Novel DNMT3A Germline Variant in a Patient with Multiple Paragangliomas and Papillary Thyroid Carcinoma
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Whole Exome Sequencing Analysis
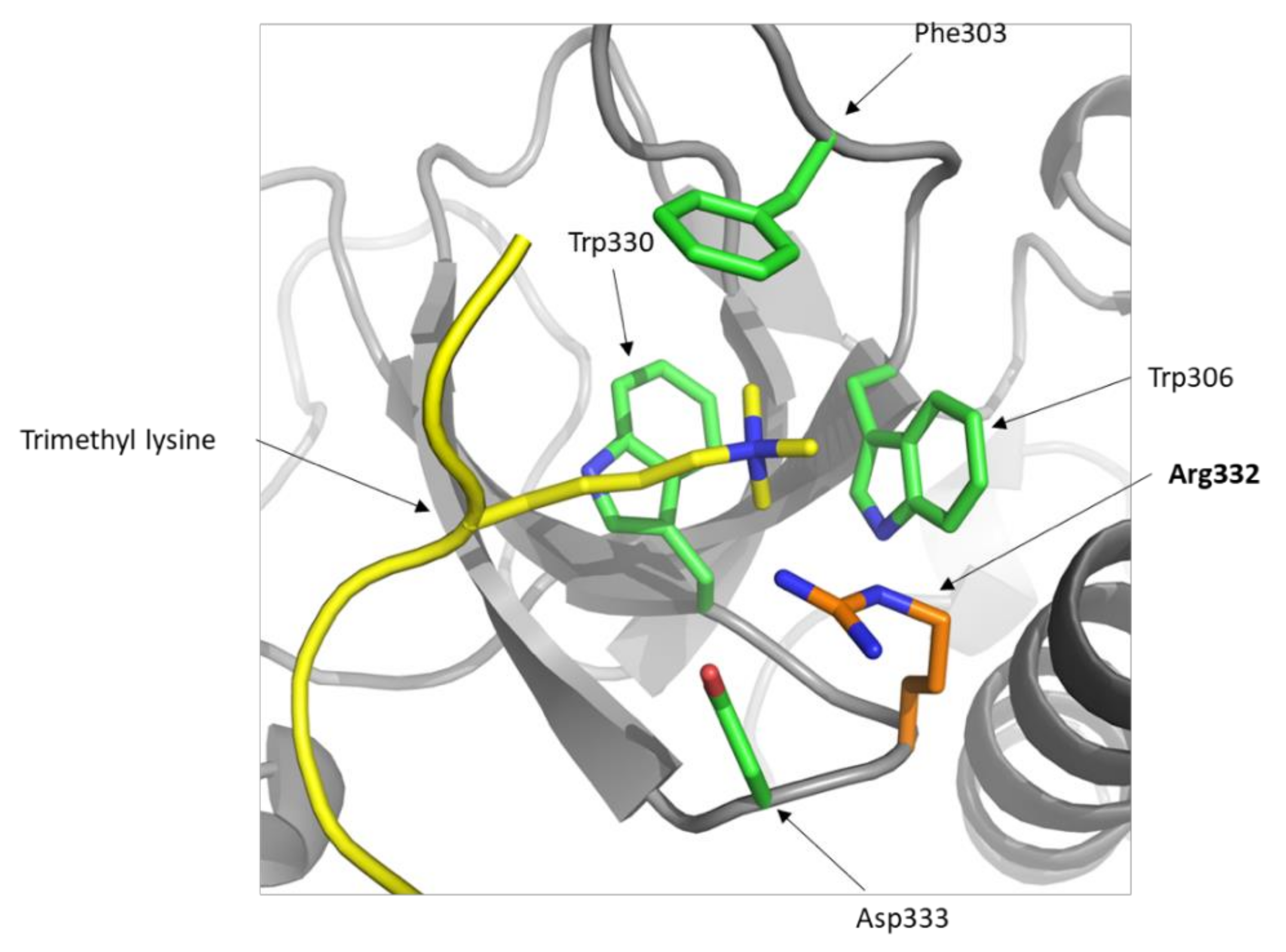
2.2. Presence of DNMT3A p.Gly332Arg in Databases and In Silico Predictions
2.3. Methylation Profiling
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Targeted Next-Generation Sequencing
4.3. In Silico Predictions
4.4. DNA Methylation Array
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fishbein, L. Pheochromocytoma/Paraganglioma: Is This a Genetic Disorder? Curr. Cardiol. Rep. 2019, 21, 104. [Google Scholar] [CrossRef]
- Remacha, L.; Pirman, D.; Mahoney, C.E.; Coloma, J.; Calsina, B.; Curras-Freixes, M.; Leton, R.; Torres-Perez, R.; Richter, S.; Pita, G.; et al. Recurrent Germline DLST Mutations in Individuals with Multiple Pheochromocytomas and Paragangliomas. Am. J. Hum. Genet. 2019, 104, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Buffet, A.; Morin, A.; Castro-Vega, L.J.; Habarou, F.; Lussey-Lepoutre, C.; Letouze, E.; Lefebvre, H.; Guilhem, I.; Haissaguerre, M.; Raingeard, I.; et al. Germline Mutations in the Mitochondrial 2-Oxoglutarate/Malate Carrier SLC25A11 Gene Confer a Predisposition to Metastatic Paragangliomas. Cancer Res. 2018, 78, 1914–1922. [Google Scholar] [CrossRef] [PubMed]
- Remacha, L.; Comino-Mendez, I.; Richter, S.; Contreras, L.; Curras-Freixes, M.; Pita, G.; Leton, R.; Galarreta, A.; Torres-Perez, R.; Honrado, E.; et al. Targeted exome sequencing of Krebs cycle genes reveals candidate cancer–predisposing mutations in pheochromocytomas and paragangliomas. Clin. Cancer Res. 2017, 23, 6315–6325. [Google Scholar] [CrossRef] [PubMed]
- Gowher, H.; Jeltsch, A. Mammalian DNA methyltransferases: New discoveries and open questions. Biochem. Soc. Trans. 2018, 46, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Yagi, M.; Kabata, M.; Tanaka, A.; Ukai, T.; Ohta, S.; Nakabayashi, K.; Shimizu, M.; Hata, K.; Meissner, A.; Yamamoto, T.; et al. Identification of distinct loci for de novo DNA methylation by DNMT3A and DNMT3B during mammalian development. Nat. Commun. 2020, 11, 3199. [Google Scholar] [CrossRef]
- Tatton-Brown, K.; Seal, S.; Ruark, E.; Harmer, J.; Ramsay, E.; Del Vecchio Duarte, S.; Zachariou, A.; Hanks, S.; O’Brien, E.; Aksglaede, L.; et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 2014, 46, 385–388. [Google Scholar] [CrossRef]
- Ley, T.J.; Ding, L.; Walter, M.J.; McLellan, M.D.; Lamprecht, T.; Larson, D.E.; Kandoth, C.; Payton, J.E.; Baty, J.; Welch, J.; et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010, 363, 2424–2433. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Rau, R.; Goodell, M.A. DNMT3A in haematological malignancies. Nat. Rev. Cancer 2015, 15, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Chaudry, S.F.; Chevassut, T.J. Epigenetic Guardian: A Review of the DNA Methyltransferase DNMT3A in Acute Myeloid Leukaemia and Clonal Haematopoiesis. BioMed Res. Int. 2017, 2017, 5473197. [Google Scholar] [CrossRef]
