Identification of VRK1 as a New Neuroblastoma Tumor Progression Marker Regulating Cell Proliferation
Abstract
Simple Summary
Abstract
1. Introduction
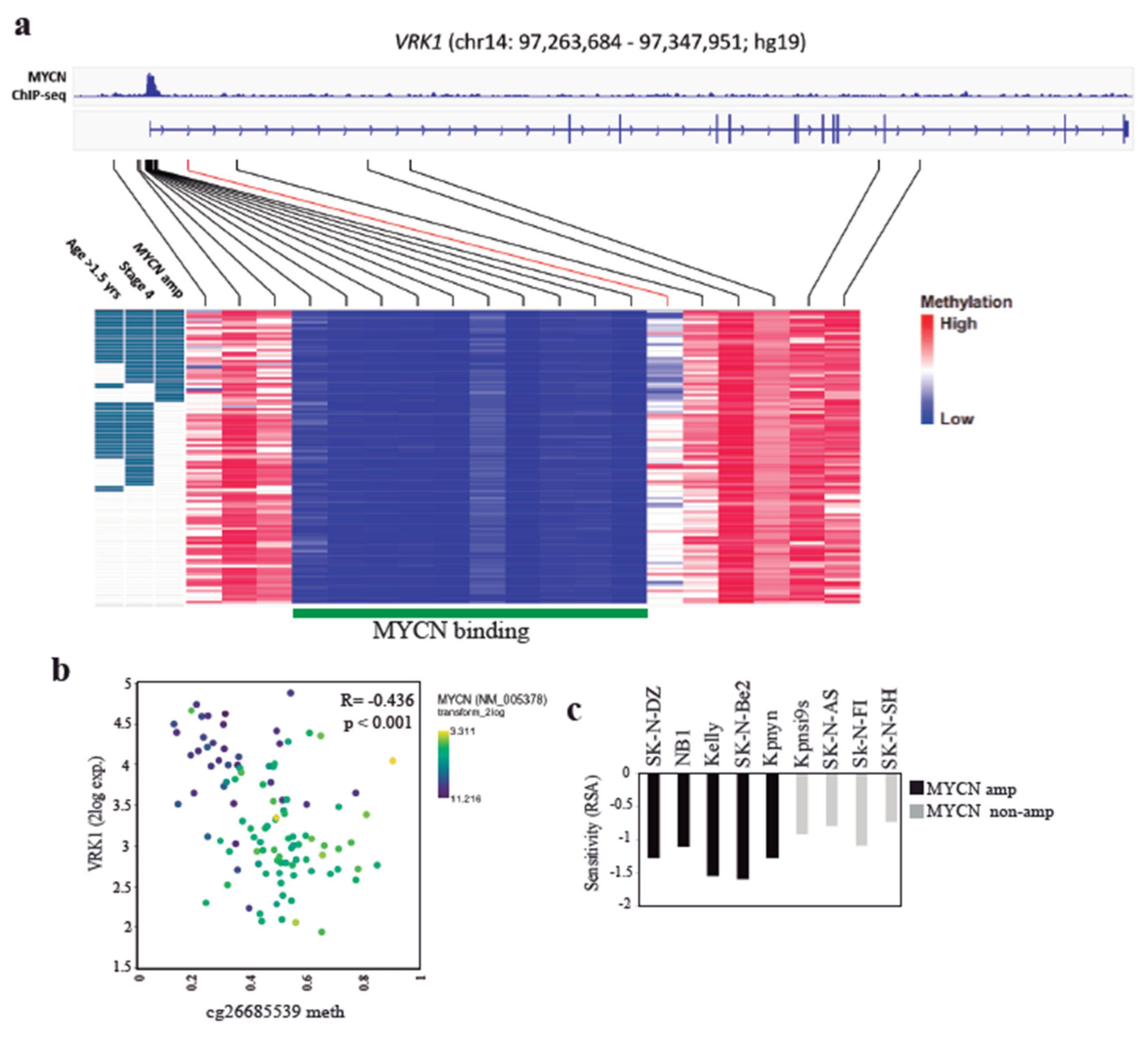
2. Results
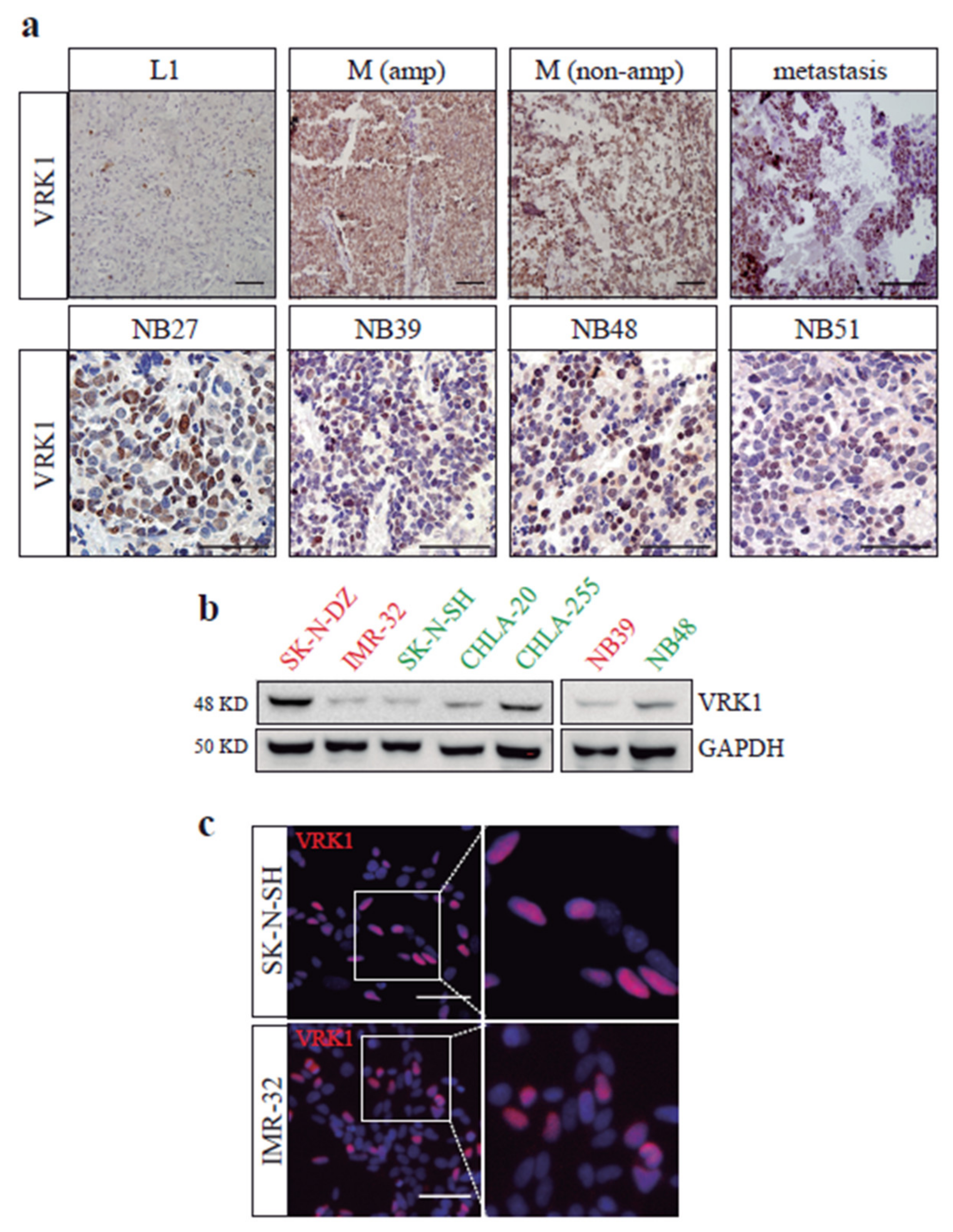
2.1. VRK1 Expression Correlates with Aggressiveness in Neuroblastoma Tumors
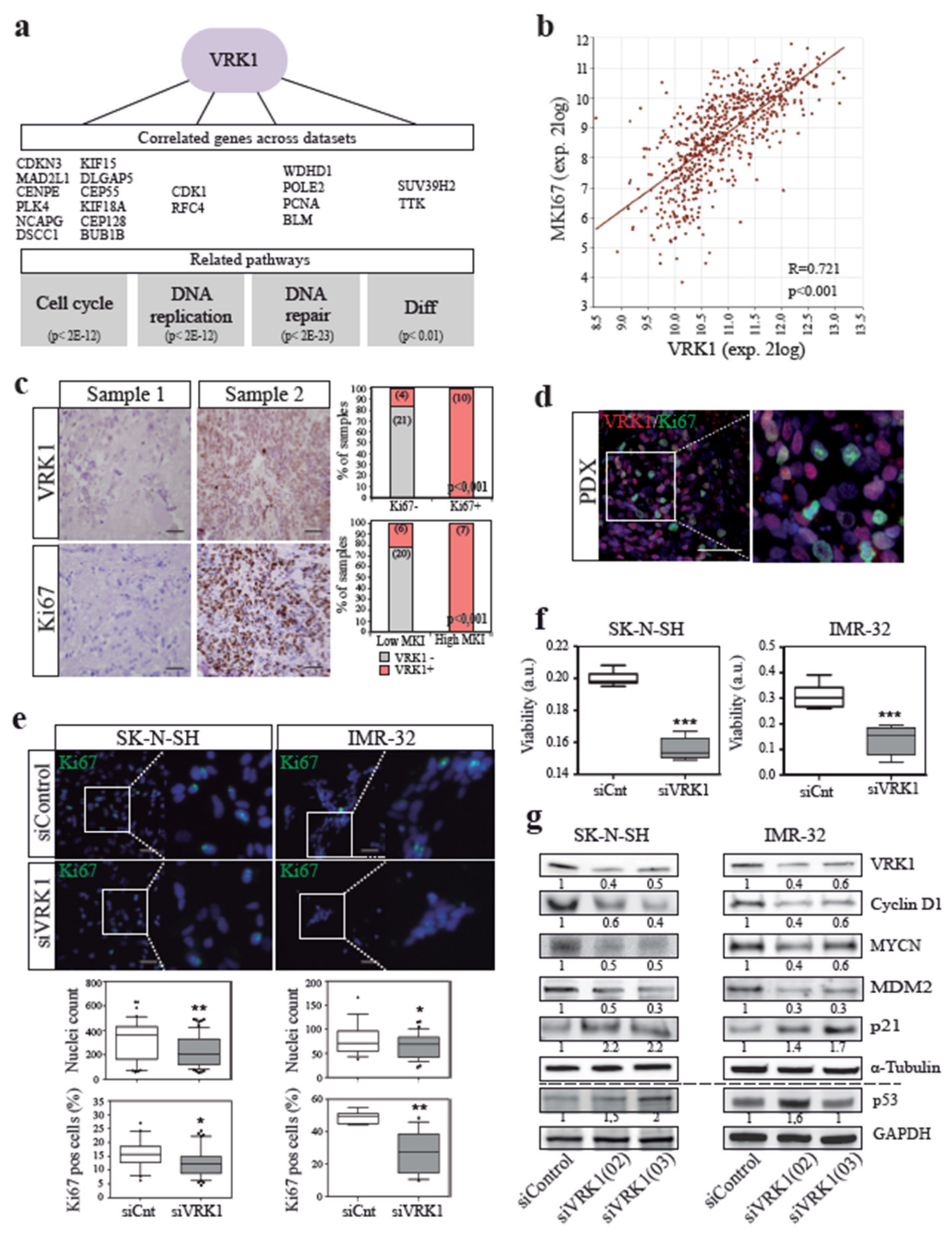
2.2. VRK1 is Associated with NB Tumor Cell Proliferation
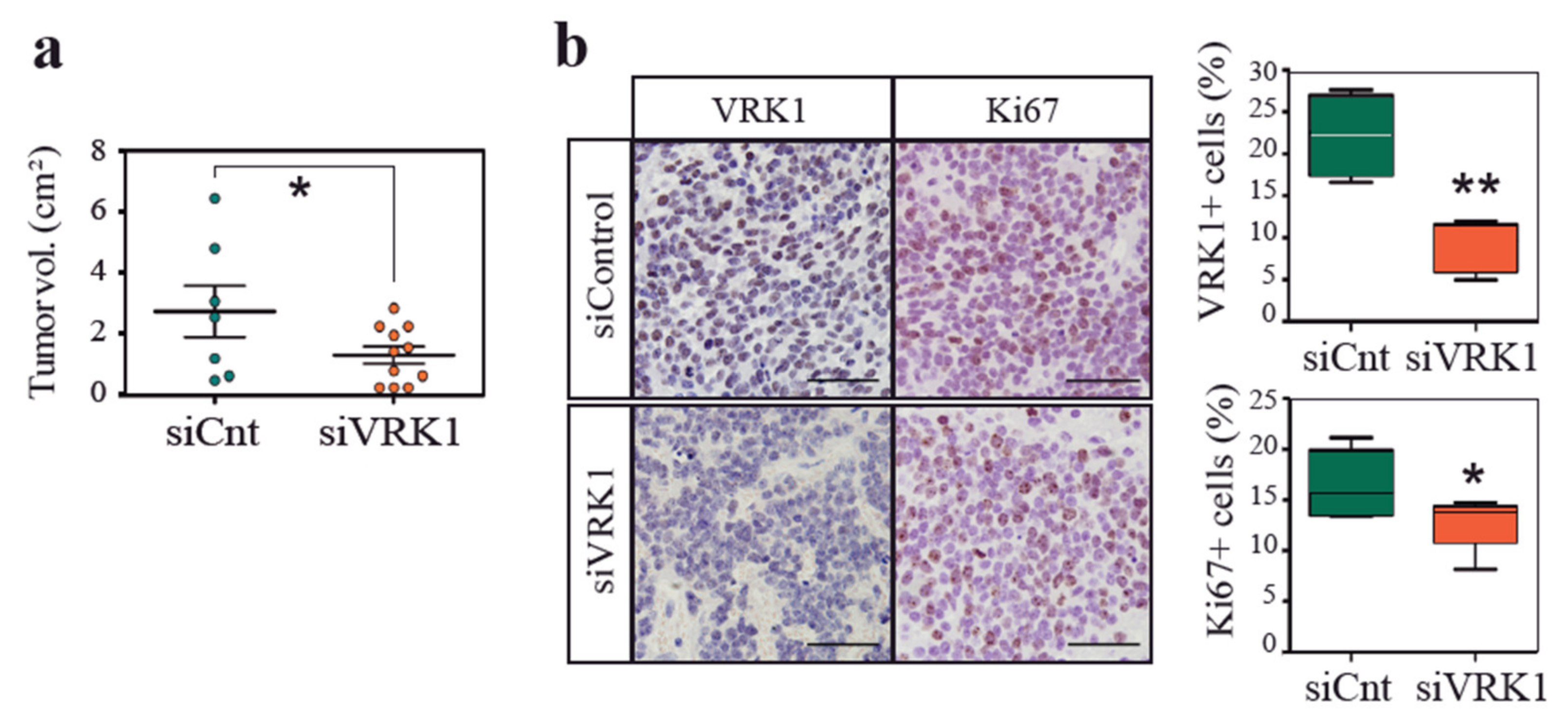
2.3. VRK1 Downregulation Impairs Neuroblastoma Tumorigenesis in a Xenograft Model
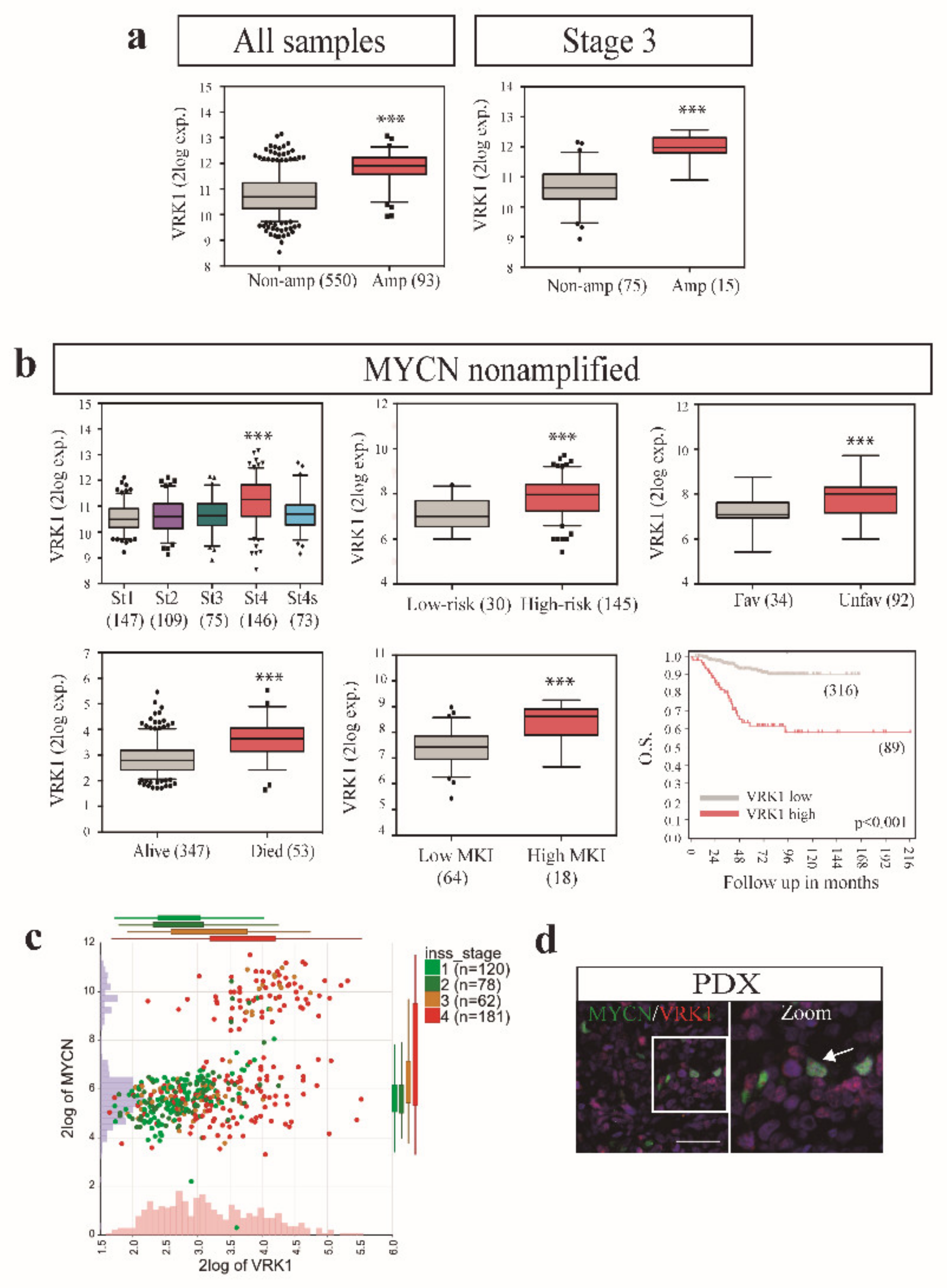
2.4. VRK1 Associates with Neuroblastoma Progression Independently of MYCN Amplification
3. Discussion
4. Materials and Methods
4.1. Human Tumor Gene Expression and Methylation Analysis
4.2. Cell Culture and siRNA Transfection
4.3. Western Blot and Cellular Assays
4.4. Tumor Xenografts
4.5. Immunofluorescence and Immunohistochemistry
4.6. Statistical Analysis
4.7. Ethics Approval and Consent to Participate
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnsen, J.I.; Dyberg, C.; Wickström, M. Neuroblastoma-A neural crest derived embryonal malignancy. Front. Mol. Neurosci. 2019, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Prim. 2016, 2, 16079. [Google Scholar] [CrossRef] [PubMed]
- Kamijo, T.; Nakagawara, A. Molecular and genetic bases of neuroblastoma. Int. J. Clin. Oncol. 2012, 17, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Van Groningen, T.; Koster, J.; Valentijn, L.J.; Zwijnenburg, D.A.; Akogul, N.; Hasselt, N.E.; Broekmans, M.; Haneveld, F.; Nowakowska, N.E.; Bras, J.; et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 2017, 49, 1261–1266. [Google Scholar] [CrossRef]
