Non-Coding RNAs: Uncharted Mediators of Thyroid Cancer Pathogenesis
Simple Summary
Abstract
1. Introduction
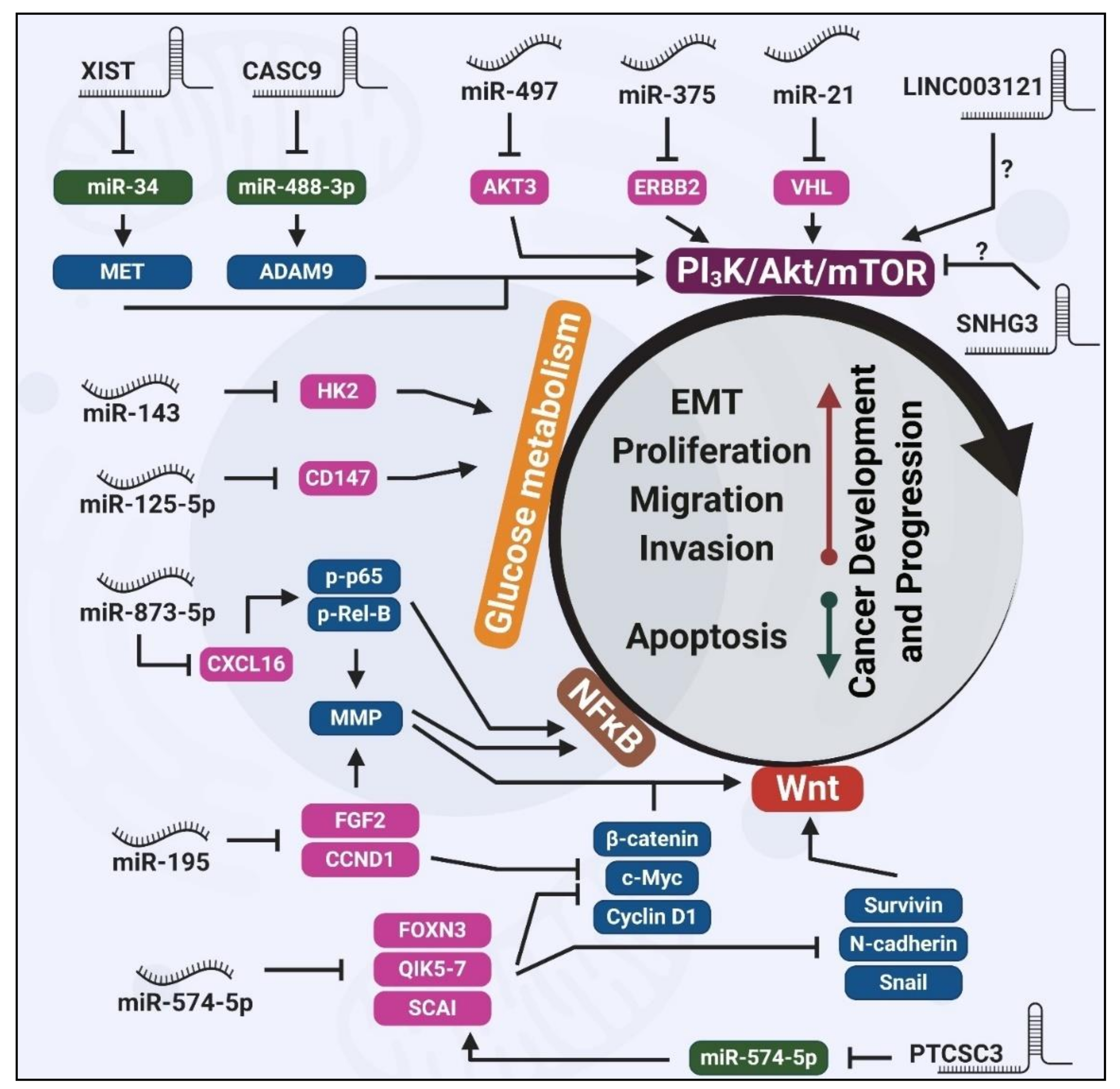
2. miRNAs Regulating Thyroid Carcinogenesis
2.1. Wnt-Mediated Tumorigenic Effects of Dysregulated miRNAs
2.2. PI3K/Akt-Mediated Tumorigenic Effects of Dysregulated miRNAs
2.3. Glucose Metabolism-Mediated Tumorigenic Effects of Dysregulated miRNAs
2.4. Dysregulated miRNAs in Other Signaling Pathways
3. lncRNAs Regulating Thyroid Carcinogenesis
3.1. Wnt-Mediated Tumorigenic Effects of Dysregulated lncRNAs
3.2. PI3K/Akt-Mediated Tumorigenic Effects of Dysregulated lncRNAs
4. circRNAs Regulating Thyroid Carcinogenesis
5. ncRNAs May Regulate the Biology of Thyroid Tumor Microenvironment
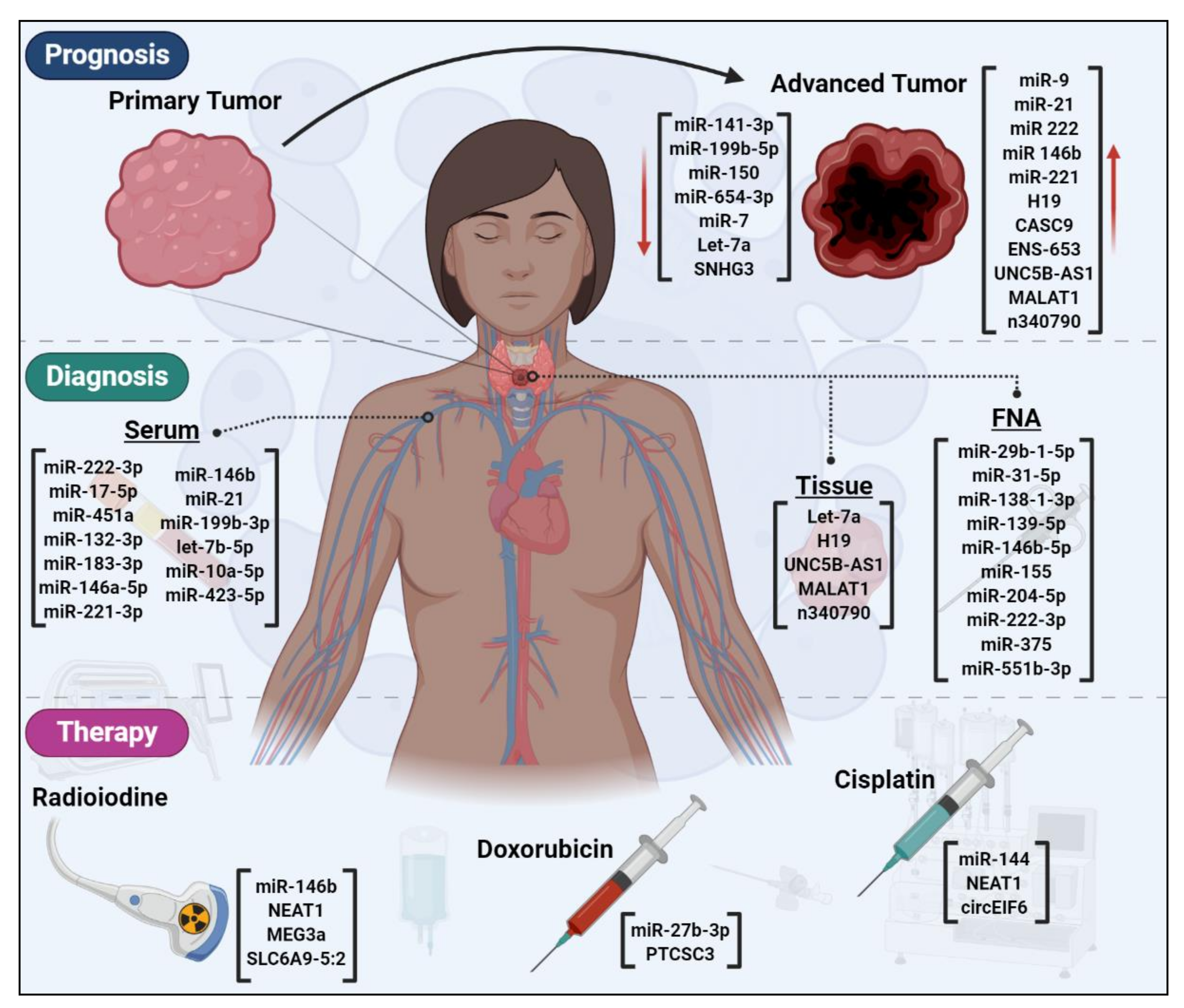
6. ncRNAs are Novel Candidates for Early Detection of Thyroid Cancer
7. ncRNAs as Prognostic Factors for Thyroid Cancer
8. ncRNAs Could Affect Thyroid Cancer Therapy
9. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAM9 | ADAM Metallopeptidase Domain 9 |
| AKT3 | RAC-γ serine/threonine-protein kinase |
| AMPK | 5′ AMP-activated Protein Kinase |
| ATC | Anaplastic Thyroid Cancer |
| BDNF | Brain-Derived Neurotrophic Factor |
| CASC9 | Cancer Susceptibility 9 |
| CCND1 | Cyclin D1 |
| CDH6 | Cadherin 6 |
| circEIF6 | Circular Eukaryotic Translation Initiation Factor 6 |
| circFOXM1 | Circular Forkhead Box Protein M1 |
| circRNA | Circular RNA |
| CXCL16 | C-X-C Motif Chemokine Ligand 16 |
| DLX6-AS1 | Distal-Less Homeobox 6-Antisense 1 |
| EGFR | Epidermal Growth Factor Receptor |
| ERBB2 | Erb-B2 Receptor Tyrosine Kinase 2 |
| ER-β | Estrogen Receptor Beta |
| EZH2 | Enhancer of Zeste Homolog 2 |
| FGF2 | Fibroblast Growth Factor |
| FGFR2 | Fibroblast Growth Factor Receptor 2 |
| FNA | Fine Needle Aspiration |
| FOXE1 | Forkhead Box E1 |
| FOXN3 | Forkhead Box N3 |
| FOXO1 | Forkhead Box O1 |
| FTC | Follicular Thyroid Carcinoma |
| HCP5 | HLA complex P5 |
| HK2 | Hexokinase 2 |
| HMGB1 | High Mobility Group Box Protein 1 |
| ITGA3 | Integrin Subunit Alpha 3 |
| lncRNA | Long Non-coding RNA |
| MALAT1 | Metastasis Associated Lung Adenocarcinoma Transcript 1 |
| MEG3a | Maternally Expressed Gene 3 |
| miRNA | microRNA |
| MMP1 | Matrix Metallopeptidase 1 |
| MTC | Medullary Thyroid Carcinoma |
| mTOR | Mechanistic Target of Rapamycin Kinase |
| ncRNA | Non-coding RNA |
| NEAT1 | Nuclear Paraspeckle Assembly Transcript 1 |
| PAK1 | p21 Activated Kinase-1 |
| PAR5 | Prader Willi/Angelman Region RNA5 |
| PDTC | Poorly differentiated thyroid carcinoma |
| PI3K | Phosphatidylinositol-4,5-Bisphosphate 3-Kinase |
| piRNA | PIWI-interacting RNA: |
| PTC | Papillary Thyroid Cancer |
| PTCSC3 | Thyroid Carcinoma Susceptibility Candidate 3 |
| PTCSCs | Papillary Thyroid Cancer Stem Cells |
| QKI5-7 | Quaking protein 5-7 |
| RAR-β | Retinoic Acid Receptor Beta |
| ROCK1 | Rho-associated Protein Kinase 1 |
| SCAI | Suppressor of Cancer Cell Invasion |
| SEER | Surveillance, Epidemiology and End Results |
| SLC6A9-5:2 | Solute Carrier Family 6 Member 9-5:2 |
| SNHG3 | Small Nucleolar RNA Host Gene 3 |
| snoRNA | Small nuclear RNA |
| SOCS1 | Suppressor Protein of Cytokine Signaling 1 |
| SPAG9 | Perm Associated Antigen 9 |
| ST6GAL2 | alpha-2, 6-sialyltransferase 2 |
| STAT3 | Signal Transducer And Activator of Transcription 3 |
| STON2 | Stonin 2 |
| TGF-α | Transforming Growth Factor Alpha |
| UNC5B-AS1 | Unc-5 Netrin Receptor B-Antisense RNA 1 |
| UPF1 | UPF1 RNA Helicase and ATPase |
| VHL | Von Hippel-Lindau Tumor Suppressor |
| XIST | X-inactive specific transcript |
| YY1 | Yin Yang 1 |
References
- Katoh, H.; Yamashita, K.; Enomoto, T.; Watanabe, M. Classification and general considerations of thyroid cancer. Ann. Clin. Pathol. 2015, 3, 1045. [Google Scholar]
- James, B.C.; Mitchell, J.M.; Jeon, H.D.; Vasilottos, N.; Grogan, R.H.; Aschebrook-Kilfoy, B. An update in international trends in incidence rates of thyroid cancer, 1973–2007. Cancer Causes Control 2018, 29, 465–473. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Davies, L.; Morris, L.; Hankey, B. Increases in thyroid cancer incidence and mortality. JAMA 2017, 318, 389–390. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Surveillance, Epidemiology and End Results (SEER) Program. Available online: https://seer.cancer.gov/statfacts/html/thyro.htm (accessed on 4 October 2020).
- Fiore, M.; Oliveri Conti, G.; Caltabiano, R.; Buffone, A.; Zuccarello, P.; Cormaci, L.; Cannizzaro, M.A.; Ferrante, M. Role of emerging environmental risk factors in thyroid cancer: A brief review. Int. J. Environ. Res. Public Health 2019, 16, 1185. [Google Scholar] [CrossRef] [PubMed]
- Thun, M.; Linet, M.S.; Cerhan, J.R.; Haiman, C.A.; Schottenfeld, D. Cancer Epidemiology and Prevention; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Pirahanchi, Y.; Jialal, I. Physiology, thyroid. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2018. [Google Scholar]
- Soundarrajan, M.; Kopp, P.A. Thyroid Hormone Biosynthesis and Physiology. In Thyroid Disease and Reproduction; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–17. [Google Scholar]
- Romei, C.; Tacito, A.; Molinaro, E.; Piaggi, P.; Cappagli, V.; Pieruzzi, L.; Matrone, A.; Viola, D.; Agate, L.; Torregrossa, L. Clinical, pathological and genetic features of anaplastic and poorly differentiated thyroid cancer: A single institute experience. Oncol. Lett. 2018, 15, 9174–9182. [Google Scholar] [CrossRef] [PubMed]
- Sipos, J.; Mazzaferri, E. Thyroid cancer epidemiology and prognostic variables. Clin. Oncol. 2010, 22, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Fugazzola, L.; Elisei, R.; Fuhrer, D.; Jarzab, B.; Leboulleux, S.; Newbold, K.; Smit, J. 2019 European Thyroid Association Guidelines for the Treatment and Follow-Up of Advanced Radioiodine-Refractory Thyroid Cancer. Eur. Thyroid J. 2019, 8, 227–245. [Google Scholar] [CrossRef]
- Haugen, B.R.; Alexander, E.K.; Bible, K.C.; Doherty, G.M.; Mandel, S.J.; Nikiforov, Y.E.; Pacini, F.; Randolph, G.W.; Sawka, A.M.; Schlumberger, M. 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid 2016, 26, 1–133. [Google Scholar] [CrossRef] [PubMed]
- Grewal, R.K.; Ho, A.; Schöder, H. Novel approaches to thyroid cancer treatment and response assessment. Semin. Nucl. Med. 2016, 46, 109–118. [Google Scholar] [CrossRef]
