Liquid Biopsy for Solid Ophthalmic Malignancies: An Updated Review and Perspectives
Abstract
Simple Summary
Abstract
1. Introduction
2. Liquid Biopsy in Uveal Melanoma (UM)
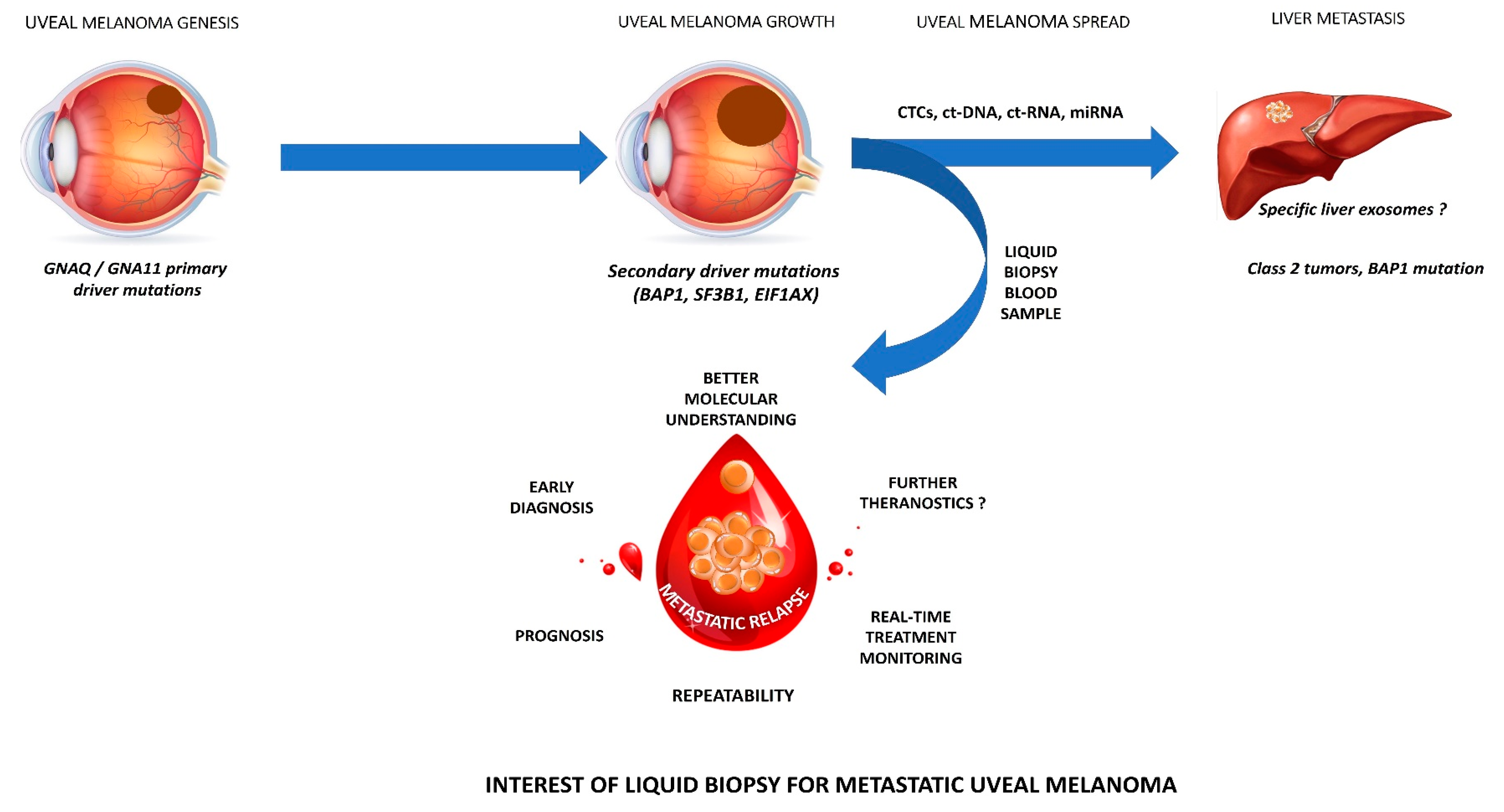
2.1. Molecular Characteristics of UM
2.2. CTCs
2.2.1. CTC Isolation and Identification
2.2.2. Main Clinical Findings
2.3. Ct-DNA and ct-RNA
2.3.1. Ct-DNA and ct-RNA Detection
2.3.2. Main Clinical Findings
2.4. Non-Coding RNAs
2.4.1. Non-Coding RNAs Detection
2.4.2. Main Clinical Findings
2.5. Tumor-Related Exosomes (TREs)
2.5.1. TRE Detection
2.5.2. Main Clinical Findings
2.6. Tumor-Educated Platelets (TEPs)
2.7. Future Perspectives: Towards a Better UM Understanding?
2.8. Limitations
2.9. Conclusion
3. Retinoblastoma (RB)
4. LB in Conjunctival Malignancies
5. LB in Choroidal Metastases
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Scarlotta, M.; Simsek, C.; Kim, A.K. Liquid Biopsy in Solid Malignancy. Genet. Test. Mol. Biomark. 2019, 23, 284–296. [Google Scholar] [CrossRef] [PubMed]
- AlAli, A.; Kletke, S.; Gallie, B.; Lam, W.-C. Retinoblastoma for Pediatric Ophthalmologists. Asia Pac. J. Ophthalmol. 2018, 7, 160–168. [Google Scholar] [CrossRef]
- León-Mateos, L.; Vieito, M.; Anido, U.; López López, R.; Muinelo Romay, L. Clinical Application of Circulating Tumour Cells in Prostate Cancer: From Bench to Bedside and Back. Int. J. Mol. Sci. 2016, 17, 1580. [Google Scholar] [CrossRef]
- Payne, K.; Brooks, J.; Spruce, R.; Batis, N.; Taylor, G.; Nankivell, P.; Mehanna, H. Circulating Tumour Cell Biomarkers in Head and Neck Cancer: Current Progress and Future Prospects. Cancers 2019, 11, 1115. [Google Scholar] [CrossRef]
- Hofman, P. Liquid biopsy for early detection of lung cancer. Curr. Opin. Oncol. 2017, 29, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.L.; Xu, L.; Polski, A.; Jubran, R.; Kuhn, P.; Kim, J.W.; Hicks, J. Aqueous Humor Is Superior to Blood as a Liquid Biopsy for Retinoblastoma. Ophthalmology 2020, 127, 552–554. [Google Scholar] [CrossRef]
- Singh, A.D.; Turell, M.E.; Topham, A.K. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology 2011, 118, 1881–1885. [Google Scholar] [CrossRef]
- Cohen, V.M.; Dinakaran, S.; Parsons, M.A.; Rennie, I.G. Transvitreal fine needle aspiration biopsy: The influence of intraocular lesion size on diagnostic biopsy result. Eye 2001, 15, 143–147. [Google Scholar] [CrossRef]
- Eide, N.; Walaas, L. Fine-needle aspiration biopsy and other biopsies in suspected intraocular malignant disease: A review. Acta Ophthalmol. 2009, 87, 588–601. [Google Scholar] [CrossRef]
- Bensoussan, E.; Thariat, J.; Maschi, C.; Delas, J.; Schouver, E.D.; Hérault, J.; Baillif, S.; Caujolle, J.-P. Outcomes after Proton Beam Therapy for Large Choroidal Melanomas in 492 Patients. Am. J. Ophthalmol. 2016, 165, 78–87. [Google Scholar] [CrossRef]
- Sellam, A.; Desjardins, L.; Barnhill, R.; Plancher, C.; Asselain, B.; Savignoni, A.; Pierron, G.; Cassoux, N. Fine Needle Aspiration Biopsy in Uveal Melanoma: Technique, Complications, and Outcomes. Am. J. Ophthalmol. 2016, 162, 28–34. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Char, D.H.; Augsburger, J.J.; Correa, Z.M.; Nudleman, E.; Aaberg, T.M.; Altaweel, M.M.; Bardenstein, D.S.; Finger, P.T.; et al. Collaborative Ocular Oncology Group report number 1: Prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology 2012, 119, 1596–1603. [Google Scholar] [CrossRef]
- Kujala, E.; Mäkitie, T.; Kivelä, T. Very long-term prognosis of patients with malignant uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4651–4659. [Google Scholar] [CrossRef]
- Shields, C.L.; Say, E.A.T.; Hasanreisoglu, M.; Saktanasate, J.; Lawson, B.M.; Landy, J.E.; Badami, A.U.; Sivalingam, M.D.; Hauschild, A.J.; House, R.J.; et al. Personalized Prognosis of Uveal Melanoma Based on Cytogenetic Profile in 1059 Patients over an 8-Year Period. Ophthalmology 2017, 124, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Goh, A.Y.; Layton, C.J. Evolving systemic targeted therapy strategies in uveal melanoma and implications for ophthalmic management: A review. Clin. Exp. Ophthalmol. 2016, 44, 509–519. [Google Scholar] [CrossRef]
- Xu, L.T.; Funchain, P.F.; Bena, J.F.; Li, M.; Tarhini, A.; Berber, E.; Singh, A.D. Uveal Melanoma Metastatic to the Liver: Treatment Trends and Outcomes. Ocul. Oncol. Pathol. 2019, 5, 323–332. [Google Scholar] [CrossRef]
- Shoushtari, A.N.; Carvajal, R.D. Treatment of Uveal Melanoma. Cancer Treat. Res. 2016, 167, 281–293. [Google Scholar] [CrossRef]