- Vega, F.M.; Colmenero-Repiso, A.; Gómez-Muñoz, M.A.; Rodríguez-Prieto, I.; Aguilar-Morante, D.; Ramírez, G.; Márquez, C.; Cabello, R.; Pardal, R. CD44-high neural crest stem-like cells are associated with tumour aggressiveness and poor survival in neuroblastoma tumours. EBioMedicine 2019, 49, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Gröbner, S.N.; Project, I.P.-S.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; et al. The landscape of genomic alterations across childhood cancers. Nat. Cell Biol. 2018, 555, 321–327. [Google Scholar] [CrossRef]
- Louis, C.U.; Shohet, J.M. Neuroblastoma: Molecular pathogenesis and therapy. Annu. Rev. Med. 2015, 66, 49–63. [Google Scholar] [CrossRef]
- Tomolonis, J.A.; Agarwal, S.; Shohet, J.M. Neuroblastoma pathogenesis: Deregulation of embryonic neural crest development. Cell Tissue Res. 2018, 372, 245–262. [Google Scholar] [CrossRef]
- Delloye-Bourgeois, C.; Castellani, V. Hijacking of embryonic programs by neural crest-derived neuroblastoma: From physiological migration to metastatic dissemination. Front. Mol. Neurosci. 2019, 12, 52. [Google Scholar] [CrossRef]
- Klerkx, E.P.F.; Lazo, P.A.; Askjaer, P. Emerging biological functions of the vaccinia-related kinase (VRK) family. Histol. Histopathol. 2009, 24, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Campillo-Marcos, I.; Lazo, P.A. Implication of the VRK1 chromatin kinase in the signaling responses to DNA damage: A therapeutic target? Cell. Mol. Life Sci. 2018, 75, 2375–2388. [Google Scholar] [CrossRef] [PubMed]