- Shen, W.; Heeley, J.M.; Carlston, C.M.; Acuna-Hidalgo, R.; Nillesen, W.M.; Dent, K.M.; Douglas, G.V.; Levine, K.L.; Bayrak-Toydemir, P.; Marcelis, C.L.; et al. The spectrum of DNMT3A variants in Tatton-Brown-Rahman syndrome overlaps with that in hematologic malignancies. Am. J. Med. Genet. A 2017, 173, 3022–3028. [Google Scholar] [CrossRef]
- Hollink, I.; van den Ouweland, A.M.W.; Beverloo, H.B.; Arentsen-Peters, S.; Zwaan, C.M.; Wagner, A. Acute myeloid leukaemia in a case with Tatton-Brown-Rahman syndrome: The peculiar DNMT3A R882 mutation. J. Med. Genet. 2017, 54, 805–808. [Google Scholar] [CrossRef]
- Tatton-Brown, K.; Zachariou, A.; Loveday, C.; Renwick, A.; Mahamdallie, S.; Aksglaede, L.; Baralle, D.; Barge-Schaapveld, D.; Blyth, M.; Bouma, M.; et al. The Tatton-Brown-Rahman Syndrome: A clinical study of 55 individuals with de novo constitutive DNMT3A variants. Wellcome Open Res. 2018, 3, 46. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Beird, H.C.; Estecio, M.; Hardikar, S.; Takahashi, K.; Bannon, S.A.; Borthakur, G.; Jabbour, E.; Gumbs, C.; Khoury, J.D.; et al. Germline DNMT3A mutation in familial acute myeloid leukaemia. Epigenetics 2020. [Google Scholar] [CrossRef]
- Remacha, L.; Curras-Freixes, M.; Torres-Ruiz, R.; Schiavi, F.; Torres-Perez, R.; Calsina, B.; Leton, R.; Comino-Mendez, I.; Roldan-Romero, J.M.; Montero-Conde, C.; et al. Gain-of-function mutations in DNMT3A in patients with paraganglioma. Genet. Med. 2018, 20, 1644–1651. [Google Scholar] [CrossRef]
- Heyn, P.; Logan, C.V.; Fluteau, A.; Challis, R.C.; Auchynnikava, T.; Martin, C.A.; Marsh, J.A.; Taglini, F.; Kilanowski, F.; Parry, D.A.; et al. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat. Genet. 2019, 51, 96–105. [Google Scholar] [CrossRef]
- Yap, D.B.; Chu, J.; Berg, T.; Schapira, M.; Cheng, S.W.; Moradian, A.; Morin, R.D.; Mungall, A.J.; Meissner, B.; Boyle, M.; et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 2011, 117, 2451–2459. [Google Scholar] [CrossRef]
- Katoh, M. Mutation spectra of histone methyltransferases with canonical SET domains and EZH2-targeted therapy. Epigenomics 2016, 8, 285–305. [Google Scholar] [CrossRef]
- Tatton-Brown, K.; Hanks, S.; Ruark, E.; Zachariou, A.; Duarte Sdel, V.; Ramsay, E.; Snape, K.; Murray, A.; Perdeaux, E.R.; Seal, S.; et al. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget 2011, 2, 1127–1133. [Google Scholar] [CrossRef]
- Wu, H.; Zeng, H.; Lam, R.; Tempel, W.; Amaya, M.F.; Xu, C.; Dombrovski, L.; Qiu, W.; Wang, Y.; Min, J. Structural and histone binding ability characterizations of human PWWP domains. PLoS ONE 2011, 6, e18919. [Google Scholar] [CrossRef]