- Bande Rodríguez, M.F.; Fernandez Marta, B.; Lago Baameiro, N.; Santiago-Varela, M.; Silva-Rodríguez, P.; Blanco-Teijeiro, M.J.; Pardo Perez, M.; Piñeiro Ces, A. Blood Biomarkers of Uveal Melanoma: Current Perspectives. Clin. Ophthalmol. 2020, 14, 157–169. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Long, M.D.; Duan, S.; Council, M.L.; Bowcock, A.M.; Harbour, J.W. Oncogenic mutations in GNAQ occur early in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2008, 49, 5230–5234. [Google Scholar] [CrossRef]
- Pandiani, C.; Béranger, G.E.; Leclerc, J.; Ballotti, R.; Bertolotto, C. Focus on cutaneous and uveal melanoma specificities. Genes Dev. 2017, 31, 724–743. [Google Scholar] [CrossRef]
- Smit, K.N.; Jager, M.J.; de Klein, A.; Kiliç, E. Uveal melanoma: Towards a molecular understanding. Prog. Retin. Eye Res. 2020, 75, 100800. [Google Scholar] [CrossRef]
- Trolet, J.; Hupé, P.; Huon, I.; Lebigot, I.; Decraene, C.; Delattre, O.; Sastre-Garau, X.; Saule, S.; Thiéry, J.-P.; Plancher, C.; et al. Genomic profiling and identification of high-risk uveal melanoma by array CGH analysis of primary tumors and liver metastases. Investig. Ophthalmol. Vis. Sci. 2009, 50, 2572–2580. [Google Scholar] [CrossRef] [PubMed]
- Harbour, J.W.; Onken, M.D.; Roberson, E.D.O.; Duan, S.; Cao, L.; Worley, L.A.; Council, M.L.; Matatall, K.A.; Helms, C.; Bowcock, A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef]
- Nahon-Esteve, S.; Martel, A.; Maschi, C.; Caujolle, J.-P.; Baillif, S.; Lassalle, S.; Hofman, P. The Molecular Pathology of Eye Tumors: A 2019 Update Main Interests for Routine Clinical Practice. Curr. Mol. Med. 2019, 19, 632–664. [Google Scholar] [CrossRef]
- Rodrigues, M.; Rais, K.A.; Salviat, F.; Algret, N.; Simaga, F.; Barnhill, R.; Gardrat, S.; Servois, V.; Mariani, P.; Piperno-Neumann, S.; et al. Association of Partial Chromosome 3 Deletion in Uveal Melanomas with Metastasis-Free Survival. JAMA Ophthalmol. 2020. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Ehlers, J.P.; Harbour, J.W. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004, 64, 7205–7209. [Google Scholar] [CrossRef]
- Hwang, W.L.; Hwang, K.L.; Miyamoto, D.T. The promise of circulating tumor cells for precision cancer therapy. Biomark. Med. 2016, 10, 1269–1285. [Google Scholar] [CrossRef] [PubMed]
- Mader, S.; Pantel, K. Liquid Biopsy: Current Status and Future Perspectives. Oncol. Res. Treat. 2017, 40, 404–408. [Google Scholar] [CrossRef]
- Anand, K.; Roszik, J.; Gombos, D.; Upshaw, J.; Sarli, V.; Meas, S.; Lucci, A.; Hall, C.; Patel, S. Pilot Study of Circulating Tumor Cells in Early-Stage and Metastatic Uveal Melanoma. Cancers 2019, 11, 856. [Google Scholar] [CrossRef]
- Tura, A.; Merz, H.; Reinsberg, M.; Lüke, M.; Jager, M.J.; Grisanti, S.; Lüke, J. Analysis of monosomy-3 in immunomagnetically isolated circulating melanoma cells in uveal melanoma patients. Pigment Cell Melanoma Res. 2016, 29, 583–589. [Google Scholar] [CrossRef]
- Bande, M.F.; Santiago, M.; Muinelo-Romay, L.; Blanco, M.J.; Mera, P.; Capeans, C.; Pardo, M.; Piñeiro, A. Detection of circulating melanoma cells in choroidal melanocytic lesions. BMC Res. Notes 2015, 8, 452. [Google Scholar] [CrossRef]
- Terai, M.; Mu, Z.; Eschelman, D.J.; Gonsalves, C.F.; Kageyama, K.; Chervoneva, I.; Orloff, M.; Weight, R.; Mastrangelo, M.J.; Cristofanilli, M.; et al. Arterial Blood, Rather Than Venous Blood, is a Better Source for Circulating Melanoma Cells. EBioMedicine 2015, 2, 1821–1826. [Google Scholar] [CrossRef]
- Tura, A.; Lüke, J.; Merz, H.; Reinsberg, M.; Lüke, M.; Jager, M.J.; Grisanti, S. Identification of circulating melanoma cells in uveal melanoma patients by dual-marker immunoenrichment. Investig. Ophthalmol. Vis. Sci. 2014, 55, 4395–4404. [Google Scholar] [CrossRef]
- Mazzini, C.; Pinzani, P.; Salvianti, F.; Scatena, C.; Paglierani, M.; Ucci, F.; Pazzagli, M.; Massi, D. Circulating tumor cells detection and counting in uveal melanomas by a filtration-based method. Cancers 2014, 6, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.-C.; Madic, J.; Mariani, P.; Piperno-Neumann, S.; Rampanou, A.; Servois, V.; Cassoux, N.; Desjardins, L.; Milder, M.; Vaucher, I.; et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int. J. Cancer 2014, 134, 1207–1213. [Google Scholar] [CrossRef]
- Pinzani, P.; Mazzini, C.; Salvianti, F.; Massi, D.; Grifoni, R.; Paoletti, C.; Ucci, F.; Molinara, E.; Orlando, C.; Pazzagli, M.; et al. Tyrosinase mRNA levels in the blood of uveal melanoma patients: Correlation with the number of circulating tumor cells and tumor progression. Melanoma Res. 2010, 20, 303–310. [Google Scholar] [CrossRef]