- Valbuena, A.; Sanz-García, M.; López-Sánchez, I.; Vega, F.M.; Lazo, P.A. Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell. Signal. 2011, 23, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Aihara, H.; Nakagawa, T.; Mizusaki, H.; Yoneda, M.; Kato, M.; Doiguchi, M.; Imamura, Y.; Higashi, M.; Ikura, T.; Hayashi, T.; et al. Histone H2A T120 Phosphorylation promotes oncogenic transformation via upregulation of cyclin D1. Mol. Cell 2016, 64, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Dobrzynska, A.; Askjaer, P. Vaccinia-related kinase 1 is required for early uterine development in Caenorhabditis elegans. Dev. Biol. 2016, 411, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Vega, F.M.; Gonzalo, P.; Gaspar, M.L.; Lazo, P.A. Expression of the VRK (vaccinia-related kinase) gene family of p53 regulators in murine hematopoietic development. FEBS Lett. 2003, 544, 176–180. [Google Scholar] [CrossRef]
- Choi, Y.H.; Park, C.-H.; Kim, W.; Ling, H.; Kang, A.; Chang, M.W.; Im, S.-K.; Jeong, H.-W.; Kong, Y.-Y.; Kim, K.-T. Vaccinia-related kinase 1 is required for the maintenance of undifferentiated spermatogonia in mouse male germ cells. PLoS ONE 2010, 5, e15254. [Google Scholar] [CrossRef]
- Vinograd-Byk, H.; Sapir, T.; Cantarero, L.; Lazo, P.A.; Zeligson, S.; Lev, D.; Lerman-Sagie, T.; Renbaum, P.; Reiner, O.; Levy-Lahad, E. The spinal muscular atrophy with pontocerebellar hypoplasia gene VRK1 regulates neuronal migration through an amyloid-β precursor protein-dependent mechanism. J. Neurosci. 2015, 35, 936–942. [Google Scholar] [CrossRef]