- Nikiforov, Y.E.; Ohori, N.P.; Hodak, S.P.; Carty, S.E.; LeBeau, S.O.; Ferris, R.L.; Yip, L.; Seethala, R.R.; Tublin, M.E.; Stang, M.T. Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: A prospective analysis of 1056 FNA samples. J. Clin. Endocrinol. Metab. 2011, 96, 3390–3397. [Google Scholar] [CrossRef]
- Nikiforov, Y.E.; Carty, S.E.; Chiosea, S.I.; Coyne, C.; Duvvuri, U.; Ferris, R.L.; Gooding, W.E.; Hodak, S.P.; LeBeau, S.O.; Ohori, N.P. Highly accurate diagnosis of cancer in thyroid nodules with follicular neoplasm/suspicious for a follicular neoplasm cytology by ThyroSeq v2 next-generation sequencing assay. Cancer 2014, 120, 3627–3634. [Google Scholar] [CrossRef]
- Santhanam, P.; Khthir, R.; Gress, T.; Elkadry, A.; Olajide, O.; Yaqub, A.; Driscoll, H. Gene expression classifier for the diagnosis of indeterminate thyroid nodules: A meta-analysis. Med. Oncol. 2016, 33, 14. [Google Scholar] [CrossRef]
- Fagin, J.A.; Wells, S.A., Jr. Biologic and clinical perspectives on thyroid cancer. New Engl. J. Med. 2016, 375, 1054–1067. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Xing, M. TERT promoter mutations in thyroid cancer. Endocr. Relat. Cancer 2016, 23, R143–R155. [Google Scholar] [CrossRef]
- Lathief, S.; Pothuloori, A.; Liu, X.; Chaidarun, S. Advances and practical use of the molecular markers for thyroid cancer. Adv. Cell. Mol. Otolaryngol. 2016, 4, 33948. [Google Scholar] [CrossRef]
- Lin, C.; Yang, L. Long noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol. 2018, 28, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Ganini, C.; Candi, E.; Melino, G. The role of noncoding RNAs in epithelial cancer. Cell Death Discov. 2020, 6, 13. [Google Scholar] [CrossRef]
- Choudhari, R.; Sedano, M.J.; Harrison, A.L.; Subramani, R.; Lin, K.Y.; Ramos, E.I.; Lakshmanaswamy, R.; Gadad, S.S. Long noncoding RNAs in cancer: From discovery to therapeutic targets. In Advances in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 2020; Volume 95, pp. 105–147. [Google Scholar]
- Sohel, M.M.H. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci. 2020, 117473. [Google Scholar] [CrossRef]
- Wright, M.W.; Bruford, E.A. Naming’junk: Human non-protein coding RNA (ncRNA) gene nomenclature. Hum. Genom. 2011, 5, 90. [Google Scholar] [CrossRef]
- Esteller, M.; Pandolfi, P.P. The epitranscriptome of noncoding RNAs in cancer. Cancer Discov. 2017, 7, 359–368. [Google Scholar] [CrossRef]
- Lü, L.; Sun, J.; Shi, P.; Kong, W.; Xu, K.; He, B.; Zhang, S.; Wang, J. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget 2017, 8, 44096. [Google Scholar] [CrossRef]
- Wang, W.-J.; Li, H.-T.; Yu, J.-P.; Han, X.-P.; Xu, Z.-P.; Li, Y.-M.; Jiao, Z.-Y.; Liu, H.-B. A competing endogenous RNA network reveals novel potential lncRNA, miRNA, and mRNA biomarkers in the prognosis of human colon adenocarcinoma. J. Surg. Res. 2019, 235, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Mai, D.; Ding, P.; Tan, L.; Zhang, J.; Pan, Z.; Bai, R.; Li, C.; Li, M.; Zhou, Y.; Tan, W. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics 2018, 8, 5213. [Google Scholar] [CrossRef]
- Qin, X.-g.; Zeng, J.-H.; Lin, P.; Mo, W.-J.; Li, Q.; Feng, Z.-B.; Luo, D.-Z.; Yang, H.; Chen, G.; Zeng, J.-J. Prognostic value of small nuclear RNAs (snRNAs) for digestive tract pan-adenocarcinomas identified by RNA sequencing data. Pathol. Res. Pract. 2019, 215, 414–426. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef]
- Yang, L. Splicing noncoding RNAs from the inside out. Wiley Interdiscip. Rev. RNA 2015, 6, 651–660. [Google Scholar] [CrossRef]
- Khorshidi, A.; Dhaliwal, P.; Yang, B.B. Noncoding RNAs in tumor angiogenesis. In The Long and Short Non-Coding RNAs in Cancer Biology; Springer: Berlin/Heidelberg, Germany, 2016; pp. 217–241. [Google Scholar]
- Tang, X.J.; Wang, W.; Hann, S.S. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie 2019, 163, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Gong, Y.; Yin, Y.; Xing, H.; Zhang, N. The multiple function of long noncoding RNAs in osteosarcoma progression, drug resistance and prognosis. Biomed. Pharmacother. 2020, 127, 110141. [Google Scholar] [CrossRef]
- Rossi, M.; Gorospe, M. Noncoding RNAs Controlling Telomere Homeostasis in Senescence and Aging. Trends Mol. Med. 2020, 26, 422–433. [Google Scholar] [CrossRef]
- Guzel, E.; Okyay, T.M.; Yalcinkaya, B.; Karacaoglu, S.; Gocmen, M.; Akcakuyu, M.H. Tumor suppressor and oncogenic role of long non-coding RNAs in cancer. North. Clin. Istanb. 2020, 7, 81. [Google Scholar] [CrossRef]
- Dhamija, S.; Diederichs, S. From junk to master regulators of invasion: LncRNA functions in migration, EMT and metastasis. Int. J. Cancer 2016, 139, 269–280. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, J.-K.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 1–18. [Google Scholar] [CrossRef]
- Liu, K.; Gao, L.; Ma, X.; Huang, J.-J.; Chen, J.; Zeng, L.; Ashby, C.R.; Zou, C.; Chen, Z.-S. Long non-coding RNAs regulate drug resistance in cancer. Mol. Cancer 2020, 19, 1–13. [Google Scholar] [CrossRef]
- Li, Z.; Ruan, Y.; Zhang, H.; Shen, Y.; Li, T.; Xiao, B. Tumor-suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019, 110, 3630. [Google Scholar] [CrossRef]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol. Cell. Pharmacol. 2011, 3, 83. [Google Scholar]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Chavdoula, E.; Hatziapostolou, M.; Polytarchou, C.; Marcu, K.B.; Papavassiliou, A.G.; Sandaltzopoulos, R.; Kolettas, E. A step-by-step microRNA guide to cancer development and metastasis. Cell. Oncol. 2017, 40, 303–339. [Google Scholar] [CrossRef]
- Dai, X.; Kaushik, A.C.; Zhang, J. The emerging role of major regulatory RNAs in cancer control. Front. Oncol. 2019, 9, 920. [Google Scholar] [CrossRef]
- Zong, Y.; Zhang, Y.; Sun, X.; Xu, T.; Cheng, X.; Qin, Y. miR-221/222 promote tumor growth and suppress apoptosis by targeting lncRNA GAS5 in breast cancer. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhan, C.; Yuan, H.; Cui, Y.; Zhang, Z. MicroRNA-603 functions as an oncogene by suppressing BRCC2 protein translation in osteosarcoma. Oncol. Rep. 2016, 35, 3257–3264. [Google Scholar] [CrossRef]