- Suesskind, D.; Ulmer, A.; Schiebel, U.; Fierlbeck, G.; Spitzer, B.; Spitzer, M.S.; Bartz-Schmidt, K.U.; Grisanti, S. Circulating melanoma cells in peripheral blood of patients with uveal melanoma before and after different therapies and association with prognostic parameters: A pilot study. Acta Ophthalmol. 2011, 89, 17–24. [Google Scholar] [CrossRef]
- Eide, N.; Faye, R.S.; Høifødt, H.K.; Øvergaard, R.; Jebsen, P.; Kvalheim, G.; Fodstad, Ø. Immunomagnetic detection of micrometastatic cells in bone marrow in uveal melanoma patients. Acta Ophthalmol. 2009, 87, 830–836. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, A.; Beutel, J.; Süsskind, D.; Hilgers, R.-D.; Ziemssen, F.; Lüke, M.; Röcken, M.; Rohrbach, M.; Fierlbeck, G.; Bartz-Schmidt, K.-U.; et al. Visualization of circulating melanoma cells in peripheral blood of patients with primary uveal melanoma. Clin. Cancer Res. 2008, 14, 4469–4474. [Google Scholar] [CrossRef]
- Kang, J.S.; Lee, T.R.; Cha, W.C.; Shin, T.G.; Sim, M.S.; Jo, I.J.; Song, K.J.; Rhee, J.E.; Jeong, Y.K. Finger necrosis after accidental radial artery puncture. Clin. Exp. Emerg. Med. 2014, 1, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Shields, C.L.; Manalac, J.; Das, C.; Ferguson, K.; Shields, J.A. Choroidal melanoma: Clinical features, classification, and top 10 pseudomelanomas. Curr. Opin. Ophthalmol. 2014, 25, 177–185. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef]
- Cabel, L.; Riva, F.; Servois, V.; Livartowski, A.; Daniel, C.; Rampanou, A.; Lantz, O.; Romano, E.; Milder, M.; Buecher, B.; et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: A proof-of-concept study. Ann. Oncol. 2017, 28, 1996–2001. [Google Scholar] [CrossRef]
- Maheswaran, S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diederichs, S.; Iafrate, A.J.; Bell, D.W.; et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008, 359, 366–377. [Google Scholar] [CrossRef]
- Charitoudis, G.; Schuster, R.; Joussen, A.M.; Keilholz, U.; Bechrakis, N.E. Detection of tumour cells in the bloodstream of patients with uveal melanoma: Influence of surgical manipulation on the dissemination of tumour cells in the bloodstream. Br. J. Ophthalmol. 2016, 100, 468–472. [Google Scholar] [CrossRef]
- Metz, C.H.; Scheulen, M.; Bornfeld, N.; Lohmann, D.; Zeschnigk, M. Ultradeep sequencing detects GNAQ and GNA11 mutations in cell-free DNA from plasma of patients with uveal melanoma. Cancer Med. 2013, 2, 208–215. [Google Scholar] [CrossRef]
- Schuster, R.; Bechrakis, N.E.; Stroux, A.; Busse, A.; Schmittel, A.; Thiel, E.; Foerster, M.H.; Keilholz, U. Prognostic relevance of circulating tumor cells in metastatic uveal melanoma. Oncology 2011, 80, 57–62. [Google Scholar] [CrossRef]
- Schuster, R.; Bechrakis, N.E.; Stroux, A.; Busse, A.; Schmittel, A.; Scheibenbogen, C.; Thiel, E.; Foerster, M.H.; Keilholz, U. Circulating tumor cells as prognostic factor for distant metastases and survival in patients with primary uveal melanoma. Clin. Cancer Res. 2007, 13, 1171–1178. [Google Scholar] [CrossRef]
- Callejo, S.A.; Antecka, E.; Blanco, P.L.; Edelstein, C.; Burnier, M.N. Identification of circulating malignant cells and its correlation with prognostic factors and treatment in uveal melanoma. A prospective longitudinal study. Eye 2007, 21, 752–759. [Google Scholar] [CrossRef]
- Boldin, I.; Langmann, G.; Richtig, E.; Schwantzer, G.; Ardjomand, N.; Wegscheider, B.; El-Shabrawi, Y. Five-year results of prognostic value of tyrosinase in peripheral blood of uveal melanoma patients. Melanoma Res. 2005, 15, 503–507. [Google Scholar] [CrossRef]
- Keilholz, U.; Goldin-Lang, P.; Bechrakis, N.E.; Max, N.; Letsch, A.; Schmittel, A.; Scheibenbogen, C.; Heufelder, K.; Eggermont, A.; Thiel, E. Quantitative detection of circulating tumor cells in cutaneous and ocular melanoma and quality assessment by real-time reverse transcriptase-polymerase chain reaction. Clin. Cancer Res. 2004, 10, 1605–1612. [Google Scholar] [CrossRef]
- Junqueira-Neto, S.; Batista, I.A.; Costa, J.L.; Melo, S.A. Liquid Biopsy beyond Circulating Tumor Cells and Cell-Free DNA. Acta Cytol. 2019, 63, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, X.; Shen, J.; Jiang, Y. MicroRNA dysregulation in uveal melanoma: A new player enters the game. Oncotarget 2015, 6, 4562–4568. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Available online: https://pubmed.ncbi.nlm.nih.gov/18817506/?from_term=micro+rna+cancer+review&from_pos=4 (accessed on 9 May 2020).