- Vinograd-Byk, H.; Renbaum, P.; Levy-Lahad, E. Vrk1 partial knockdown in mice results in reduced brain weight and mild motor dysfunction, and indicates neuronal VRK1 target pathways. Sci. Rep. 2018, 8, 11265. [Google Scholar] [CrossRef]
- Santos, C.R.; Rodríguez-Pinilla, M.; Vega, F.M.; Rodríguez-Peralto, J.L.; Blanco, S.; Sevilla, A.; Valbuena, A.; Hernandez, A.S.; Van Wijnen, A.J.; Li, F.; et al. VRK1 Signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol. Cancer Res. 2006, 4, 177–185. [Google Scholar] [CrossRef]
- Valbuena, A.; López-Sánchez, I.; Vega, F.M.; Sevilla, A.; Sanz-García, M.; Blanco, S.; Lazo, P.A. Identification of a dominant epitope in human vaccinia-related kinase 1 (VRK1) and detection of different intracellular subpopulations. Arch. Biochem. Biophys. 2007, 465, 219–226. [Google Scholar] [CrossRef]
- Lee, N.; Kwon, J.-H.; Kim, Y.B.; Kim, S.-H.; Park, S.J.; Xu, W.; Jung, H.-Y.; Kim, K.-T.; Wang, H.J.; Choi, K.Y. Vaccinia-related kinase 1 promotes hepatocellular carcinoma by controlling the levels of cell cycle regulators associated with G1/S transition. Oncotarget 2015, 6, 30130–30148. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-C.; Cao, K.; Xiao, Z.-H.; Qiao, L.; Wang, X.-Q.; Shang, B.; Jia, Y.; Wang, Z. VRK1 promotes cisplatin resistance by up-regulating c-MYC via c-Jun activation and serves as a therapeutic target in esophageal squamous cell carcinoma. Oncotarget 2017, 8, 65642–65658. [Google Scholar] [CrossRef] [PubMed]