- Geli, J.; Kiss, N.; Karimi, M.; Lee, J.J.; Backdahl, M.; Ekstrom, T.J.; Larsson, C. Global and regional CpG methylation in pheochromocytomas and abdominal paragangliomas: Association to malignant behavior. Clin. Cancer Res. 2008, 14, 2551–2559. [Google Scholar] [CrossRef]
- Toledo, R.A.; Qin, Y.; Cheng, Z.M.; Gao, Q.; Iwata, S.; Silva, G.M.; Prasad, M.L.; Ocal, I.T.; Rao, S.; Aronin, N.; et al. Recurrent Mutations of Chromatin-Remodeling Genes and Kinase Receptors in Pheochromocytomas and Paragangliomas. Clin. Cancer Res. 2016, 22, 2301–2310. [Google Scholar] [CrossRef]
- Fishbein, L.; Khare, S.; Wubbenhorst, B.; DeSloover, D.; D’Andrea, K.; Merrill, S.; Cho, N.W.; Greenberg, R.A.; Else, T.; Montone, K.; et al. Whole-exome sequencing identifies somatic ATRX mutations in pheochromocytomas and paragangliomas. Nat. Commun. 2015, 6, 6140. [Google Scholar] [CrossRef]
- Sendzikaite, G.; Hanna, C.W.; Stewart-Morgan, K.R.; Ivanova, E.; Kelsey, G. A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun. 2019, 10, 1884. [Google Scholar] [CrossRef]
- Dukatz, M.; Holzer, K.; Choudalakis, M.; Emperle, M.; Lungu, C.; Bashtrykov, P.; Jeltsch, A. H3K36me2/3 Binding and DNA Binding of the DNA Methyltransferase DNMT3A PWWP Domain Both Contribute to its Chromatin Interaction. J. Mol. Biol. 2019, 431, 5063–5074. [Google Scholar] [CrossRef]
- Ketkar, S.; Verdoni, A.M.; Smith, A.M.; Bangert, C.V.; Leight, E.R.; Chen, D.Y.; Brune, M.K.; Helton, N.M.; Hoock, M.; George, D.R.; et al. Remethylation of Dnmt3a (-/-) hematopoietic cells is associated with partial correction of gene dysregulation and reduced myeloid skewing. Proc. Natl. Acad. Sci. USA 2020, 117, 3123–3134. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, A.R.; Maroofian, R.; Salter, C.G.; Chioza, B.A.; Cross, H.E.; Patton, M.A.; Dempster, E.; Temple, I.K.; Mackay, D.J.G.; Rezwan, F.I.; et al. Growth disrupting mutations in epigenetic regulatory molecules are associated with abnormalities of epigenetic aging. Genome Res. 2019, 29, 1057–1066. [Google Scholar] [CrossRef]
- Choufani, S.; Gibson, W.T.; Turinsky, A.L.; Chung, B.H.Y.; Wang, T.; Garg, K.; Vitriolo, A.; Cohen, A.S.A.; Cyrus, S.; Goodman, S.; et al. DNA Methylation Signature for EZH2 Functionally Classifies Sequence Variants in Three PRC2 Complex Genes. Am. J. Hum. Genet. 2020, 106, 596–610. [Google Scholar] [CrossRef]
- Kaasinen, E.; Kuismin, O.; Rajamaki, K.; Ristolainen, H.; Aavikko, M.; Kondelin, J.; Saarinen, S.; Berta, D.G.; Katainen, R.; Hirvonen, E.A.M.; et al. Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat. Commun. 2019, 10, 1252. [Google Scholar] [CrossRef]
- Kaner, J.; Desai, P.; Mencia-Trinchant, N.; Guzman, M.L.; Roboz, G.J.; Hassane, D.C. Clonal Hematopoiesis and Premalignant Diseases. Cold Spring Harb. Perspect. Med. 2019, 10, a035675. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Genovese, G.; Kahler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef]