- Xin, X.; Zhang, Y.; Ling, F.; Wang, L.; Sheng, X.; Qin, L.; Zhao, X. Identification of a nine-miRNA signature for the prognosis of Uveal Melanoma. Exp. Eye Res. 2019, 180, 242–249. [Google Scholar] [CrossRef]
- Stark, M.S.; Gray, E.S.; Isaacs, T.; Chen, F.K.; Millward, M.; McEvoy, A.; Zaenker, P.; Ziman, M.; Soyer, H.P.; Glasson, W.J.; et al. A Panel of Circulating MicroRNAs Detects Uveal Melanoma With High Precision. Transl. Vis. Sci. Technol. 2019, 8, 12. [Google Scholar] [CrossRef]
- Ragusa, M.; Barbagallo, C.; Statello, L.; Caltabiano, R.; Russo, A.; Puzzo, L.; Avitabile, T.; Longo, A.; Toro, M.D.; Barbagallo, D.; et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: Pathological and diagnostic implications. Cancer Biol. Ther. 2015, 16, 1387–1396. [Google Scholar] [CrossRef]
- Achberger, S.; Aldrich, W.; Tubbs, R.; Crabb, J.W.; Singh, A.D.; Triozzi, P.L. Circulating immune cell and microRNA in patients with uveal melanoma developing metastatic disease. Mol. Immunol. 2014, 58, 182–186. [Google Scholar] [CrossRef]
- Joshi, P.; Kooshki, M.; Aldrich, W.; Varghai, D.; Zborowski, M.; Singh, A.D.; Triozzi, P.L. Expression of natural killer cell regulatory microRNA by uveal melanoma cancer stem cells. Clin. Exp. Metastasis 2016, 33, 829–838. [Google Scholar] [CrossRef]
- Russo, A.; Caltabiano, R.; Longo, A.; Avitabile, T.; Franco, L.M.; Bonfiglio, V.; Puzzo, L.; Reibaldi, M. Increased Levels of miRNA-146a in Serum and Histologic Samples of Patients with Uveal Melanoma. Front. Pharmacol. 2016, 7, 424. [Google Scholar] [CrossRef]
- Gómez-Pérez, A.M.; Cornejo Pareja, I.M.; García Alemán, J.; Coín Aragüez, L.; Sebastián Ochoa, A.; Alcaide Torres, J.; Molina Vega, M.; Clu Fernández, C.; Mancha Doblas, I.; Tinahones, F.J. New molecular biomarkers in differentiated thyroid carcinoma: Impact of miR-146, miR-221 and miR-222 levels in the evolution of the disease. Clin. Endocrinol. 2019, 91, 187–194. [Google Scholar] [CrossRef]
- Aksenenko, M.; Palkina, N.; Komina, A.; Tashireva, L.; Ruksha, T. Differences in microRNA expression between melanoma and healthy adjacent skin. BMC Dermatol. 2019, 19, 1. [Google Scholar] [CrossRef]
- Triozzi, P.L.; Achberger, S.; Aldrich, W.; Singh, A.D.; Grane, R.; Borden, E.C. The association of blood angioregulatory microRNA levels with circulating endothelial cells and angiogenic proteins in patients receiving dacarbazine and interferon. J. Transl. Med. 2012, 10, 241. [Google Scholar] [CrossRef]
- Worley, L.A.; Long, M.D.; Onken, M.D.; Harbour, J.W. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008, 18, 184–190. [Google Scholar] [CrossRef]
- Yan, D.; Zhou, X.; Chen, X.; Hu, D.-N.; Dong, X.D.; Wang, J.; Lu, F.; Tu, L.; Qu, J. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Investig. Ophthalmol. Vis. Sci. 2009, 50, 1559–1565. [Google Scholar] [CrossRef]
- Dong, F.; Lou, D. MicroRNA-34b/c suppresses uveal melanoma cell proliferation and migration through multiple targets. Mol. Vis. 2012, 18, 537–546. [Google Scholar]
- Eedunuri, V.K.; Rajapakshe, K.; Fiskus, W.; Geng, C.; Chew, S.A.; Foley, C.; Shah, S.S.; Shou, J.; Mohamed, J.S.; Coarfa, C.; et al. miR-137 Targets p160 Steroid Receptor Coactivators SRC1, SRC2, and SRC3 and Inhibits Cell Proliferation. Mol. Endocrinol. 2015, 29, 1170–1183. [Google Scholar] [CrossRef]
- Chen, X.; Wang, J.; Shen, H.; Lu, J.; Li, C.; Hu, D.-N.; Dong, X.D.; Yan, D.; Tu, L. Epigenetics, microRNAs, and carcinogenesis: Functional role of microRNA-137 in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2011, 52, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Cheli, Y.; Giuliano, S.; Guiliano, S.; Botton, T.; Rocchi, S.; Hofman, V.; Hofman, P.; Bahadoran, P.; Bertolotto, C.; Ballotti, R. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene 2011, 30, 2307–2318. [Google Scholar] [CrossRef]
- Wu, S.; Chen, H.; Han, N.; Zhang, C.; Yan, H. Long Noncoding RNA PVT1 Silencing Prevents the Development of Uveal Melanoma by Impairing MicroRNA-17-3p-Dependent MDM2 Upregulation. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4904–4914. [Google Scholar] [CrossRef]
- Xu, H.; Gong, J.; Liu, H. High expression of lncRNA PVT1 independently predicts poor overall survival in patients with primary uveal melanoma. PLoS ONE 2017, 12, e0189675. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; He, J.; Yang, Z.; Ge, S.; Zhang, H.; Zhong, Q.; Fan, X. ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy 2019, 1–14. [Google Scholar] [CrossRef]
- Zheng, X.; Tang, H.; Zhao, X.; Sun, Y.; Jiang, Y.; Liu, Y. Long non-coding RNA FTH1P3 facilitates uveal melanoma cell growth and invasion through miR-224-5p. PLoS ONE 2017, 12, e0184746. [Google Scholar] [CrossRef]
- Sun, L.; Sun, P.; Zhou, Q.-Y.; Gao, X.; Han, Q. Long noncoding RNA MALAT1 promotes uveal melanoma cell growth and invasion by silencing of miR-140. Am. J. Transl. Res. 2016, 8, 3939–3946. [Google Scholar]