- Salzano, M.; Vázquez-Cedeira, M.; Sanz-García, M.; Valbuena, A.; Blanco, S.; Fernández, I.F.; Lazo, P.A. Vaccinia-related kinase 1 (VRK1) confers resistance to DNA-damaging agents in human breast cancer by affecting DNA damage response. Oncotarget 2014, 5, 1770–1778. [Google Scholar] [CrossRef]
- Campillo-Marcos, I.; Lazo, P.A. Olaparib and ionizing radiation trigger a cooperative DNA-damage repair response that is impaired by depletion of the VRK1 chromatin kinase. J. Exp. Clin. Cancer Res. 2019, 38, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.S.; Song, Y.K.; Durinck, S.; Chen, Q.-R.; Cheuk, A.T.C.; Tsang, P.; Zhang, Q.; Thiele, C.J.; Slack, A.; Shohet, J.; et al. The MYCN oncogene is a direct target of miR-34a. Oncogene 2008, 27, 5204–5213. [Google Scholar] [CrossRef]
- Vega, F.M.; Sevilla, A.; Lazo, P.A. p53 Stabilization and accumulation induced by human vaccinia-related kinase 1. Mol. Cell. Biol. 2004, 24, 10366–10380. [Google Scholar] [CrossRef]
- Hovestadt, V.; Jones, D.T.W.; Picelli, S.; Wang, W.; Kool, M.; Northcott, P.A.; Sultan, M.; Stachurski, K.; Ryzhova, M.; Warnatz, H.-J.; et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nat. Cell Biol. 2014, 510, 537–541. [Google Scholar] [CrossRef]
- Lee, J.W.; Son, M.H.; Cho, H.W.; Ma, Y.E.; Yoo, K.H.; Sung, K.W.; Koo, H.H. Clinical significance of MYCN amplification in patients with high-risk neuroblastoma. Pediatr. Blood Cancer 2018, 65, e27257. [Google Scholar] [CrossRef]