- Izzo, F.; Lee, S.C.; Poran, A.; Chaligne, R.; Gaiti, F.; Gross, B.; Murali, R.R.; Deochand, S.D.; Ang, C.; Jones, P.W.; et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat. Genet. 2020, 52, 378–387. [Google Scholar] [CrossRef]
- Young, A.L.; Tong, R.S.; Birmann, B.M.; Druley, T.E. Clonal hematopoiesis and risk of acute myeloid leukemia. Haematologica 2019, 104, 2410–2417. [Google Scholar] [CrossRef]
- Watson, C.J.; Papula, A.L.; Poon, G.Y.P.; Wong, W.H.; Young, A.L.; Druley, T.E.; Fisher, D.S.; Blundell, J.R. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 2020, 367, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Siraj, A.K.; Pratheeshkumar, P.; Parvathareddy, S.K.; Bu, R.; Masoodi, T.; Iqbal, K.; Al-Rasheed, M.; Al-Dayel, F.; Al-Sobhi, S.S.; Alzahrani, A.S.; et al. Prognostic significance of DNMT3A alterations in Middle Eastern papillary thyroid carcinoma. Eur. J. Cancer 2019, 117, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.C.; Zhu, W.D.; Ma, X.Y.; Ni, H.; Zhong, E.J.; Shao, Y.W.; Yu, J.; Gu, D.M.; Ji, S.D.; Xu, H.D.; et al. Mutations of genes including DNMT3A detected by next-generation sequencing in thyroid cancer. Cancer Biol. Ther. 2019, 20, 240–246. [Google Scholar] [CrossRef]
- Duployez, N.; Goursaud, L.; Fenwarth, L.; Bories, C.; Marceau-Renaut, A.; Boyer, T.; Fournier, E.; Nibourel, O.; Roche-Lestienne, C.; Huet, G.; et al. Familial myeloid malignancies with germline TET2 mutation. Leukemia 2020, 34, 1450–1453. [Google Scholar] [CrossRef]
- Chmielik, E.; Rusinek, D.; Oczko-Wojciechowska, M.; Jarzab, M.; Krajewska, J.; Czarniecka, A.; Jarzab, B. Heterogeneity of Thyroid Cancer. Pathobiology 2018, 85, 117–129. [Google Scholar] [CrossRef]
- Pozdeyev, N.; Gay, L.M.; Sokol, E.S.; Hartmaier, R.; Deaver, K.E.; Davis, S.; French, J.D.; Borre, P.V.; LaBarbera, D.V.; Tan, A.C.; et al. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin. Cancer Res. 2018, 24, 3059–3068. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, Z.X.; Jiang, Z.H.; Wang, X.S. Identification of genomic features associated with immunotherapy response in gastrointestinal cancers. World J. Gastrointest. Oncol. 2019, 11, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Bendl, J.; Stourac, J.; Salanda, O.; Pavelka, A.; Wieben, E.D.; Zendulka, J.; Brezovsky, J.; Damborsky, J. PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014, 10, e1003440. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Rondelet, G.; Dal Maso, T.; Willems, L.; Wouters, J. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J. Struct. Biol. 2016, 194, 357–367. [Google Scholar] [CrossRef]
- Bibikova, M.; Le, J.; Barnes, B.; Saedinia-Melnyk, S.; Zhou, L.; Shen, R.; Gunderson, K.L. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics 2009, 1, 177–200. [Google Scholar] [CrossRef]