- Lu, Q.; Zhao, N.; Zha, G.; Wang, H.; Tong, Q.; Xin, S. LncRNA HOXA11-AS Exerts Oncogenic Functions by Repressing p21 and miR-124 in Uveal Melanoma. DNA Cell Biol. 2017, 36, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Ma, R.; Ren, H.; Qian, J. Genome-Wide Analysis of Uveal Melanoma Metastasis-Associated LncRNAs and Their Functional Network. DNA Cell Biol. 2018, 37, 99–108. [Google Scholar] [CrossRef]
- Weidle, U.H.; Birzele, F.; Kollmorgen, G.; Rüger, R. The Multiple Roles of Exosomes in Metastasis. Cancer Genom. Proteom. 2017, 14, 1–15. [Google Scholar] [CrossRef]
- Eldh, M.; Olofsson Bagge, R.; Lässer, C.; Svanvik, J.; Sjöstrand, M.; Mattsson, J.; Lindnér, P.; Choi, D.-S.; Gho, Y.S.; Lötvall, J. MicroRNA in exosomes isolated directly from the liver circulation in patients with metastatic uveal melanoma. BMC Cancer 2014, 14, 962. [Google Scholar] [CrossRef]
- Giovannucci, E.; Egan, K.M.; Hunter, D.J.; Stampfer, M.J.; Colditz, G.A.; Willett, W.C.; Speizer, F.E. Aspirin and the risk of colorectal cancer in women. N. Engl. J. Med. 1995, 333, 609–614. [Google Scholar] [CrossRef]
- Simon, T.G.; Duberg, A.-S.; Aleman, S.; Chung, R.T.; Chan, A.T.; Ludvigsson, J.F. Association of Aspirin with Hepatocellular Carcinoma and Liver-Related Mortality. N. Engl. J. Med. 2020, 382, 1018–1028. [Google Scholar] [CrossRef]
- In’t Veld, S.G.; Wurdinger, T. Tumor-educated platelets. Blood 2019, 133, 2359–2364. [Google Scholar] [CrossRef]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef]
- Goddard, E.T.; Bozic, I.; Riddell, S.R.; Ghajar, C.M. Dormant tumour cells, their niches and the influence of immunity. Nat. Cell Biol. 2018, 20, 1240–1249. [Google Scholar] [CrossRef]
- Blanco, P.L.; Lim, L.A.; Miyamoto, C.; Burnier, M.N. Uveal melanoma dormancy: An acceptable clinical endpoint? Melanoma Res. 2012, 22, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Sollier, E.; Go, D.E.; Che, J.; Gossett, D.R.; O’Byrne, S.; Weaver, W.M.; Kummer, N.; Rettig, M.; Goldman, J.; Nickols, N.; et al. Size-selective collection of circulating tumor cells using Vortex technology. Lab Chip 2014, 14, 63–77. [Google Scholar] [CrossRef]
- Eide, N.; Faye, R.S.; Høifødt, H.K.; Sandvik, L.; Qvale, G.A.; Faber, R.; Jebsen, P.; Kvalheim, G.; Fodstad, Ø. The Results of Stricter Inclusion Criteria in an Immunomagnetic Detection Study of Micrometastatic Cells in Bone Marrow of Uveal Melanoma Patients-Relevance for Dormancy. Pathol. Oncol. Res. 2019, 25, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.N.; Bhowmick, N.A. Role of EMT in Metastasis and Therapy Resistance. J. Clin. Med. 2016, 5, 17. [Google Scholar] [CrossRef]
- Kahlert, U.D.; Joseph, J.V.; Kruyt, F.A.E. EMT- and MET-related processes in nonepithelial tumors: Importance for disease progression, prognosis, and therapeutic opportunities. Mol. Oncol. 2017, 11, 860–877. [Google Scholar] [CrossRef]
- Marshall, J.-C.; Nantel, A.; Blanco, P.; Ash, J.; Cruess, S.R.; Burnier, M.N. Transcriptional profiling of human uveal melanoma from cell lines to intraocular tumors to metastasis. Clin. Exp. Metastasis 2007, 24, 353–362. [Google Scholar] [CrossRef]
- Sun, J.; Xi, H.-Y.; Shao, Q.; Liu, Q.-H. Biomarkers in retinoblastoma. Int. J. Ophthalmol. 2020, 13, 325–341. [Google Scholar] [CrossRef]
- Munier, F.L.; Gaillard, M.-C.; Balmer, A.; Soliman, S.; Podilsky, G.; Moulin, A.P.; Beck-Popovic, M. Intravitreal chemotherapy for vitreous disease in retinoblastoma revisited: From prohibition to conditional indications. Br. J. Ophthalmol. 2012, 96, 1078–1083. [Google Scholar] [CrossRef]
- Soliman, S.E.; Racher, H.; Zhang, C.; MacDonald, H.; Gallie, B.L. Genetics and Molecular Diagnostics in Retinoblastoma–An Update. Asia Pac. J. Ophthalmol. 2017, 6, 197–207. [Google Scholar] [CrossRef]
- Soliman, S.E.; Wan, M.J.; Heon, E.; Hazrati, L.-N.; Gallie, B. Retinoblastoma versus advanced Coats’ disease: Is enucleation the answer? Ophthalmic Genet. 2017, 38, 291–293. [Google Scholar] [CrossRef]
- Berry, J.L.; Xu, L.; Kooi, I.; Murphree, A.L.; Prabakar, R.K.; Reid, M.; Stachelek, K.; Le, B.H.A.; Welter, L.; Reiser, B.J.; et al. Genomic cfDNA Analysis of Aqueous Humor in Retinoblastoma Predicts Eye Salvage: The Surrogate Tumor Biopsy for Retinoblastoma. Mol. Cancer Res. 2018, 16, 1701–1712. [Google Scholar] [CrossRef]
- Xu, L.; Polski, A.; Prabakar, R.K.; Reid, M.W.; Chevez-Barrios, P.; Jubran, R.; Kim, J.W.; Kuhn, P.; Cobrinik, D.; Hicks, J.; et al. Chromosome 6p Amplification in Aqueous Humor Cell-Free DNA Is a Prognostic Biomarker for Retinoblastoma Ocular Survival. Mol. Cancer Res. 2020, 18, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- Gerrish, A.; Stone, E.; Clokie, S.; Ainsworth, J.R.; Jenkinson, H.; McCalla, M.; Hitchcott, C.; Colmenero, I.; Allen, S.; Parulekar, M.; et al. Non-invasive diagnosis of retinoblastoma using cell-free DNA from aqueous humour. Br. J. Ophthalmol. 2019. [Google Scholar] [CrossRef]
- Kothari, P.; Marass, F.; Yang, J.L.; Stewart, C.M.; Stephens, D.; Patel, J.; Hasan, M.; Jing, X.; Meng, F.; Enriquez, J.; et al. Cell-free DNA profiling in retinoblastoma patients with advanced intraocular disease: An MSKCC experience. Cancer Med. 2020. [Google Scholar] [CrossRef]