- Valbuena, A.; López-Sánchez, I.; Lazo, P.A. Human VRK1 is an early response gene and its loss causes a block in cell cycle progression. PLoS ONE 2008, 3, e1642. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; He, S.; Xu, X.; Huang, Y.; Tang, J.; Wu, Y.; Miao, X.; He, Y.; Wang, Q.; et al. Expression of vaccinia-related kinase 1 (VRK1) accelerates cell proliferation but overcomes cell adhesion mediated drug resistance (CAM-DR) in multiple myeloma. Hematology 2016, 21, 603–612. [Google Scholar] [CrossRef]
- Kim, W.; Chakraborty, G.; Kim, S.; Shin, J.; Park, C.-H.; Jeong, M.-W.; Bharatham, N.; Yoon, H.S.; Kim, K.-T. Macro histone H2A1.2 (MacroH2A1) protein suppresses mitotic kinase VRK1 during interphase. J. Biol. Chem. 2011, 287, 5278–5289. [Google Scholar] [CrossRef] [PubMed]
- Couñago, R.M.; Allerston, C.K.; Savitsky, P.; Azevedo, H.; Godoi, P.H.; Wells, C.I.; Mascarello, A.; Gama, F.H.D.S.; Massirer, K.B.; Zuercher, W.J.; et al. Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Valbuena, A.; Vega, F.M.; Blanco, S.; Lazo, P.A. p53 Downregulates its activating vaccinia-related kinase 1, forming a new autoregulatory loop. Mol. Cell. Biol. 2006, 26, 4782–4793. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Henssen, A.G.; Althoff, K.; Odersky, A.; Beckers, A.; Koche, R.; Speleman, F.; Schäfers, S.; Bell, E.; Nortmeyer, M.; Westermann, F.; et al. Targeting MYCN-driven transcription by BET-bromodomain inhibition. Clin. Cancer Res. 2015, 22, 2470–2481. [Google Scholar] [CrossRef] [PubMed]
- Puissant, A.; Frumm, S.M.; Alexe, G.; Bassil, C.F.; Qi, J.; Chanthery, Y.H.; Nekritz, E.A.; Zeid, R.; Gustafson, W.C.; Greninger, P.; et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013, 3, 308–323. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.; Castel, V.; Castelberry, R.P.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar] [CrossRef]