- Zeng, B.; Li, Y.; Jiang, F.; Wei, C.; Chen, G.; Zhang, W.; Zhao, W.; Yu, D. LncRNA GAS5 suppresses proliferation, migration, invasion, and epithelial-mesenchymal transition in oral squamous cell carcinoma by regulating the miR-21/PTEN axis. Exp. Cell Res. 2019, 374, 365–373. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- He, H.; Jazdzewski, K.; Li, W.; Liyanarachchi, S.; Nagy, R.; Volinia, S.; Calin, G.A.; Liu, C.-g.; Franssila, K.; Suster, S. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 19075–19080. [Google Scholar] [CrossRef]
- Zembska, A.; Jawiarczyk-Przybyłowska, A.; Wojtczak, B.; Bolanowski, M. MicroRNA expression in the progression and aggressiveness of papillary thyroid carcinoma. Anticancer Res. 2019, 39, 33–40. [Google Scholar] [CrossRef]
- Pishkari, S.; Paryan, M.; Hashemi, M.; Baldini, E.; Mohammadi-Yeganeh, S. The role of microRNAs in different types of thyroid carcinoma: A comprehensive analysis to find new miRNA supplementary therapies. J. Endocrinol. Investig. 2018, 41, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Czajka, A.A.; Wojcicka, A.; Kubiak, A.; Kotlarek, M.; Bakuła-Zalewska, E.; Koperski, Ł.; Wiechno, W.; Jażdżewski, K. Family of microRNA-146 regulates RARβ in papillary thyroid carcinoma. PLoS ONE 2016, 11, e0151968. [Google Scholar] [CrossRef]
- Zhang, W.; Ji, W.; Zhao, X. MiR-155 promotes anaplastic thyroid cancer progression by directly targeting SOCS1. BMC Cancer 2019, 19, 1093. [Google Scholar] [CrossRef]
- Zang, C.; Sun, J.; Liu, W.; Chu, C.; Jiang, L.; Ge, R. miRNA-21 promotes cell proliferation and invasion via VHL/PI3K/AKT in papillary thyroid carcinoma. Hum. Cell 2019, 32, 428–436. [Google Scholar] [CrossRef]
- Wang, X.; Lu, X.; Geng, Z.; Yang, G.; Shi, Y. LncRNA PTCSC3/miR-574-5p governs cell proliferation and migration of papillary thyroid carcinoma via Wnt/β-catenin signaling. J. Cell. Biochem. 2017, 118, 4745–4752. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Xu, W.; Kong, D. Icariin inhibits cell proliferation, migration and invasion by down-regulation of microRNA-625-3p in thyroid cancer cells. Biomed. Pharmacother. 2019, 109, 2456–2463. [Google Scholar] [CrossRef]
- Wang, X.-Z.; Hang, Y.-K.; Liu, J.-B.; Hou, Y.-Q.; Wang, N.; Wang, M.-J. Over-expression of microRNA-375 inhibits papillary thyroid carcinoma cell proliferation and induces cell apoptosis by targeting ERBB2. J. Pharmacol. Sci. 2016, 130, 78–84. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, Q.; Qi, Y.-H.; Wang, H.; Yang, L.; Fan, Q.-Y. Effects of long non-coding RNA H19 and microRNA let7a expression on thyroid cancer prognosis. Exp. Mol. Pathol. 2017, 103, 71–77. [Google Scholar] [CrossRef]
- Huang, P.; Mao, L.-F.; Zhang, Z.-P.; Lv, W.-W.; Feng, X.-P.; Liao, H.-J.; Dong, C.; Kaluba, B.; Tang, X.-F.; Chang, S. Down-regulated miR-125a-5p promotes the reprogramming of glucose metabolism and cell malignancy by increasing levels of CD147 in thyroid cancer. Thyroid 2018, 28, 613–623. [Google Scholar] [CrossRef]
- Fu, Y.; Zheng, H.; Zhang, D.; Zhou, L.; Sun, H. MicroRNA-1266 suppresses papillary thyroid carcinoma cell metastasis and growth via targeting FGFR2. Eur. Rev. Med. Pharm. Sci. 2018, 22, 3430–3438. [Google Scholar]
- Fang, M.; Huang, W.; Wu, X.; Gao, Y.; Ou, J.; Zhang, X.; Li, Y. MiR-141-3p suppresses tumor growth and metastasis in papillary thyroid cancer via targeting Yin Yang 1. Anat. Rec. 2019, 302, 258–268. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Zhang, H.; Zhang, J.; Zhang, Y.; Li, S.; Qin, L.; Yang, Z.; Xiong, J. Effects of miR-144 on the sensitivity of human anaplastic thyroid carcinoma cells to cisplatin by autophagy regulation. Cancer Biol. Ther. 2018, 19, 484–496. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, Y.; Gong, Y.; Dong, T.; Zhang, B.; Gao, W. miR-148a inhibits self-renewal of thyroid cancer stem cells via repressing INO80 expression. Oncol. Rep. 2016, 36, 3387–3396. [Google Scholar] [CrossRef]
- Cheng, L.; Zhou, R.; Chen, M.; Feng, L.; Li, H. MicroRNA-150 targets Rho-associated protein kinase 1 to inhibit cell proliferation, migration and invasion in papillary thyroid carcinoma. Mol. Med. Rep. 2017, 16, 2217–2224. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Hong, S.; Yu, S.; Huang, Y.; Chen, S.; Liu, Y.; Zhang, Q.; Li, Y.; Xiao, H. MiR-195 inhibits tumor growth and metastasis in papillary thyroid carcinoma cell lines by targeting CCND1 and FGF2. Int. J. Endocrinol. 2017, 2017, 6180425. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Xu, Y.; Qin, G.; Liu, C.; Yan, Y.; Zhang, H. miR-199b-5p-Stonin 2 axis regulates metastases and epithelial-to-mesenchymal transition of papillary thyroid carcinoma. IUBMB Life 2019, 71, 28–40. [Google Scholar] [CrossRef]
- Liu, H.; Chen, X.; Lin, T.; Chen, X.; Yan, J.; Jiang, S. MicroRNA-524-5p suppresses the progression of papillary thyroid carcinoma cells via targeting on FOXE1 and ITGA3 in cell autophagy and cycling pathways. J. Cell. Physiol. 2019, 234, 18382–18391. [Google Scholar] [CrossRef]
- Yue, K.; Wang, X.; Wu, Y.; Zhou, X.; He, Q.; Duan, Y. microRNA-7 regulates cell growth, migration and invasion via direct targeting of PAK1 in thyroid cancer. Mol. Med. Rep. 2016, 14, 2127–2134. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, W.; Wang, C.; Ai, Z. miR-873-5p Inhibits Cell Migration and Invasion of Papillary Thyroid Cancer via Regulation of CXCL16. Oncotargets Ther. 2020, 13, 1037. [Google Scholar] [CrossRef]
- Guo, F.; Hou, X.; Sun, Q. MicroRNA-9-5p functions as a tumor suppressor in papillary thyroid cancer via targeting BRAF. Oncol. Lett. 2018, 16, 6815–6821. [Google Scholar] [CrossRef] [PubMed]
- Sastre-Perona, A.; Santisteban, P. Role of the wnt pathway in thyroid cancer. Front. Endocrinol. 2012, 3, 31. [Google Scholar] [CrossRef]
- Fuziwara, C.S.; Kimura, E.T. How does microRNA modulate Wnt/β-catenin signaling in thyroid oncogenesis? Ann. Transl. Med. 2020, 8, 266. [Google Scholar] [CrossRef]
- Ely, K.A.; Bischoff, L.A.; Weiss, V.L. Wnt signaling in thyroid homeostasis and carcinogenesis. Genes 2018, 9, 204. [Google Scholar] [CrossRef]
- Zhang, Z.-J.; Xiao, Q.; Li, X.-Y. MicroRNA-574-5p directly targets FOXN3 to mediate thyroid cancer progression via Wnt/β-catenin signaling pathway. Pathol. Res. Pract. 2020, 216, 152939. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.; Xiao, Q.; Wang, Z. MiR-574-5p mediates the cell cycle and apoptosis in thyroid cancer cells via Wnt/β-catenin signaling by repressing the expression of Quaking proteins. Oncol. Lett. 2018, 15, 5841–5848. [Google Scholar] [CrossRef]