- Ghiam, B.K.; Xu, L.; Berry, J.L. Aqueous Humor Markers in Retinoblastoma, a Review. Transl. Vis. Sci. Technol. 2019, 8, 13. [Google Scholar] [CrossRef]
- Beta, M.; Venkatesan, N.; Vasudevan, M.; Vetrivel, U.; Khetan, V.; Krishnakumar, S. Identification and Insilico Analysis of Retinoblastoma Serum microRNA Profile and Gene Targets towards Prediction of Novel Serum Biomarkers. Bioinform. Biol. Insights 2013, 7, 21–34. [Google Scholar] [CrossRef]
- Scholz, S.L.; Cosgarea, I.; Süßkind, D.; Murali, R.; Möller, I.; Reis, H.; Leonardelli, S.; Schilling, B.; Schimming, T.; Hadaschik, E.; et al. NF1 mutations in conjunctival melanoma. Br. J. Cancer 2018, 118, 1243–1247. [Google Scholar] [CrossRef]
- Kaštelan, S.; Gverović Antunica, A.; Beketić Orešković, L.; Salopek Rabatić, J.; Kasun, B.; Bakija, I. Conjunctival Melanoma-Epidemiological Trends and Features. Pathol. Oncol. Res. 2018, 24, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Maiorano, B.A.; Pagliara, M.M.; Sammarco, M.G.; Dosa, T.; Martini, M.; Rindi, G.; Bria, E.; Blasi, M.A.; Tortora, G.; et al. Dabrafenib and Trametinib in BRAF Mutant Metastatic Conjunctival Melanoma. Front. Oncol. 2019, 9, 232. [Google Scholar] [CrossRef]
- Sagiv, O.; Thakar, S.D.; Kandl, T.J.; Ford, J.; Sniegowski, M.C.; Hwu, W.-J.; Esmaeli, B. Immunotherapy With Programmed Cell Death 1 Inhibitors for 5 Patients With Conjunctival Melanoma. JAMA Ophthalmol. 2018, 136, 1236–1241. [Google Scholar] [CrossRef]
- Finger, P.T.; Pavlick, A.C. Checkpoint inhibition immunotherapy for advanced local and systemic conjunctival melanoma: A clinical case series. J. Immunother. Cancer 2019, 7, 83. [Google Scholar] [CrossRef]
- Huang, S.K.; Hoon, D.S.B. Liquid biopsy utility for the surveillance of cutaneous malignant melanoma patients. Mol. Oncol. 2016, 10, 450–463. [Google Scholar] [CrossRef]
- Kenawy, N.; Garrick, A.; Heimann, H.; Coupland, S.E.; Damato, B.E. Conjunctival squamous cell neoplasia: The Liverpool Ocular Oncology Centre experience. Graefes Arch. Clin. Exp. Ophthalmol. 2015, 253, 143–150. [Google Scholar] [CrossRef]
- Gichuhi, S.; Ohnuma, S.; Sagoo, M.S.; Burton, M.J. Pathophysiology of ocular surface squamous neoplasia. Exp. Eye Res. 2014, 129, 172–182. [Google Scholar] [CrossRef]
- Santoni, A.; Thariat, J.; Maschi, C.; Herault, J.; Baillif, S.; Lassalle, S.; Peyrichon, M.L.; Salleron, J.; Caujolle, J.-P. Management of Invasive Squamous Cell Carcinomas of the Conjunctiva. Am. J. Ophthalmol. 2019, 200, 1–9. [Google Scholar] [CrossRef]
- Mathis, T.; Jardel, P.; Loria, O.; Delaunay, B.; Nguyen, A.-M.; Lanza, F.; Mosci, C.; Caujolle, J.-P.; Kodjikian, L.; Thariat, J. New concepts in the diagnosis and management of choroidal metastases. Prog. Retin. Eye Res. 2019, 68, 144–176. [Google Scholar] [CrossRef]
- Konstantinidis, L.; Rospond-Kubiak, I.; Zeolite, I.; Heimann, H.; Groenewald, C.; Coupland, S.E.; Damato, B. Management of patients with uveal metastases at the Liverpool Ocular Oncology Centre. Br. J. Ophthalmol. 2014, 98, 92–98. [Google Scholar] [CrossRef]
- Shields, C.L.; Shields, J.A.; Gross, N.E.; Schwartz, G.P.; Lally, S.E. Survey of 520 eyes with uveal metastases. Ophthalmology 1997, 104, 1265–1276. [Google Scholar] [CrossRef]
- Bouhlel, L.; Hofman, V.; Maschi, C.; Ilié, M.; Allégra, M.; Marquette, C.-H.; Audigier-Valette, C.; Thariat, J.; Hofman, P. The liquid biopsy: A tool for a combined diagnostic and theranostic approach for care of a patient with late-stage lung carcinoma presenting with bilateral ocular metastases. Expert Rev. Anticancer Ther. 2017, 17, 1087–1092. [Google Scholar] [CrossRef]
- Daxecker, F.; Zirm, M. Diagnostic value of determining carcino-embryonic antigens in the aqueous humor (author’s transl). Klin. Mon. Augenheilkd. 1980, 177, 768–771. [Google Scholar] [CrossRef]
| Antibody Marker | Molecular Marker |
|---|---|
| Melan-A | Tyrosinase |
| HMW-MAA | GNAQ, GNA11 |
| GP 100 | BAP 1 |
| Authors | Study Population | Number of Patients | CTC Isolation Method and Device | CTC Identification | Mean CTC (Range) | Main Findings | Follow-Up: Months (Range) |
|---|---|---|---|---|---|---|---|
| Anand et al. [29] | Primary and metastatic UM | 39 patients 20 primary UM 19 metastatic UM | Immunomagnetism Cellsearch | Cellsearch protocol: DAPI+ HMW-MAA+ CD146+ CD45- CD34- | 5.9 (1–38) | At initial sampling: CTC detected in 14 out of 39 (36%) patients. CTC detected in 6/20 (30%) primary UM and 8/19 (42%) metastatic UM During the follow-up period: CTC detected in 21/39 (54%) of patients CTC were more likely detected in Class 2 UM (83%) | 16.4 |
| Tura et al. [30] | Primary UM | 44 UM patients | Immuno-FISH isolation | NKIC3 and MCSP antibodies | Median: 2.4 (0–10.2) Median CTC in Monosomy 3 patients: 3.4 (0.7–10.2) Median CTC without Monosomy 3: 1.2 (0.3–8.4) | CTC detected in 40/444 (91%) patients Monosomy 3 detected in 23/40 (58%) patients Monosomy 3 on CTC associated with a higher TNM stage (T3) | 48 |
