A Gene Expression High-Throughput Screen (GE-HTS) for Coordinated Detection of Functionally Similar Effectors in Cancer
Simple Summary
Abstract
1. Introduction
2. Gene Expression-Based High-Throughput Screens (GE-HTS) Using Chemical Libraries
3. Screening Strategies to Identify Functional Similar Proteins in Ras-Driven Cancers
3.1. Identifying Vulnerabilities in Ras-Driven Cancers Using RNAi or CRISPR Screening
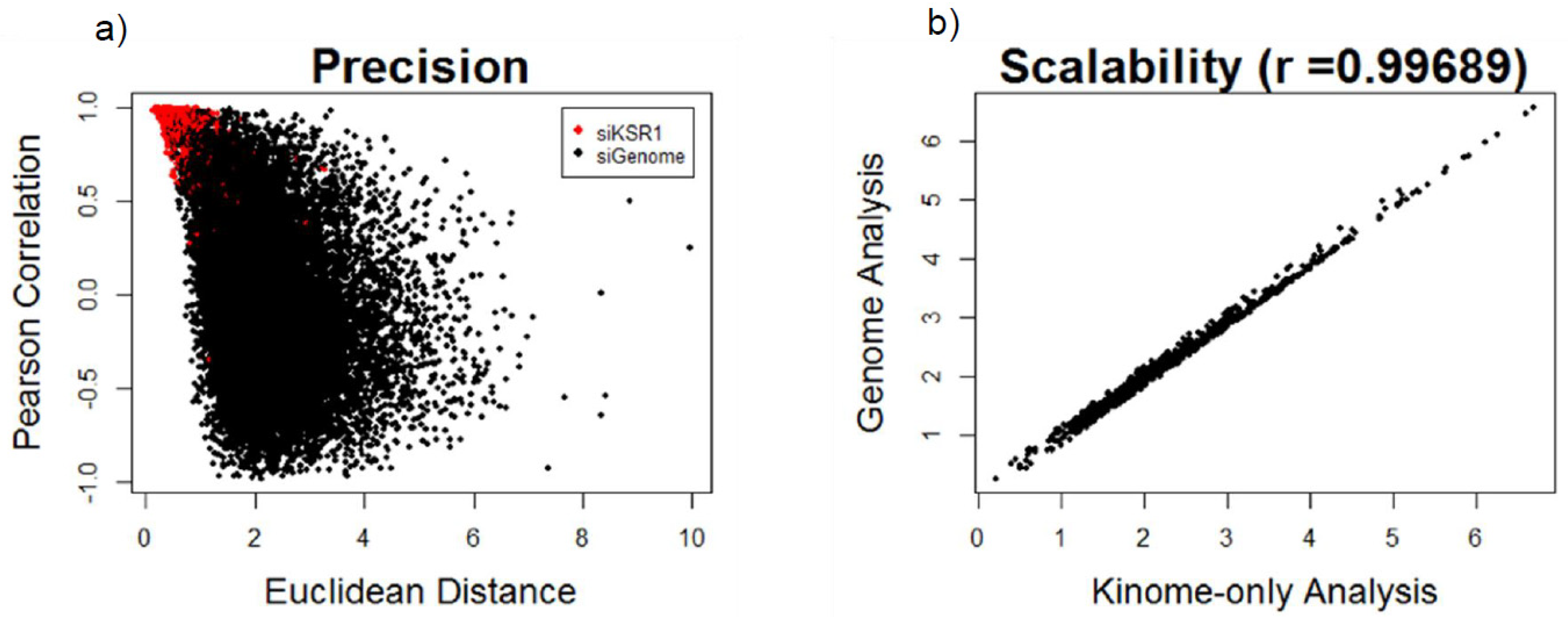
3.2. Identifying Novel Functional Protein Similarities in Ras-Driven Cancers Using FUSION
3.3. Identifying the Method of Action of Uncharacterized Natural Products in Ras-Driven Cancers Using FUSION
4. Guidelines for Designing, Performing, and Analyzing Future FUSION Screens
4.1. Batch Effects and Normalization
4.2. Minimizing Cost
4.3. Off-Target Effects and False Positives
5. The Future of FUSION
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef]
- Diehl, P.; Tedesco, D.; Chenchik, A. Use of RNAi screens to uncover resistance mechanisms in cancer cells and identify synthetic lethal interactions. Drug Discov. Today Technol. 2014, 11, 11–18. [Google Scholar] [CrossRef]
- Hinze, L.; Pfirrmann, M.; Karim, S.; Degar, J.; McGuckin, C.; Vinjamur, D.; Sacher, J.; Stevenson, K.E.; Neuberg, D.S.; Orellana, E.; et al. Synthetic Lethality of Wnt Pathway Activation and Asparaginase in Drug-Resistant Acute Leukemias. Cancer Cell 2019, 35, 664–676.e7. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef]
- Stegmaier, K.; Ross, K.N.; Colavito, S.A.; O’Malley, S.; Stockwell, B.R.; Golub, T.R. Gene expression-based high-throughput screening (GE-HTS) and application to leukemia differentiation. Nat. Genet. 2004, 36, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Zhang, F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015, 16, 299–311. [Google Scholar] [CrossRef]
- Inglese, J.; Johnson, R.L.; Simeonov, A.; Xia, M.; Zheng, W.; Austin, C.P.; Auld, D.S. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007, 3, 466–479. [Google Scholar] [CrossRef]
- Carpenter, A.E. Image-based chemical screening. Nat. Chem. Biol. 2007, 3, 461–465. [Google Scholar] [CrossRef]
- Boutros, M.; Ahringer, J. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 2008, 9, 554–566. [Google Scholar] [CrossRef]
- Kiger, A.A.; Baum, B.; Jones, S.; Jones, M.R.; Coulson, A.; Echeverri, C.; Perrimon, N. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2003, 2, 27. [Google Scholar] [CrossRef]
- Fisher, K.W.; Das, B.; Kim, H.S.; Clymer, B.K.; Gehring, D.; Smith, D.R.; Costanzo-Garvey, D.L.; Fernandez, M.R.; Brattain, M.G.; Kelly, D.L.; et al. AMPK Promotes Aberrant PGC1β Expression to Support Human Colon Tumor Cell Survival. Mol. Cell. Biol. 2015, 35, 3866–3879. [Google Scholar] [CrossRef] [PubMed]