- Feige, J.; Cherradi, N. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocr. Relat. Cancer 2013, 20, 579–594. [Google Scholar]
- Liu, B.; Qu, J.; Xu, F.; Guo, Y.; Wang, Y.; Yu, H.; Qian, B. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget 2015, 6, 9445. [Google Scholar] [CrossRef]
- Ujifuku, K.; Mitsutake, N.; Takakura, S.; Matsuse, M.; Saenko, V.; Suzuki, K.; Hayashi, K.; Matsuo, T.; Kamada, K.; Nagata, I. MiR-195, miR-455-3p and miR-10a∗ are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 2010, 296, 241–248. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Y.; Ning, Y.-E.; Gu, H.; Tong, Y.; Wang, N. MiR-195-5p Inhibits Malignant Progression of Cervical Cancer by Targeting YAP1. Oncotargets Ther. 2020, 13, 931. [Google Scholar] [CrossRef]
- Balmeh, N.; Tabatabaeian, H.; Asgari, M.; Mokhtarian, R.; Abharian, P.H.; Azadeh, M.; Ghaedi, K. miR-195 down-regulation is a distinctive biomarker of HER2 positive state in breast cancer. Gene Rep. 2020, 100703. [Google Scholar] [CrossRef]
- Jung, Y.-S.; Park, J.-I. Wnt signaling in cancer: Therapeutic targeting of Wnt signaling beyond β-catenin and the destruction complex. Exp. Mol. Med. 2020, 52, 183–191. [Google Scholar] [CrossRef]
- Noorolyai, S.; Shajari, N.; Baghbani, E.; Sadreddini, S.; Baradaran, B. The relation between PI3K/AKT signalling pathway and cancer. Gene 2019, 698, 120–128. [Google Scholar] [CrossRef]
- Nozhat, Z.; Hedayati, M. PI3K/AKT pathway and its mediators in thyroid carcinomas. Mol. Diagn. Ther. 2016, 20, 13–26. [Google Scholar] [CrossRef]
- Zhang, J.-G.; Wang, J.-J.; Zhao, F.; Liu, Q.; Jiang, K.; Yang, G.-h. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin. Chim. Acta 2010, 411, 846–852. [Google Scholar] [CrossRef]
- Li, L.-Q.; Li, X.-L.; Wang, L.; Du, W.-J.; Guo, R.; Liang, H.-H.; Liu, X.; Liang, D.-S.; Lu, Y.-J.; Shan, H.-L. Matrine inhibits breast cancer growth via miR-21/PTEN/Akt pathway in MCF-7 cells. Cell. Physiol. Biochem. 2012, 30, 631–641. [Google Scholar] [CrossRef] [PubMed]
- McClelland, A.D.; Herman-Edelstein, M.; Komers, R.; Jha, J.C.; Winbanks, C.E.; Hagiwara, S.; Gregorevic, P.; Kantharidis, P.; Cooper, M.E. miR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin. Sci. 2015, 129, 1237–1249. [Google Scholar] [CrossRef]
- Haugen, D.; Akslen, L.; Varhaug, J.A.; Lillehaug, J. Expression of c-erb B-2 protein in papillary thyroid carcinomas. Br. J. Cancer 1992, 65, 832–837. [Google Scholar] [CrossRef]
- Montero-Conde, C.; Ruiz-Llorente, S.; Dominguez, J.M.; Knauf, J.A.; Viale, A.; Sherman, E.J.; Ryder, M.; Ghossein, R.A.; Rosen, N.; Fagin, J.A. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 2013, 3, 520–533. [Google Scholar] [CrossRef]
- Lv, Y.; Sui, F.; Ma, J.; Ren, X.; Yang, Q.; Zhang, Y.; Guan, H.; Shi, B.; Hou, P.; Ji, M. Increased expression of EHF contributes to thyroid tumorigenesis through transcriptionally regulating HER2 and HER3. Oncotarget 2016, 7, 57978. [Google Scholar] [CrossRef]
- Zaballos, M.A.; Santisteban, P. FOXO1 controls thyroid cell proliferation in response to TSH and IGF-I and is involved in thyroid tumorigenesis. Mol. Endocrinol. 2013, 27, 50–62. [Google Scholar] [CrossRef]
- Rena, G.; Prescott, A.R.; Guo, S.; Cohen, P.; Unterman, T.G. Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targetting. Biochem. J. 2001, 354, 605–612. [Google Scholar] [CrossRef]
- Wong, K.; Di Cristofano, F.; Ranieri, M.; De Martino, D.; Di Cristofano, A. PI3K/mTOR inhibition potentiates and extends palbociclib activity in anaplastic thyroid cancer. Endocr. Relat. Cancer 2019, 26, 425–436. [Google Scholar] [CrossRef]
- Coelho, R.G.; Fortunato, R.S.; Carvalho, D.P. Metabolic reprogramming in thyroid carcinoma. Front. Oncol. 2018, 8, 82. [Google Scholar] [CrossRef]
- Suh, H.Y.; Choi, H.; Paeng, J.C.; Cheon, G.J.; Chung, J.-K.; Kang, K.W. Comprehensive gene expression analysis for exploring the association between glucose metabolism and differentiation of thyroid cancer. BMC Cancer 2019, 19, 1260. [Google Scholar] [CrossRef]
- Miao, Y.; Zhang, L.-F.; Zhang, M.; Guo, R.; Liu, M.-F.; Li, B. Therapeutic Delivery of miR-143 Targeting Tumor Metabolism in Poorly Differentiated Thyroid Cancer Xenografts and Efficacy Evaluation Using 18F-FDG MicroPET-CT. Hum. Gene Ther. 2019, 30, 882–892. [Google Scholar] [CrossRef]
- Hay, N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 2016, 16, 635. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Chandel, N.S. Targeting glucose metabolism for cancer therapy. J. Exp. Med. 2012, 209, 211–215. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Matsubara, K.; Qian, G.-S.; Jackson, P.; Groopman, J.D.; Manning, J.E.; Harris, C.C.; Herman, J.G. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat. Genet. 2001, 28, 29–35. [Google Scholar] [CrossRef]
- Oshimo, Y.; Kuraoka, K.; Nakayama, H.; Kitadai, Y.; Yoshida, K.; Chayama, K.; Yasui, W. Epigenetic inactivation of SOCS-1 by CpG island hypermethylation in human gastric carcinoma. Int. J. Cancer 2004, 112, 1003–1009. [Google Scholar] [CrossRef]
- Chen, C.Y.; Tsay, W.; Tang, J.L.; Shen, H.L.; Lin, S.W.; Huang, S.Y.; Yao, M.; Chen, Y.C.; Shen, M.C.; Wang, C.H. SOCS1 methylation in patients with newly diagnosed acute myeloid leukemia. Genes Chromosomes Cancer 2003, 37, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Liau, N.P.; Laktyushin, A.; Lucet, I.S.; Murphy, J.M.; Yao, S.; Whitlock, E.; Callaghan, K.; Nicola, N.A.; Kershaw, N.J.; Babon, J.J. The molecular basis of JAK/STAT inhibition by SOCS1. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Danial, N.N.; Rothman, P. JAK-STAT signaling activated by Abl oncogenes. Oncogene 2000, 19, 2523–2531. [Google Scholar] [CrossRef]
- Jin, S.; Borkhuu, O.; Bao, W.; Yang, Y.-T. Signaling pathways in thyroid cancer and their therapeutic implications. J. Clin. Med. Res. 2016, 8, 284. [Google Scholar] [CrossRef]
- Braun, J.; Hoang-Vu, C.; Dralle, H.; Hüttelmaier, S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene 2010, 29, 4237–4244. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes-Rojas, C.; Hernandez, A.J.; Sarma, K.; Lee, J.T. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell 2014, 55, 171–185. [Google Scholar] [CrossRef]