| Bande et al. [31] | Primary UM Uveal naevi | 12 patients 8 primary UM 4 uveal naevi | Immunomagnetism CellSearch | Cellsearch protocol: DAPI+ HMW-MAA+ CD146+ CD45- CD34- | UM: 1 (0–3) | CTC detected in 50% of UM patients and 0% in uveal naevi No relationship between CTC detection and the UM clinical-pathological features | 25 (16–27) |
| Terai et al. [32] | Metastatic UM | 17 patients 10 hepatic metastases 7 extra hepatic metastases | Immunomagnetism CellSearch | Cellsearch protocol: DAPI+ HMW-MAA+ CD146+ CD45- CD34- | Arterial: median: 5 (1–168) Venous: median: 1 (0–5) | No morphological difference between CTC collected through the arterial and venous route Arterial blood: CTC detection in 100% of cases Venous blood: CTC detection in 52.9% of cases No correlation between CTC number and number and size of metastases | None |
| Tura et al. [33] | Primary UM | 31 patients | Immunomagnetism Immunobeads | 2 antibodies: NKI/C3, NKI/beteb | Median: 3.5 (0–10.2) | CTC detected in 29/31 (93.6%) of patients No correlation between the CTC count and clinical parameters | None |
| Mazzini et al. [34] | Primary UM Metastatic UM Uveal nevi | 31 UM 10 uveal nevi | Isolation by size ISET | Antibodies anti S100, anti MART-1 and anti-tyrosinase | Median 8 (2–50) | CTC detected in 17/31 (55%) of UM patients. No CTC detected in uveal nevi patients No correlation between clinical and biological parameters and CTC positivity Detection of >10 CTC associated with a larger basal diameter, tumor height, disease free survival, and OS | 24–60 |
| Bidard et al. [35] | Metastatic UM | 40 patients | For CTC detection: Immunomagentism Cellsearch For Ct-DNA detection: BiPAP technique with 3 mutations screening: GNAQ c.626A > T, GNAQ c.626A > C and GNA11 c.626A > T | Cellsearch protocol: DAPI+ HMW-MAA+ CD146+ CD45- CD34- | 0 CTC: 70% ≥ 1 CTC: 30% 1 CTC: 10% 3 CTC: 15% 12 CTC: 2.5% 20 CTC: 2.5% DNA quantity: Median: 4.1 ng/mL (0.5–512) | Liver miliary associated with higher ct-DNA levels and CTC counts Correlation between CTC, ct-DNA, and tumor volume assessed by liver MRI Univariate analysis: CTC and ct-DNA positivity associated with PFS and OS Multivariate analysis: Only ct-DNA was associated with PFS and OS | 8 (median) |
| Pinzani et al. [36] | Primary UM Healthy Controls | 41 primary UM 16 controls | mRNA detected by RT-PCR (41 patients) CTC: Isolation by size using ISET device (16 patients) Blood samples repeated every 6 months | CTC morphology: cell size > 16-micron, nucleocytoplasmic ratio > 50%, irregular nuclear shape, hyperchromatic nucleus, and basophilic cytoplasm | PCR: median: 0.8 cell equivalent /mL of blood (0.1–14.4) ISET: 5.8, 2.33, 2.00, 1.25, and 0.75 CTC/ml | RT-PCR positivity in 20/41 (49%) of patients among at least one of the blood samples PCR positivity associated with decreased PFS and OS CTC detected in 5/16 (31%) patients Tyrosinase level correlated with CTC detection | 55 |
| Suesskind et al. [37] | Primary UM | 81 primary UM 94 samples before /after treatment | Immunomagnetism MACS | MCSP antibody | Preoperative median CTC count: 1 (1–8) Post-treatment: median CTC count: 7.5 (1–26) | CTC count before and after treatment (enucleation =7, radiotherapy stereotaxic =49, endoresection =19, brachytherapy =15, thermotherapy = 4) Before treatment: CTC detected in 13/94 (14%) of patients After treatment: CTC detected in 9/94 (10%) of patients No significant difference in terms of the CTC count before and after treatment No relationship between the CTC positivity and patient characteristics and metastatic status | 16 (median) |
| Eide et al. [38] | Primary UM | 328 patients | Immunomagnetism | Several anti-melanoma antibodies (9.2.27 antimelanoma-associated antibody, IgG1 Ep-1 antibody, 376.96 antibody) | Median cells number: 50 (1–500) | CTC detected in 4/328 (1,6%) patients Tumor cells detected in 98/328 (29.9%) patients in bone marrow No relationship between bone marrow tumor detection and further metastatic spread | 60 |
| Ulmer et al. [39] | Primary UM Healthy controls | 52 primary UM before treatment 20 healthy controls | Immunomagnetism MACS | MCSP antibody | Median: 2.5 (1–5) for 50 ml | CTC detected in 10/52 (19%) of patients No CTC detected in controls CTC positivity associated with ciliary body invasion, advanced local tumor stage, and anterior tumor localization Multivariate analysis: Only ciliary body involvement associated with CTC positivity | None |
| Authors | Study Population | Number of Patients | Ct-DNA/ct-RNA Detection | Main Findings | Follow-Up: Months (Range) |
|---|---|---|---|---|---|
| Charitoudis et al. [46] | Primary UM undergoing surgery | 202 patients | RT-PCR screening tyrosinase and MELAN-A/MART-1 | RT-PCR tyrosinase positive in 2/184 (1.1%) patients before and 4/180 (2.2%) patients after surgery RT-PCR MELAN-A/MART-1 positive in 20/184 (10.9%) before and in 25/180 (13.9%) patients after surgery RT-PCR results on MELAN-A/MART-1 and Tyrosinase levels were not affected by surgical manipulation | 24 |