- Potts, M.B.; Kim, H.S.; Fisher, K.W.; Hu, Y.; Carrasco, Y.P.; Bulut, G.B.; Ou, Y.H.; Herrera-Herrera, M.L.; Cubillos, F.; Mendiratta, S.; et al. Using functional signature ontology (FUSION) to identify mechanisms of action for natural products. Sci. Signal. 2013, 6, ra90. [Google Scholar] [CrossRef] [PubMed]
- McCall, J.L.; Gehring, D.; Clymer, B.K.; Fisher, K.W.; Das, B.; Kelly, D.L.; Kim, H.; White, M.A.; Lewis, R.E. KSR1 and EPHB4 Regulate Myc and PGC1beta to Promote Survival of Human Colon Tumors. Mol. Cell. Biol. 2016, 36, 2246–2261. [Google Scholar] [CrossRef]
- Neilsen, B.K.; Chakraborty, B.; McCall, J.L.; Frodyma, D.E.; Sleightholm, R.L.; Fisher, K.W.; Lewis, R.E. WDR5 supports colon cancer cells by promoting methylation of H3K4 and suppressing DNA damage. BMC Cancer 2018, 18, 673. [Google Scholar] [CrossRef]
- Das, B.; Neilsen, B.K.; Fisher, K.W.; Gehring, D.; Hu, Y.; Volle, D.J.; Kim, H.S.; McCall, J.L.; Kelly, D.L.; MacMillan, J.B.; et al. A Functional Signature Ontology (FUSION) screen detects an AMPK inhibitor with selective toxicity toward human colon tumor cells. Sci. Rep. 2018, 8, 3770. [Google Scholar] [CrossRef]
- McMillan, E.A.; Kwon, G.; Clemenceau, J.R.; Fisher, K.W.; Vaden, R.M.; Shaikh, A.F.; Neilsen, B.K.; Kelly, D.; Potts, M.B.; Sung, Y.J.; et al. A Genome-wide Functional Signature Ontology Map and Applications to Natural Product Mechanism of Action Discovery. Cell Chem. Biol. 2019, 26, 1380–1392 e1386. [Google Scholar] [CrossRef]
- Neilsen, B.K.; Frodyma, D.E.; McCall, J.L.; Fisher, K.W.; Lewis, R.E. ERK-mediated TIMELESS expression suppresses G2/M arrest in colon cancer cells. PLoS ONE 2019, 14, e0209224. [Google Scholar] [CrossRef]
- Hight, S.K.; Kurita, K.L.; McMillan, E.A.; Bray, W.; Clark, T.N.; Shaikh, A.F.; Jake Haeckl, F.P.; Carnevale-Neto, F.; La, S.; Lohith, A.; et al. High-Throughput Functional Annotation of Natural Products by Integrated Activity Profiling. bioRxiv 2019. [Google Scholar] [CrossRef]
- Vaden, R.M.; Oswald, N.W.; Potts, M.B.; MacMillan, J.B.; White, M.A. FUSION-Guided Hypothesis Development Leads to the Identification of N6,N6-Dimethyladenosine, a Marine-Derived AKT Pathway Inhibitor. Mar. Drugs 2017, 15, 75. [Google Scholar] [CrossRef]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.T. Natural products and Pharma 2011: Strategic changes spur new opportunities. Nat. Prod. Rep. 2011, 28, 1783–1789. [Google Scholar] [CrossRef] [PubMed]
- Antipova, A.A.; Stockwell, B.R.; Golub, T.R. Gene expression-based screening for inhibitors of PDGFR signaling. Genome Biol. 2008, 9, R47. [Google Scholar] [CrossRef] [PubMed]