- Monti, S.; Tamayo, P.; Mesirov, J.; Golub, T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Mach. Learn. 2003, 52, 91–118. [Google Scholar] [CrossRef]
- Reich, M.; Ohm, K.; Angelo, M.; Tamayo, P.; Mesirov, J.P. GeneCluster 2.0: An advanced toolset for bioarray analysis. Bioinformatics 2004, 20, 1797–1798. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]


| Tissue | Number Samples Mutated | Samples Tested | % Samples with Mutations |
|---|---|---|---|
| Endometrium | 7 | 958 | 0.730688935 |
| Thyroid | 5 | 2151 | 0.232450023 |
| Large intestine | 8 | 4480 | 0.178571429 |
| Urinary tract | 2 | 1204 | 0.166112957 |
| Bone | 1 | 724 | 0.138121547 |
| AML | 18 | 13,565 | 0.132694434 |
| Skin | 2 | 1780 | 0.112359551 |
| Kidney | 3 | 2790 | 0.107526882 |
| Hematopoietic and lymphoid (≠ AML) | 13 | 14,357 | 0.090548165 |
| Lung | 4 | 4931 | 0.081119448 |
| Breast | 4 | 5131 | 0.077957513 |
| Biliary tract | 1 | 1474 | 0.067842605 |
| Central nervous system | 2 | 3246 | 0.061614295 |
| Esophagus | 1 | 1668 | 0.059952038 |
| Upper aerodigestive tract | 1 | 1795 | 0.055710306 |
| Liver | 1 | 2370 | 0.042194093 |
| Prostate | 1 | 2869 | 0.03485535 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mellid, S.; Coloma, J.; Calsina, B.; Monteagudo, M.; Roldán-Romero, J.M.; Santos, M.; Leandro-García, L.J.; Lanillos, J.; Martínez-Montes, Á.M.; Rodríguez-Antona, C.; et al. Novel DNMT3A Germline Variant in a Patient with Multiple Paragangliomas and Papillary Thyroid Carcinoma. Cancers 2020, 12, 3304. https://doi.org/10.3390/cancers12113304
Mellid S, Coloma J, Calsina B, Monteagudo M, Roldán-Romero JM, Santos M, Leandro-García LJ, Lanillos J, Martínez-Montes ÁM, Rodríguez-Antona C, et al. Novel DNMT3A Germline Variant in a Patient with Multiple Paragangliomas and Papillary Thyroid Carcinoma. Cancers. 2020; 12(11):3304. https://doi.org/10.3390/cancers12113304
Chicago/Turabian StyleMellid, Sara, Javier Coloma, Bruna Calsina, María Monteagudo, Juan M. Roldán-Romero, María Santos, Luis J. Leandro-García, Javier Lanillos, Ángel M. Martínez-Montes, Cristina Rodríguez-Antona, and et al. 2020. "Novel DNMT3A Germline Variant in a Patient with Multiple Paragangliomas and Papillary Thyroid Carcinoma" Cancers 12, no. 11: 3304. https://doi.org/10.3390/cancers12113304
APA StyleMellid, S., Coloma, J., Calsina, B., Monteagudo, M., Roldán-Romero, J. M., Santos, M., Leandro-García, L. J., Lanillos, J., Martínez-Montes, Á. M., Rodríguez-Antona, C., Montero-Conde, C., Martínez-López, J., Ayala, R., Matias-Guiu, X., Robledo, M., & Cascón, A. (2020). Novel DNMT3A Germline Variant in a Patient with Multiple Paragangliomas and Papillary Thyroid Carcinoma. Cancers, 12(11), 3304. https://doi.org/10.3390/cancers12113304