- Guil, S.; Soler, M.; Portela, A.; Carrère, J.; Fonalleras, E.; Gómez, A.; Villanueva, A.; Esteller, M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat. Struct. Mol. Biol. 2012, 19, 664. [Google Scholar] [CrossRef]
- Mohammad, F.; Mondal, T.; Guseva, N.; Pandey, G.K.; Kanduri, C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 2010, 137, 2493–2499. [Google Scholar] [CrossRef]
- Merry, C.R.; Forrest, M.E.; Sabers, J.N.; Beard, L.; Gao, X.-H.; Hatzoglou, M.; Jackson, M.W.; Wang, Z.; Markowitz, S.D.; Khalil, A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015, 24, 6240–6253. [Google Scholar] [CrossRef]
- Arab, K.; Park, Y.J.; Lindroth, A.M.; Schäfer, A.; Oakes, C.; Weichenhan, D.; Lukanova, A.; Lundin, E.; Risch, A.; Meister, M. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell 2014, 55, 604–614. [Google Scholar] [CrossRef]
- Lister, N.; Shevchenko, G.; Walshe, J.L.; Groen, J.; Johnsson, P.; Vidarsdóttir, L.; Grander, D.; Ataide, S.F.; Morris, K.V. The molecular dynamics of long noncoding RNA control of transcription in PTEN and its pseudogene. Proc. Natl. Acad. Sci. USA 2017, 114, 9942–9947. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Zeng, T.; Ni, H.; Yu, Y.; Zhang, M.; Wu, M.; Wang, Q.; Wang, L.; Xu, S.; Xu, Z.; Xu, C. BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs. J. Chem. Neuroanat. 2019, 98, 87–96. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef]
- Peng, W.-X.; Koirala, P.; Mo, Y.-Y. Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Spizzo, R.; Almeida, M.I.; Colombatti, A.; Calin, G.A. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene 2012, 31, 4577–4587. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Gao, H. Long noncoding RNA CASC9 promotes the proliferation and metastasis of papillary thyroid cancer via sponging miR-488-3p. Cancer Med. 2020, 9, 1830–1841. [Google Scholar] [CrossRef]
- Zhong, Z.; Wu, Y.; Luo, J.; Hu, X.; Yuan, Z.; Li, G.; Wang, Y.; Yao, G.; Ge, X. Knockdown of long noncoding RNA DLX6-AS1 inhibits migration and invasion of thyroid cancer cells by upregulating UPF1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10867–10873. [Google Scholar] [PubMed]
- Song, B.; Li, R.; Zuo, Z.; Tan, J.; Liu, L.; Ding, D.; Lu, Y.; Hou, D. LncRNA ENST00000539653 acts as an oncogenic factor via MAPK signalling in papillary thyroid cancer. BMC Cancer 2019, 19, 297. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, Y.; Zhang, K.; Liu, X.; Dai, Y.; Jiao, X. Targeted inhibition of long non-coding RNA H19 blocks anaplastic thyroid carcinoma growth and metastasis. Bioengineered 2019, 10, 306–315. [Google Scholar] [CrossRef]
- Li, M.; Chai, H.-F.; Peng, F.; Meng, Y.-T.; Zhang, L.-Z.; Zhang, L.; Zou, H.; Liang, Q.-L.; Li, M.-M.; Mao, K.-G. Estrogen receptor β upregulated by lncRNA-H19 to promote cancer stem-like properties in papillary thyroid carcinoma. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Liang, L.; Xu, J.; Wang, M.; Xu, G.; Zhang, N.; Wang, G.; Zhao, Y. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Dai, Y.; Miao, Y.; Zhu, Q.; Gao, M.; Hao, F. Expression of long non-coding RNA H19 predicts distant metastasis in minimally invasive follicular thyroid carcinoma. Bioengineered 2019, 10, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Guo, X.; Zang, M.; Wang, P.; Xue, S.; Chen, G. Long non-coding RNA LINC00152 promotes cell growth and invasion of papillary thyroid carcinoma by regulating the miR-497/BDNF axis. J. Cell. Physiol. 2019, 234, 1336–1345. [Google Scholar] [CrossRef]
- Li, X.; Zhong, W.; Xu, Y.; Yu, B.; Liu, H. Silencing of lncRNA LINC00514 inhibits the malignant behaviors of papillary thyroid cancer through miR-204–3p/CDC23 axis. Biochem. Biophys. Res. Commun. 2019, 508, 1145–1148. [Google Scholar] [CrossRef]
- Gugnoni, M.; Manicardi, V.; Torricelli, F.; Sauta, E.; Bellazzi, R.; Manzotti, G.; Vitale, E.; de Biase, D.; Piana, S.; Ciarrocchi, A. Linc00941 is a novel TGFβ target that primes papillary thyroid cancer metastatic behavior by regulating the expression of Cadherin 6. Thyroid 2020. [Google Scholar] [CrossRef]
- Liu, J.; Dong, H.; Yang, Y.; Qian, Y.; Liu, J.; Li, Z.; Guan, H.; Chen, Z.; Li, C.; Zhang, K. Upregulation of long noncoding RNA MALAT1 in papillary thyroid cancer and its diagnostic value. Future Oncol. 2018, 14, 3015–3022. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bhandari, A.; Niu, J.; Yang, F.; Xia, E.; Yao, Z.; Jin, Y.; Zheng, Z.; Lv, S.; Wang, O. The lncRNA UNC5B-AS1 promotes proliferation, migration, and invasion in papillary thyroid cancer cell lines. Hum. Cell 2019, 32, 334–342. [Google Scholar] [CrossRef]
- Liu, H.; Deng, H.; Zhao, Y.; Li, C.; Liang, Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J. Exp. Clin. Cancer Res. 2018, 37, 1–12. [Google Scholar] [CrossRef]
- Jiao, X.; Lu, J.; Huang, Y.; Zhang, J.; Zhang, H.; Zhang, K. Long non-coding RNA H19 may be a marker for prediction of prognosis in the follow-up of patients with papillary thyroid cancer. Cancer Biomark. 2019, 26, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Liu, K.; Zhu, H.; Zhang, J. Long noncoding RNA LINC003121 inhibits proliferation and invasion of thyroid cancer cells by suppression of the phosphatidylinositol-3-kinase (PI3K)/Akt signaling pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 4592. [Google Scholar] [CrossRef]
- Pellecchia, S.; Sepe, R.; Decaussin-Petrucci, M.; Ivan, C.; Shimizu, M.; Coppola, C.; Testa, D.; Calin, G.A.; Fusco, A.; Pallante, P. The long non-coding RNA prader willi/angelman region RNA5 (PAR5) is downregulated in anaplastic thyroid carcinomas where it acts as a tumor suppressor by reducing EZH2 activity. Cancers 2020, 12, 235. [Google Scholar] [CrossRef]
- Wang, X.-M.; Liu, Y.; Fan, Y.-X.; Liu, Z.; Yuan, Q.-L.; Jia, M.; Geng, Z.-S.; Gu, L.; Lu, X.-B. LncRNA PTCSC3 affects drug resistance of anaplastic thyroid cancer through STAT3/INO80 pathway. Cancer Biol. Ther. 2018, 19, 590–597. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, Z.; Xu, L.; Sun, L.; Song, H.; Yin, H.; He, F. lncRNA SNHG3 acts as a novel Tumor Suppressor and regulates Tumor Proliferation and Metastasis via AKT/mTOR/ERK pathway in Papillary Thyroid Carcinoma. J. Cancer 2020, 11, 3492. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhong, Z.; Huang, M.; Tian, Q.; Jiang, R.; Chen, J. lncRNA H19 regulates epithelial–mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1887–1899. [Google Scholar] [CrossRef]