- Peck, D.; Crawford, E.D.; Ross, K.N.; Stegmaier, K.; Golub, T.R.; Lamb, J. A method for high-throughput gene expression signature analysis. Genome Biol. 2006, 7, R61. [Google Scholar] [CrossRef]
- Luo, J.; Emanuele, M.J.; Li, D.; Creighton, C.J.; Schlabach, M.R.; Westbrook, T.F.; Wong, K.K.; Elledge, S.J. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 2009, 137, 835–848. [Google Scholar] [CrossRef]
- Steckel, M.; Molina-Arcas, M.; Weigelt, B.; Marani, M.; Warne, P.H.; Kuznetsov, H.; Kelly, G.; Saunders, B.; Howell, M.; Downward, J.; et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 2012, 22, 1227–1245. [Google Scholar] [CrossRef]
- Scholl, C.; Frohling, S.; Dunn, I.F.; Schinzel, A.C.; Barbie, D.A.; Kim, S.Y.; Silver, S.J.; Tamayo, P.; Wadlow, R.C.; Ramaswamy, S.; et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 2009, 137, 821–834. [Google Scholar] [CrossRef]
- Singh, A.; Sweeney, M.F.; Yu, M.; Burger, A.; Greninger, P.; Benes, C.; Haber, D.A.; Settleman, J. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell 2012, 148, 639–650. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Cheng, K.A.; Hata, A.N.; Faber, A.C.; Ebi, H.; Coffee, E.M.; Greninger, P.; Brown, R.D.; Godfrey, J.T.; Cohoon, T.J.; et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 2013, 23, 121–128. [Google Scholar] [CrossRef]
- Stephen, A.G.; Esposito, D.; Bagni, R.K.; McCormick, F. Dragging ras back in the ring. Cancer Cell 2014, 25, 272–281. [Google Scholar] [CrossRef]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef]
- Xue, J.Y.; Zhao, Y.; Aronowitz, J.; Mai, T.T.; Vides, A.; Qeriqi, B.; Kim, D.; Li, C.; de Stanchina, E.; Mazutis, L.; et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 2020, 577, 421–425. [Google Scholar] [CrossRef]
- Whitehurst, A.W.; Bodemann, B.O.; Cardenas, J.; Ferguson, D.; Girard, L.; Peyton, M.; Minna, J.D.; Michnoff, C.; Hao, W.; Roth, M.G.; et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 2007, 446, 815–819. [Google Scholar] [CrossRef]
- Agrotis, A.; Ketteler, R. A new age in functional genomics using CRISPR/Cas9 in arrayed library screening. Front. Genet. 2015, 6, 300. [Google Scholar] [CrossRef] [PubMed]
- Cullis, J.; Meiri, D.; Sandi, M.J.; Radulovich, N.; Kent, O.A.; Medrano, M.; Mokady, D.; Normand, J.; Larose, J.; Marcotte, R.; et al. The RhoGEF GEF-H1 is required for oncogenic RAS signaling via KSR-1. Cancer Cell 2014, 25, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Downward, J. RAS Synthetic Lethal Screens Revisited: Still Seeking the Elusive Prize? Clin. Cancer Res. 2015, 21, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014, 343, 80–84. [Google Scholar] [CrossRef]