- Cohn, S.L.; Pearson, A.D.J.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG task force report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef]
- Henrich, K.-O.; Bender, S.; Saadati, M.; Dreidax, D.; Gartlgruber, M.; Shao, C.; Herrmann, C.; Wiesenfarth, M.; Parzonka, M.; Wehrmann, L.; et al. Integrative genome-scale analysis identifies epigenetic mechanisms of transcriptional deregulation in unfavorable neuroblastomas. Cancer Res. 2016, 76, 5523–5537. [Google Scholar] [CrossRef]
- Zeid, R.; Lawlor, M.A.; Poon, E.; Reyes, J.M.; Fulciniti, M.; Lopez, M.A.; Scott, T.G.; Nabet, B.; Erb, M.A.; Winter, G.E.; et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 2018, 50, 515–523. [Google Scholar] [CrossRef]
- Iii, E.R.M.; De Weck, A.; Schlabach, M.R.; Billy, E.; Mavrakis, K.J.; Hoffman, G.; Belur, D.; Castelletti, D.; Frias, E.; Gampa, K.; et al. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 2017, 170, 577–592. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [PubMed]
- Monclair, T.; Brodeur, G.M.; Ambros, P.F.; Brisse, H.J.; Cecchetto, G.; Holmes, K.; Kaneko, M.; London, W.B.; Matthay, K.K.; Nuchtern, J.G.; et al. The International Neuroblastoma Risk Group (INRG) staging system: An INRG task force report. J. Clin. Oncol. 2009, 27, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Allred, D.C.; Harvey, J.M.; Berardo, M.; Clark, G.M. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod. Pathol. 