- Zhou, W.; Ye, X.-L.; Xu, J.; Cao, M.-G.; Fang, Z.-Y.; Li, L.-Y.; Guan, G.-H.; Liu, Q.; Qian, Y.-H.; Xie, D. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal. 2017, 10, eaak9557. [Google Scholar] [CrossRef]
- Peng, F.; Li, T.-T.; Wang, K.-L.; Xiao, G.-Q.; Wang, J.-H.; Zhao, H.-D.; Kang, Z.-J.; Fan, W.-J.; Zhu, L.-L.; Li, M. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2018, 8, e2569. [Google Scholar] [CrossRef]
- Vernucci, M.; Cerrato, F.; Besnard, N.; Casola, S.; Pedone, P.V.; Bruni, C.B.; Riccio, A. The H19 endodermal enhancer is required for Igf2 activation and tumor formation in experimental liver carcinogenesis. Oncogene 2000, 19, 6376–6385. [Google Scholar] [CrossRef]
- Liu, W.; Xu, Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/beta-catenin pathway and indicates a poor prognosis. Eur. Rev. Med. Pharm. Sci. 2019, 23, 7905–7912. [Google Scholar]
- Shen, Z.; Zhou, L.; Zhang, C.; Xu, J. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020, 468, 88–101. [Google Scholar] [CrossRef]
- Sun, Y.; Li, X.; Chen, A.; Shi, W.; Wang, L.; Yi, R.; Qiu, J. circPIP5K1A serves as a competitive endogenous RNA contributing to ovarian cancer progression via regulation of miR-661/IGFBP5 signaling. J. Cell. Biochem. 2019, 120, 19406–19414. [Google Scholar] [CrossRef]
- Zhou, G.; Zhang, G.; Yuan, Z.; Pei, R.; Liu, D. Has_circ_0008274 promotes cell proliferation and invasion involving AMPK/mTOR signaling pathway in papillary thyroid carcinoma. Eur. Rev. Med. Pharm. Sci. 2018, 22, 8772–8780. [Google Scholar]
- Liu, F.; Zhang, J.; Qin, L.; Yang, Z.; Xiong, J.; Zhang, Y.; Li, R.; Li, S.; Wang, H.; Yu, B. Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging 2018, 10, 3806. [Google Scholar] [CrossRef]
- Ye, M.; Hou, H.; Shen, M.; Dong, S.; Zhang, T. Circular RNA circFOXM1 plays a role in papillary thyroid carcinoma by sponging miR-1179 and regulating HMGB1 expression. Mol. Ther. Nucleic Acids 2020, 19, 741–750. [Google Scholar] [CrossRef]
- Cork, G.K.; Thompson, J.; Slawson, C. Real talk: The inter-play between the mTOR, AMPK, and hexosamine biosynthetic pathways in cell signaling. Front. Endocrinol. 2018, 9, 522. [Google Scholar] [CrossRef] [PubMed]
- Allinen, M.; Beroukhim, R.; Cai, L.; Brennan, C.; Lahti-Domenici, J.; Huang, H.; Porter, D.; Hu, M.; Chin, L.; Richardson, A. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 2004, 6, 17–32. [Google Scholar] [CrossRef]
- Moroishi, T.; Hayashi, T.; Pan, W.-W.; Fujita, Y.; Holt, M.V.; Qin, J.; Carson, D.A.; Guan, K.-L. The Hippo pathway kinases LATS1/2 suppress cancer immunity. Cell 2016, 167, 1525–1539. [Google Scholar] [CrossRef]
- Locati, M.; Mantovani, A.; Sica, A. Macrophage activation and polarization as an adaptive component of innate immunity. In Advances in Immunology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 120, pp. 163–184. [Google Scholar]
- Baer, C.; Squadrito, M.L.; Iruela-Arispe, M.L.; De Palma, M. Reciprocal interactions between endothelial cells and macrophages in angiogenic vascular niches. Exp. Cell Res. 2013, 319, 1626–1634. [Google Scholar] [CrossRef]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-associated macrophages: An accomplice in solid tumor progression. J. Biomed. Sci. 2019, 26, 1–13. [Google Scholar] [CrossRef]
- Adeegbe, D.O.; Nishikawa, H. Natural and induced T regulatory cells in cancer. Front. Immunol. 2013, 4, 190. [Google Scholar] [CrossRef]
- Li, J.-H.; Zhang, S.-Q.; Qiu, X.-G.; Zhang, S.-J.; Zheng, S.-H.; Zhang, D.-H. Long non-coding RNA NEAT1 promotes malignant progression of thyroid carcinoma by regulating miRNA-214. Int. J. Oncol. 2017, 50, 708–716. [Google Scholar] [CrossRef]
- Han, D.; Fang, Y.; Guo, Y.; Hong, W.; Tu, J.; Wei, W. The emerging role of long non-coding RNAs in tumor-associated macrophages. J. Cancer 2019, 10, 6738. [Google Scholar] [CrossRef]
- Huang, J.K.; Ma, L.; Song, W.H.; Lu, B.Y.; Huang, Y.B.; Dong, H.M.; Ma, X.K.; Zhu, Z.Z.; Zhou, R. LncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J. Cell. Biochem. 2017, 118, 4821–4830. [Google Scholar] [CrossRef]
- Amit, M.; Rudnicki, Y.; Binenbaum, Y.; Trejo-Leider, L.; Cohen, J.T.; Gil, Z. Defining the outcome of patients with delayed diagnosis of differentiated thyroid cancer. Laryngoscope 2014, 124, 2837–2840. [Google Scholar] [CrossRef]
- WHO. Guide to Cancer Early Diagnosis; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Cooper, D.S.; Doherty, G.M.; Haugen, B.R.; Kloos, R.T.; Lee, S.L.; Mandel, S.J.; Mazzaferri, E.L.; McIver, B.; Pacini, F.; Schlumberger, M. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association (ATA) guidelines taskforce on thyroid nodules and differentiated thyroid cancer. Thyroid 2009, 19, 1167–1214. [Google Scholar] [CrossRef]
- Sclabas, G.M.; Staerkel, G.A.; Shapiro, S.E.; Fornage, B.D.; Sherman, S.I.; Vassillopoulou-Sellin, R.; Lee, J.E.; Evans, D.B. Fine-needle aspiration of the thyroid and correlation with histopathology in a contemporary series of 240 patients. Am. J. Surg. 2003, 186, 702–710. [Google Scholar] [CrossRef]
- Yang, J.; Schnadig, V.; Logrono, R.; Wasserman, P.G. Fine-needle aspiration of thyroid nodules: A study of 4703 patients with histologic and clinical correlations. Cancer Cytopathol. 2007, 111, 306–315. [Google Scholar] [CrossRef]
- Cibas, E.S.; Ali, S.Z. The 2017 Bethesda system for reporting thyroid cytopathology. Thyroid 2017, 27, 1341–1346. [Google Scholar] [CrossRef]
- Lum, J.N.M. Thyroid FNA: A retrospective audit of 1541 cases at NUH, Singapore. In Proceedings of the NUH Cytopathology Workshop 2014, Singapore, 23–25 May 2014; NUH: Singapore, 2014. [Google Scholar]
- Sistrunk, J.; Alexander, S.; Marc, F.; Johnson, T.; Norman, F.; Philip, G.; Richard, G.; Edward, G. Clinical performance of multiplatform mutation panel microRNA risk classifier in indeterminate thyroid nodules. J. Am. Soc. Cytopathol. 2020, 9, 232–241. [Google Scholar] [CrossRef]
- Albarel, F.; Conte-Devolx, B.; Oliver, C. From nodule to differentiated thyroid carcinoma: Contributions of molecular analysis in 2012. In Annales D’endocrinologie; Elsevier Masson: Paris, France; pp. 155–164.