- Yau, E.H.; Kummetha, I.R.; Lichinchi, G.; Tang, R.; Zhang, Y.; Rana, T.M. Genome-Wide CRISPR Screen for Essential Cell Growth Mediators in Mutant KRAS Colorectal Cancers. Cancer Res. 2017, 77, 6330–6339. [Google Scholar] [CrossRef]
- Neilsen, B.K.; Kelly, D.L.; Chakraborty, B.; Kim, H.S.; White, M.A.; Lewis, R.E.; Fisher, K.W. High-throughput identification of protein functional similarities using a gene-expression-based siRNA screen. Sci. Data 2020, 7, 27. [Google Scholar] [CrossRef]
- Rancati, G.; Moffat, J.; Typas, A.; Pavelka, N. Emerging and evolving concepts in gene essentiality. Nat. Rev. Genet. 2018, 19, 34–49. [Google Scholar] [CrossRef]
- Cacace, A.M.; Michaud, N.R.; Therrien, M.; Mathes, K.; Copeland, T.; Rubin, G.M.; Morrison, D.K. Identification of constitutive and ras-inducible phosphorylation sites of KSR: Implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol. Cell. Biol. 1999, 19, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Downward, J. KSR: A novel player in the RAS pathway. Cell 1995, 83, 831–834. [Google Scholar] [CrossRef]
- Kortum, R.L.; Lewis, R.E. The Molecular Scaffold KSR1 Regulates the Proliferative and Oncogenic Potential of Cells. Mol. Cell. Biol. 2004, 24, 4407–4416. [Google Scholar] [CrossRef] [PubMed]
- Therrien, M.; Michaud, N.R.; Rubin, G.M.; Morrison, D.K. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996, 10, 2684–2695. [Google Scholar] [CrossRef] [PubMed]
- Kortum, R.L.; Johnson, H.J.; Costanzo, D.L.; Volle, D.J.; Razidlo, G.L.; Fusello, A.M.; Shaw, A.S.; Lewis, R.E. The molecular scaffold kinase suppressor of Ras 1 is a modifier of RasV12-induced and replicative senescence. Mol. Cell. Biol. 2006, 26, 2202–2214. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Burack, W.R.; Stock, J.L.; Kortum, R.; Chaika, O.V.; Afkarian, M.; Muller, W.J.; Murphy, K.M.; Morrison, D.K.; Lewis, R.E.; et al. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol. Cell. Biol. 2002, 22, 3035–3045. [Google Scholar] [CrossRef]
- Lozano, J.; Xing, R.; Cai, Z.; Jensen, H.L.; Trempus, C.; Mark, W.; Cannon, R.; Kolesnick, R. Deficiency of Kinase Suppressor of Ras1 Prevents Oncogenic Ras Signaling in Mice. Cancer Res. 2003, 63, 4232–4238. [Google Scholar]
- Neilsen, B.K. Functional Signature Ontology-Based Identification and Validation of Novel Therapeutic Targets and Natural Products for the Treatment of Cancer. Ph.D. Thesis, University of Nebraska Medical Center, Omaha, NE, USA, 2018. [Google Scholar]
- PubChem [Internet]. PubChem Bioassay Record for AID 1259424; Source: Lewis Laboratory, University of Nebraska Medical Center; National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD, USA, 2004. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/1259424 (accessed on 8 September 2020).