1998, 11, 155–168. [Google Scholar] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colmenero-Repiso, A.; Gómez-Muñoz, M.A.; Rodríguez-Prieto, I.; Amador-Álvarez, A.; Henrich, K.-O.; Pascual-Vaca, D.; Okonechnikov, K.; Rivas, E.; Westermann, F.; Pardal, R.; et al. Identification of VRK1 as a New Neuroblastoma Tumor Progression Marker Regulating Cell Proliferation. Cancers 2020, 12, 3465. https://doi.org/10.3390/cancers12113465
Colmenero-Repiso A, Gómez-Muñoz MA, Rodríguez-Prieto I, Amador-Álvarez A, Henrich K-O, Pascual-Vaca D, Okonechnikov K, Rivas E, Westermann F, Pardal R, et al. Identification of VRK1 as a New Neuroblastoma Tumor Progression Marker Regulating Cell Proliferation. Cancers. 2020; 12(11):3465. https://doi.org/10.3390/cancers12113465
Chicago/Turabian StyleColmenero-Repiso, Ana, María A. Gómez-Muñoz, Ismael Rodríguez-Prieto, Aida Amador-Álvarez, Kai-Oliver Henrich, Diego Pascual-Vaca, Konstantin Okonechnikov, Eloy Rivas, Frank Westermann, Ricardo Pardal, and et al. 2020. "Identification of VRK1 as a New Neuroblastoma Tumor Progression Marker Regulating Cell Proliferation" Cancers 12, no. 11: 3465. https://doi.org/10.3390/cancers12113465
APA StyleColmenero-Repiso, A., Gómez-Muñoz, M. A., Rodríguez-Prieto, I., Amador-Álvarez, A., Henrich, K.-O., Pascual-Vaca, D., Okonechnikov, K., Rivas, E., Westermann, F., Pardal, R., & Vega, F. M. (2020). Identification of VRK1 as a New Neuroblastoma Tumor Progression Marker Regulating Cell Proliferation. Cancers, 12(11), 3465. https://doi.org/10.3390/cancers12113465