- Ferrari, S.M.; Fallahi, P.; Ruffilli, I.; Elia, G.; Ragusa, F.; Paparo, S.R.; Ulisse, S.; Baldini, E.; Giannini, R.; Miccoli, P. Molecular testing in the diagnosis of differentiated thyroid carcinomas. Gland Surg. 2018, 7, S19. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Narkar, A.; Mukhopadhyaya, R.; Kane, S.; D’Cruz, A.; Rajan, M. BRAF V600E mutation in papillary thyroid carcinoma: Significant association with node metastases and extra thyroidal invasion. Endocr. Pathol. 2012, 23, 83–93. [Google Scholar] [CrossRef]
- Guan, H.; Ji, M.; Bao, R.; Yu, H.; Wang, Y.; Hou, P.; Zhang, Y.; Shan, Z.; Teng, W.; Xing, M. Association of high iodine intake with the T1799A BRAF mutation in papillary thyroid cancer. J. Clin. Endocrinol. Metab. 2009, 94, 1612–1617. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.-K.; Im, S.-Y.; Kang, Y.-J.; Lee, H.; Jung, E.-S.; Kang, C.-S.; Bae, J.-S.; Choi, Y.-J. Mutational patterns and novel mutations of the BRAF gene in a large cohort of Korean patients with papillary thyroid carcinoma. Thyroid 2012, 22, 791–797. [Google Scholar] [CrossRef]
- Network, C.G.A.R. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar]
- Romei, C.; Fugazzola, L.; Puxeddu, E.; Frasca, F.; Viola, D.; Muzza, M.; Moretti, S.; Luisa Nicolosi, M.; Giani, C.; Cirello, V. Modifications in the papillary thyroid cancer gene profile over the last 15 years. J. Clin. Endocrinol. Metab. 2012, 97, E1758–E1765. [Google Scholar] [CrossRef]
- Goh, X.; Lum, J.; Yang, S.P.; Chionh, S.B.; Koay, E.; Chiu, L.; Parameswaran, R.; Ngiam, K.Y.; Loh, T.K.S.; Nga, M.E. BRAF mutation in papillary thyroid cancer—Prevalence and clinical correlation in a South-East Asian cohort. Clin. Otolaryngol. 2019, 44, 114–123. [Google Scholar] [CrossRef]
- Arita, T.; Ichikawa, D.; Konishi, H.; Komatsu, S.; Shiozaki, A.; Shoda, K.; Kawaguchi, T.; Hirajima, S.; Nagata, H.; Kubota, T. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013, 33, 3185–3193. [Google Scholar]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Ferracin, M.; Veronese, A.; Negrini, M. Micromarkers: MiRNAs in cancer diagnosis and prognosis. Expert Rev. Mol. Diagn. 2010, 10, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhu, D.; Wu, L.; He, M.; Zhou, X.; Zhang, L.; Zhang, H.; Wang, W.; Zhu, J.; Cheng, W. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol. Prev. Biomark. 2017, 26, 188–196. [Google Scholar] [CrossRef]
- Sartori, D.A.; Chan, D.W. Biomarkers in prostate cancer: What’s new? Curr. Opin. Oncol. 2014, 26, 259. [Google Scholar] [CrossRef]
- Labourier, E.; Shifrin, A.; Busseniers, A.E.; Lupo, M.A.; Manganelli, M.L.; Andruss, B.; Wylie, D.; Beaudenon-Huibregtse, S. Molecular testing for miRNA, mRNA, and DNA on fine-needle aspiration improves the preoperative diagnosis of thyroid nodules with indeterminate cytology. J. Clin. Endocrinol. Metab. 2015, 100, 2743–2750. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, H.; Leng, X.; Hao, F.; Wang, L. miR-146b measurement in FNA to distinguish papillary thyroid cancer from benign thyroid masses. Br. J. Biomed. Sci. 2018, 75, 43–45. [Google Scholar] [CrossRef]
- Zhang, A.; Wang, C.; Lu, H.; Chen, X.; Ba, Y.; Zhang, C.; Zhang, C.-Y. Altered Serum MicroRNA Profile May Serve as an Auxiliary Tool for Discriminating Aggressive Thyroid Carcinoma from Nonaggressive Thyroid Cancer and Benign Thyroid Nodules. Dis. Markers 2019, 2019, 3717683. [Google Scholar] [CrossRef] [PubMed]
- Rosignolo, F.; Sponziello, M.; Giacomelli, L.; Russo, D.; Pecce, V.; Biffoni, M.; Bellantone, R.; Lombardi, C.P.; Lamartina, L.; Grani, G. Identification of thyroid-associated serum microRNA profiles and their potential use in thyroid cancer follow-up. J. Endocr. Soc. 2017, 1, 3–13. [Google Scholar]
- Zhang, Y.; Pan, J.; Xu, D.; Yang, Z.; Sun, J.; Sun, L.; Wu, Y.; Qiao, H. Combination of serum microRNAs and ultrasound profile as predictive biomarkers of diagnosis and prognosis for papillary thyroid microcarcinoma. Oncol. Rep. 2018, 40, 3611–3624. [Google Scholar] [CrossRef]
- Graham, M.E.R.; Hart, R.D.; Douglas, S.; Makki, F.M.; Pinto, D.; Butler, A.L.; Bullock, M.; Rigby, M.H.; Trites, J.R.; Taylor, S.M. Serum microRNA profiling to distinguish papillary thyroid cancer from benign thyroid masses. J. Otolaryngol. Head Neck Surg. 2015, 44, 33. [Google Scholar] [CrossRef]
- Ye, W.; Deng, X.; Fan, Y. Exosomal miRNA423-5p mediated oncogene activity in papillary thyroid carcinoma: A potential diagnostic and biological target for cancer therapy. Neoplasma 2019, 66, 516–523. [Google Scholar] [CrossRef]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. Ejifcc 2019, 30, 114. [Google Scholar]
- Jackson, S.; Kumar, G.; Banizs, A.B.; Toney, N.; Silverman, J.F.; Narick, C.M.; Finkelstein, S.D. Incremental utility of expanded mutation panel when used in combination with microRNA classification in indeterminate thyroid nodules. Diagn. Cytopathol. 2020, 48, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Sistrunk, J.W.; Shifrin, A.; Frager, M.; Bardales, R.H.; Thomas, J.; Fishman, N.; Goldberg, P.; Guttler, R.; Grant, E. Clinical impact of testing for mutations and microRNAs in thyroid nodules. Diagn. Cytopathol. 2019, 47, 758–764. [Google Scholar] [CrossRef]
- Schlosser, K.; Hanson, J.; Villeneuve, P.J.; Dimitroulakos, J.; McIntyre, L.; Pilote, L.; Stewart, D.J. Assessment of circulating LncRNAs under physiologic and pathologic conditions in humans reveals potential limitations as biomarkers. Sci. Rep. 2016, 6, 36596. [Google Scholar] [CrossRef]
- Xu, W.; Zhou, G.; Wang, H.; Liu, Y.; Chen, B.; Chen, W.; Lin, C.; Wu, S.; Gong, A.; Xu, M. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int. J. Cancer 2020, 146, 2901–2912. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Duan, W.; Yan, S.; Xie, Y.; Wang, C. Circulating long non-coding RNA colon cancer-associated transcript 2 protected by exosome as a potential biomarker for colorectal cancer. Biomed. Pharmacother. 2019, 113, 108758. [Google Scholar] [CrossRef]
- Abedini, P.; Fattahi, A.; Agah, S.; Talebi, A.; Beygi, A.H.; Amini, S.M.; Mirzaei, A.; Akbari, A. Expression analysis of circulating plasma long noncoding RNAs in colorectal cancer: The relevance of lncRNAs ATB and CCAT1 as potential clinical hallmarks. J. Cell. Physiol. 2019, 234, 22028–22033. [Google Scholar] [CrossRef]
- Özgür, E.; Ferhatoğlu, F.; Şen, F.; Saip, P.; Gezer, U. Circulating lncRNA H19 may be a useful marker of response to neoadjuvant chemotherapy in breast cancer. Cancer Biomark. 2020, 27, 11–17. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, W.; Liu, W.; Kong, X.; Li, L.; He, J.; Wang, D.; Zhang, M.; Zhou, G.; Xu, W. Circulating lncRNA ABHD11-AS1 serves as a biomarker for early pancreatic cancer diagnosis. J. Cancer 2019, 10, 3746. [Google Scholar] [CrossRef]
- Yu, J.; Ding, W.B.; Wang, M.C.; Guo, X.G.; Xu, J.; Xu, Q.G.; Yang, Y.; Sun, S.H.; Liu, J.F.; Qin, L.X. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int. J. Cancer 2020, 146, 1754–1763. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.-Y.; Wang, J.; Ouyang, S.-B.; Huang, Z.-K.; Liao, L. Salivary circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as novel biomarkers for the diagnosis of Oral squamous cell carcinoma. Cell. Physiol. Biochem. 2018, 47, 2511–2521. [Google Scholar] [CrossRef]
- Liu, Y.-T.; Han, X.-H.; Xing, P.-Y.; Hu, X.-S.; Hao, X.-Z.; Wang, Y.; Li, J.-L.; Zhang, Z.-S.; Yang, Z.-H.; Shi, Y.-K. Circular RNA profiling identified as a biomarker for predicting the efficacy of gefitinib therapy for non-small cell lung cancer. J. Thorac. Dis. 2019, 11, 1779. [Google Scholar] [CrossRef]