| Metz et al. [47] | Primary and metastatic UM | 28 patients | PCR screening GNAQ Q209 (298 bp), GNAQ R183 (212 bp), GNA11 Q209 (150 bp), and GNA11 R183 (249 bp) | Oncogenic GNAQ/GNA11 mutations identified in ct-DNA of 9 out of 22 (41%) metastatic patients. Ct-DNA correlated with the metastatic status ct-DNA detected in younger patients with larger metastases | None |
| Schuster et al. [48] | Metastatic UM | 68 patients | RT-PCR screening tyrosinase and MELAN-A/MART 1 | RT-PCR positive in 43/68 (63%) patients 31 patients positive for tyrosinase 40 patients positive for MELAN-A /MART 1 28 patients positive for both RT-PCR positivity associated with poorer PFS and OS | 10 (median) |
| Schuster et al. [49] | Primary UM | 110 patients | RT-PCR screening tyrosinase, MELAN-A/ MART1 | RT-PCR positive in 11/110 (10%) patients (5 tyrosinase, 5 MALAN-A/MART1, 1 both) No correlation between RT-PCR positivity and clinical features Univariate analysis: The relationship between RT-PCR positivity and time to progression and OS RT-PCR positivity indicated an increased risk of metastasis and disease-specific mortality | 22 (median) |
| Callejo et al. [50] | Primary UM | 30 patients | RT-PCR screening tyrosinase, Melan-A | RT-PCR positive in 29/30 (97%) patients (119 visits, 1360 samples, 2720 PCR performed) No correlation between RT-PCR positivity, tumor size and treatment | NR |
| Boldin et al. [51] | Primary UM | 41 patients | RT-PCR screening tyrosinase | RT-PCR positive in 16/41 (39%) patients at baseline 11/16 (69%) patients initially positive were negative after treatment RT-PCR positivity associated with decreased 5-year OS RT-PCR positivity not correlated with tumor size and histology | 60–66 |
| Keilholz et al. [52] | Primary and metastatic UM | 61 patients 21 primary UM 40 metastatic UM | RT-PCR screening tyrosinase, MELAN-A/MART-1 and GP100 | Primary UM: tyrosinase detected in 3 (12.5%) patients, MELAN/MART detected in 1 (4%) patient and GP100 detected in 1 (4%) patient. Metastatic UM: Tyrosinase detected in 24 (60%) patients, Melan/MART 31 (77%) patients and GP100 in 4/26 (15%) patients GP100 positive in 4/40 (10%) samples. Accuracy detection rates: Tyrosinase > Melan > GP100 | 6 |
| LB Feature | Advantages | Disadvantages |
|---|---|---|
| CTC | ● Allows a better understanding of the metastatic process by screening genetical mutations and surface biomarkers ● Allows laboratory cell culture and further in vivo investigations | ● Lack of consensus concerning pre- and post-analytic processes ● May be less reliable than ct-DNA, according to Bidard et al. |
| Ct-DNA | ● More reliable and standardized techniques compared to CTC ● More stable than ct-RNA | ● Less instructive than CTC in understanding the underlying tumorigenesis ● GNAQ and GNA11 mutations are not found in all UM |
| Ct-RNA | ● Detection by reliable techniques (RT-PCR) | ● Instability (degradation by RNAase) Low abundance ● Half-life very low |
| miRNA | ● Longer half-life, especially when encapsulated ● More stable compared to ct-DNA and ct-RNA ● Detected by reliable techniques (RT-PCR) | ● Lack of consensus regarding pre- and post-analytic processes ● Conflicting results regarding the role of certain mi-RNAs |
| TRE | ● Stable ● Long half-life ● Possibility to investigate mi-RNA, DNA, RNA, as well as surface markers | ● Lack of consensus regarding exosome definition (different definitions based on the size to distinguish exosomes from other small extracellular vesicles) ● Lack of available studies ● Lack of process standardization |
| TEP | ● Promising preliminary results in other solid malignancies ● TEPs are easily obtained and processed ● Available in large amounts | ● Lack of studies into UM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martel, A.; Baillif, S.; Nahon-esteve, S.; Gastaud, L.; Bertolotto, C.; Roméo, B.; Mograbi, B.; Lassalle, S.; Hofman, P. Liquid Biopsy for Solid Ophthalmic Malignancies: An Updated Review and Perspectives. Cancers 2020, 12, 3284. https://doi.org/10.3390/cancers12113284
Martel A, Baillif S, Nahon-esteve S, Gastaud L, Bertolotto C, Roméo B, Mograbi B, Lassalle S, Hofman P. Liquid Biopsy for Solid Ophthalmic Malignancies: An Updated Review and Perspectives. Cancers. 2020; 12(11):3284. https://doi.org/10.3390/cancers12113284
Chicago/Turabian StyleMartel, Arnaud, Stephanie Baillif, Sacha Nahon-esteve, Lauris Gastaud, Corine Bertolotto, Barnabé Roméo, Baharia Mograbi, Sandra Lassalle, and Paul Hofman. 2020. "Liquid Biopsy for Solid Ophthalmic Malignancies: An Updated Review and Perspectives" Cancers 12, no. 11: 3284. https://doi.org/10.3390/cancers12113284
APA StyleMartel, A., Baillif, S., Nahon-esteve, S., Gastaud, L., Bertolotto, C., Roméo, B., Mograbi, B., Lassalle, S., & Hofman, P. (2020). Liquid Biopsy for Solid Ophthalmic Malignancies: An Updated Review and Perspectives. Cancers, 12(11), 3284. https://doi.org/10.3390/cancers12113284