- Hardie, D.G.; Carling, D.; Gamblin, S.J. AMP-activated protein kinase: Also regulated by ADP? Trends Biochem. Sci. 2011, 36, 470–477. [Google Scholar] [CrossRef]
- Costanzo-Garvey, D.L.; Pfluger, P.T.; Dougherty, M.K.; Stock, J.L.; Boehm, M.; Chaika, O.; Fernandez, M.R.; Fisher, K.; Kortum, R.L.; Hong, E.-G.; et al. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab. 2009, 10, 366–378. [Google Scholar] [CrossRef]
- Fernandez, M.R.; Henry, M.D.; Lewis, R.E. Kinase suppressor of Ras 2 (KSR2) regulates tumor cell transformation via AMPK. Mol. Cell. Biol. 2012, 32, 3718–3731. [Google Scholar] [CrossRef] [PubMed]
- Pearce, L.R.; Atanassova, N.; Banton, M.C.; Bottomley, B.; van der Klaauw, A.A.; Revelli, J.-P.; Hendricks, A.; Keogh, J.M.; Henning, E.; Doree, D.; et al. KSR2 Mutations Are Associated with Obesity, Insulin Resistance, and Impaired Cellular Fuel Oxidation. Cell 2013, 155, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Mohr, S.E.; Smith, J.A.; Shamu, C.E.; Neumuller, R.A.; Perrimon, N. RNAi screening comes of age: Improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol. 2014, 15, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Meier, R.; Casanova, A.; Kreibich, S.; Daga, N.; Andritschke, D.; Dilling, S.; Ramo, P.; Emmenlauer, M.; Kaufmann, A.; et al. Specific inhibition of diverse pathogens in human cells by synthetic microRNA-like oligonucleotides inferred from RNAi screens. Proc. Natl. Acad. Sci. USA 2014, 111, 4548–4553. [Google Scholar] [CrossRef]
- Sigoillot, F.D.; Lyman, S.; Huckins, J.F.; Adamson, B.; Chung, E.; Quattrochi, B.; King, R.W. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods 2012, 9, 363–366. [Google Scholar] [CrossRef]
- Adams, R.; Nicke, B.; Pohlenz, H.D.; Sohler, F. Deciphering Seed Sequence Based Off-Target Effects in a Large-Scale RNAi Reporter Screen for E-Cadherin Expression. PLoS ONE 2015, 10, e0137640. [Google Scholar] [CrossRef]
- Yilmazel, B.; Hu, Y.; Sigoillot, F.; Smith, J.A.; Shamu, C.E.; Perrimon, N.; Mohr, S.E. Online GESS: Prediction of miRNA-like off-target effects in large-scale RNAi screen data by seed region analysis. BMC Bioinform. 2014, 15, 192. [Google Scholar] [CrossRef]
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 2014, 11, 333–337. [Google Scholar] [CrossRef]
- El-Hachem, N.; Gendoo, D.M.A.; Ghoraie, L.S.; Safikhani, Z.; Smirnov, P.; Chung, C.; Deng, K.; Fang, A.; Birkwood, E.; Ho, C.; et al. Integrative Cancer Pharmacogenomics to Infer Large-Scale Drug Taxonomy. Cancer Res. 2017, 77, 3057–3069. [Google Scholar] [CrossRef]
- Rohban, M.H.; Abbasi, H.S.; Singh, S.; Carpenter, A.E. Capturing single-cell heterogeneity via data fusion improves image-based profiling. Nat. Commun. 2019, 10, 2082. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Montanez-Sauri, S.I.; Sung, K.E.; Berthier, E.; Beebe, D.J. Enabling screening in 3D microenvironments: Probing matrix and stromal effects on the morphology and proliferation of T47D breast carcinoma cells. Integr. Biol. 2013, 5, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.L.; Baird, A.M.; Vaz, G.; Urquhart, A.J.; Senge, M.; Richard, D.J.; O’Byrne, K.J.; Davies, A.M. Drug Discovery Approaches Utilizing Three-Dimensional Cell Culture. Assay Drug Dev. Technol. 2016, 14, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef]
- Han, K.; Pierce, S.E.; Li, A.; Spees, K.; Anderson, G.R.; Seoane, J.A.; Lo, Y.H.; Dubreuil, M.; Olivas, M.; Kamber, R.A.; et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature 2020, 580, 136–141. [Google Scholar] [CrossRef] [PubMed]