- Ortholan, C.; Puissegur, M.-P.; Ilie, M.; Barbry, P.; Mari, B.; Hofman, P. MicroRNAs and lung cancer: New oncogenes and tumor suppressors, new prognostic factors and potential therapeutic targets. Curr. Med. Chem. 2009, 16, 1047–1061. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef]
- Hu, Y.; Tian, H.; Xu, J.; Fang, J.-Y. Roles of competing endogenous RNAs in gastric cancer. Brief. Funct. Genom. 2016, 15, 266–273. [Google Scholar] [CrossRef]
- Donghi, R.; Longoni, A.; Pilotti, S.; Michieli, P.; Della Porta, G.; Pierotti, M.A. Gene p53 mutations are restricted to poorly differentiated and undifferentiated carcinomas of the thyroid gland. J. Clin. Investig. 1993, 91, 1753–1760. [Google Scholar] [CrossRef]
- Handkiewicz-Junak, D.; Czarniecka, A.; Jarząb, B. Molecular prognostic markers in papillary and follicular thyroid cancer: Current status and future directions. Mol. Cell. Endocrinol. 2010, 322, 8–28. [Google Scholar] [CrossRef]
- Passler, C.; Scheuba, C.; Prager, G.; Kaczirek, K.; Kaserer, K.; Zettinig, G.; Niederle, B. Prognostic factors of papillary and follicular thyroid cancer: Differences in an iodine-replete endemic goiter region. Endocr. Relat. Cancer 2004, 11, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Sondermann, A.; Andreghetto, F.M.; Moulatlet, A.C.B.; da Silva Victor, E.; de Castro, M.G.; Nunes, F.D.; Brandão, L.G.; Severino, P. MiR-9 and miR-21 as prognostic biomarkers for recurrence in papillary thyroid cancer. Clin. Exp. Metastasis 2015, 32, 521–530. [Google Scholar] [CrossRef]
- Dai, L.; Wang, Y.; Chen, L.; Zheng, J.; Li, J.; Wu, X. MiR-221, a potential prognostic biomarker for recurrence in papillary thyroid cancer. World J. Surg. Oncol. 2017, 15, 11. [Google Scholar] [CrossRef]
- Geraldo, M.V.; Nakaya, H.I.; Kimura, E.T. Down-regulation of 14q32-encoded miRNAs and tumor suppressor role for miR-654-3p in papillary thyroid cancer. Oncotarget 2017, 8, 9597. [Google Scholar] [CrossRef]
- Haghpanah, V.; Fallah, P.; Tavakoli, R.; Naderi, M.; Samimi, H.; Soleimani, M.; Larijani, B. Antisense-miR-21 enhances differentiation/apoptosis and reduces cancer stemness state on anaplastic thyroid cancer. Tumor Biol. 2016, 37, 1299–1308. [Google Scholar] [CrossRef]
- Frezzetti, D.; De Menna, M.; Zoppoli, P.; Guerra, C.; Ferraro, A.; Bello, A.; De Luca, P.; Calabrese, C.; Fusco, A.; Ceccarelli, M. Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene 2011, 30, 275–286. [Google Scholar] [CrossRef]
- Pennelli, G.; Galuppini, F.; Barollo, S.; Cavedon, E.; Bertazza, L.; Fassan, M.; Guzzardo, V.; Pelizzo, M.R.; Rugge, M.; Mian, C. The PDCD4/miR-21 pathway in medullary thyroid carcinoma. Hum. Pathol. 2015, 46, 50–57. [Google Scholar] [CrossRef]
- Durante, C.; Haddy, N.; Baudin, E.; Leboulleux, S.; Hartl, D.; Travagli, J.; Caillou, B.; Ricard, M.; Lumbroso, J.; De Vathaire, F. Long-term outcome of 444 patients with distant metastases from papillary and follicular thyroid carcinoma: Benefits and limits of radioiodine therapy. J. Clin. Endocrinol. Metab. 2006, 91, 2892–2899. [Google Scholar] [CrossRef]
- Schlumberger, M.; Tubiana, M.; Florent De, V.; Hill, C.; Paule, G.; Jean-Paul, T.; Philippe, F.; Jean, L.; Bernard, C.; Claude, P. Long-term results of treatment of 283 patients with lung and bone metastases from differentiated thyroid carcinoma. J. Clin. Endocrinol. Metab. 1986, 63, 960–967. [Google Scholar]
- Buffet, C.; Wassermann, J.; Hecht, F.; Leenhardt, L.; Dupuy, C.; Groussin, L.; Lussey-Lepoutre, C. Redifferentiation of radioiodine-refractory thyroid cancers. Endocr. Relat. Cancer 2020, 27, R113–R132. [Google Scholar] [CrossRef]
- Riesco-Eizaguirre, G.; Rodríguez, I.; De la Vieja, A.; Costamagna, E.; Carrasco, N.; Nistal, M.; Santisteban, P. The BRAFV600E oncogene induces transforming growth factor β secretion leading to sodium iodide symporter repression and increased malignancy in thyroid cancer. Cancer Res. 2009, 69, 8317–8325. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, A.; Scarberry, D.; Green, J.A.; Zhang, X.; Selmi-Ruby, S.; Jhiang, S.M. Modulation of thyroidal radioiodide uptake by oncological pipeline inhibitors and Apigenin. Oncotarget 2015, 6, 31792. [Google Scholar] [CrossRef]
- Pak, K.; Shin, S.; Kim, S.-J.; Kim, I.-J.; Chang, S.; Koo, P.; Kwak, J.; Kim, J.-H. Response of retinoic acid in patients with radioactive iodine-refractory thyroid Cancer: A meta-analysis. Oncol. Res. Treat. 2018, 41, 100–104. [Google Scholar] [CrossRef]
- Fu, H.; Cheng, L.; Jin, Y.; Cheng, L.; Liu, M.; Chen, L. MAPK Inhibitors Enhance HDAC Inhibitor-Induced Redifferentiation in Papillary Thyroid Cancer Cells Harboring BRAFV600E: An In Vitro Study. Mol. Ther. Oncolytics 2019, 12, 235–245. [Google Scholar] [CrossRef]
- Rosenbaum-Krumme, S.J.; Freudenberg, L.S.; Jentzen, W.; Bockisch, A.; Nagarajah, J. Effects of rosiglitazone on radioiodine negative and progressive differentiated thyroid carcinoma as assessed by 124I PET/CT imaging. Clin. Nucl. Med. 2012, 37, e47–e52. [Google Scholar] [CrossRef]
- Li, L.; Lv, B.; Chen, B.; Guan, M.; Sun, Y.; Li, H.; Zhang, B.; Ding, C.; He, S.; Zeng, Q. Inhibition of miR-146b expression increases radioiodine-sensitivity in poorly differential thyroid carcinoma via positively regulating NIS expression. Biochem. Biophys. Res. Commun. 2015, 462, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Riesco-Eizaguirre, G.; Wert-Lamas, L.; Perales-Patón, J.; Sastre-Perona, A.; Fernández, L.P.; Santisteban, P. The miR-146b-3p/PAX8/NIS regulatory circuit modulates the differentiation phenotype and function of thyroid cells during carcinogenesis. Cancer Res. 2015, 75, 4119–4130. [Google Scholar] [CrossRef] [PubMed]
- Damanakis, A.I.; Eckhardt, S.; Wunderlich, A.; Roth, S.; Wissniowski, T.T.; Bartsch, D.K.; Di Fazio, P. MicroRNAs let7 expression in thyroid cancer: Correlation with their deputed targets HMGA2 and SLC5A5. J. Cancer Res. Clin. Oncol. 2016, 142, 1213–1220. [Google Scholar] [CrossRef]
- Sahin Lacin, E.E.; Karakas, Y.; Yalcin, S. Metastatic medullary thyroid cancer: A dramatic response to a systemic chemotherapy (temozolomide and capecitabine) regimen. Oncotargets Ther. 2015, 8, 1039. [Google Scholar]
- Chu, E.; Sartorelli, A. Cancer chemotherapy. Basic Clin. Pharmacol. 2004, 10, 878–907. [Google Scholar]
- Stein, R.; Chen, S.; Reed, L.; Richel, H.; Goldenberg, D.M. Combining radioimmunotherapy and chemotherapy for treatment of medullary thyroid carcinoma: Effectiveness of dacarbazine. Cancer 2002, 94, 51–61. [Google Scholar] [CrossRef]
- Wilson, P.; Millar, B.; Brierley, J. The management of advanced thyroid cancer. Clin. Oncol. 2004, 16, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Lessin, L.S.; Min, M. Chemotherapy for thyroid cancer. In Thyroid Cancer; Springer: Berlin/Heidelberg, Germany, 2000; pp. 179–182. [Google Scholar]
- Gottlieb, J.A.; Hill, C.S., Jr. Chemotherapy of thyroid cancer with adriamycin: Experience with 30 patients. New Engl. J. Med. 1974, 290, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Brose, M.S.; Nutting, C.M.; Jarzab, B.; Elisei, R.; Siena, S.; Bastholt, L.; De La Fouchardiere, C.; Pacini, F.; Paschke, R.; Shong, Y.K. Sorafenib in radioactive iodine-refractory, locally advanced or metastatic differentiated thyroid cancer: A randomised, double-blind, phase 3 trial. Lancet 2014, 384, 319–328. [Google Scholar] [CrossRef]
- Schlumberger, M.; Tahara, M.; Wirth, L.J.; Robinson, B.; Brose, M.S.; