | 67.7 | 12.4 | 67.5 | 9.6 | |
| Asbestos Exp. (norm) | 1.4 | 1.5 | 1.5 | 1.9 | |
| CD8T (%) | 2.9 | 4.5 | 3 | 3.4 | |
| CD4T (%) | 6.8 | 5.3 | 8.8 | 5.4 | |
| Natural Killer (%) | 4.9 | 4.9 | 6.3 | 4.1 | |
| B cell (%) | 6.1 | 2.8 | 6.4 | 2.7 | |
| Monocytes (%) | 8.1 | 4.1 | 7.6 | 4.4 | |
| Granulocytes (%) | 75 | 13 | 72 | 10 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cugliari, G.; Catalano, C.; Guarrera, S.; Allione, A.; Casalone, E.; Russo, A.; Grosso, F.; Ferrante, D.; Viberti, C.; Aspesi, A.; et al. DNA Methylation of FKBP5 as Predictor of Overall Survival in Malignant Pleural Mesothelioma. Cancers 2020, 12, 3470. https://doi.org/10.3390/cancers12113470
Cugliari G, Catalano C, Guarrera S, Allione A, Casalone E, Russo A, Grosso F, Ferrante D, Viberti C, Aspesi A, et al. DNA Methylation of FKBP5 as Predictor of Overall Survival in Malignant Pleural Mesothelioma. Cancers. 2020; 12(11):3470. https://doi.org/10.3390/cancers12113470
Chicago/Turabian StyleCugliari, Giovanni, Chiara Catalano, Simonetta Guarrera, Alessandra Allione, Elisabetta Casalone, Alessia Russo, Federica Grosso, Daniela Ferrante, Clara Viberti, Anna Aspesi, and et al. 2020. "DNA Methylation of FKBP5 as Predictor of Overall Survival in Malignant Pleural Mesothelioma" Cancers 12, no. 11: 3470. https://doi.org/10.3390/cancers12113470
APA StyleCugliari, G., Catalano, C., Guarrera, S., Allione, A., Casalone, E., Russo, A., Grosso, F., Ferrante, D., Viberti, C., Aspesi, A., Sculco, M., Pirazzini, C., Libener, R., Mirabelli, D., Magnani, C., Dianzani, I., & Matullo, G. (2020). DNA Methylation of FKBP5 as Predictor of Overall Survival in Malignant Pleural Mesothelioma. Cancers, 12(11), 3470. https://doi.org/10.3390/cancers12113470