| Screen Strategy | Key Findings | Identified NP/Protein | Proposed Mechanism | Reference |
|---|---|---|---|---|
| Functionally related proteins to KSR1 | AMPKγ1 as an essential driver of PGC1β expression and colon cancer cell survival. | AMPKγ | α2β2γ1 isoform of AMPK promotes aberrant expression of tumor-specific PGC1β and ERRα expression in colon cancer. | Fisher and Das et al. [12] |
| EPHB4 supports colon cancer cell survival through regulation of Myc and PGC1β mRNA levels. | EPHB4 | KSR1 promotes EPHB4 expression by protecting it from lysosome-dependent degradation. EPHB4 kinase inhibitor AZ12672857 is selectively toxic to colon cancer cells. | McCall and Gehring et al. [14] | |
| Increased ERK signaling through oncogenic Ras promotes TIMELESS overexpression in cancer promoting cancer cell proliferation. | TIMELESS | TIMELESS depletion induces cell cycle checkpoint-induced G2/M arrest limiting cell proliferation. | Neilsen and Frodyma et al. [18] | |
| Natural product fractions | SRMS, BMP2K, and natural products SN-B-019-cmp1 as autophagy modulators. | Isolated from Streptomyces bacillaris strain, SN-B-019-cmp1 | SN-B-019-cmp1 and bafilomycin D block autophagasome maturation. BMP2K blocks basal autophagy, while SRMS functions through mTOR to inhibit autophagy. | Potts, Kim, and Fisher et al. [13] |
| SN-B-004 and 5′-hydroxy-staurosporine (5-OH-S) as a novel AMPK inhibitor. | Streptomyces bacillaris strain SN-B-004 | SN-B-004 and 5-OH-S are competitive AMPK inhibitors and decreased levels of AMPK downstream targets ACC and Raptor. AMPK inhibition via 5-OH-S is selectively toxic to colon cancer cells. | Das and Neilsen et al. [16] | |
| De novo network of targets of uncharacterized natural products. | SN-A-022-6 | SN-A-022-6 clustered with ERRα inhibitor XCT790 and suppressed mitochondrial oxidative capacity in lung cancer cells. | McMillan and Kwon et al. [17] | |
| Marine-derived natural product as a potent AKT inhibitor. | N6,N6-dimethyladenosine | SN-A-024-A and N6,N6-dimethyladenosine behave as small molecule inhibitors of AKT signaling. | Vaden et al. [20] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, C.; Huisman, D.H.; Vieira, H.M.; Frodyma, D.E.; Neilsen, B.K.; Chakraborty, B.; Hight, S.K.; White, M.A.; Fisher, K.W.; Lewis, R.E. A Gene Expression High-Throughput Screen (GE-HTS) for Coordinated Detection of Functionally Similar Effectors in Cancer. Cancers 2020, 12, 3143. https://doi.org/10.3390/cancers12113143
Rao C, Huisman DH, Vieira HM, Frodyma DE, Neilsen BK, Chakraborty B, Hight SK, White MA, Fisher KW, Lewis RE. A Gene Expression High-Throughput Screen (GE-HTS) for Coordinated Detection of Functionally Similar Effectors in Cancer. Cancers. 2020; 12(11):3143. https://doi.org/10.3390/cancers12113143
Chicago/Turabian StyleRao, Chaitra, Dianna H. Huisman, Heidi M. Vieira, Danielle E. Frodyma, Beth K. Neilsen, Binita Chakraborty, Suzie K. Hight, Michael A. White, Kurt W. Fisher, and Robert E. Lewis. 2020. "A Gene Expression High-Throughput Screen (GE-HTS) for Coordinated Detection of Functionally Similar Effectors in Cancer" Cancers 12, no. 11: 3143. https://doi.org/10.3390/cancers12113143
APA StyleRao, C., Huisman, D. H., Vieira, H. M., Frodyma, D. E., Neilsen, B. K., Chakraborty, B., Hight, S. K., White, M. A., Fisher, K. W., & Lewis, R. E. (2020). A Gene Expression High-Throughput Screen (GE-HTS) for Coordinated Detection of Functionally Similar Effectors in Cancer. Cancers, 12(11), 3143. https://doi.org/10.3390/cancers12113143








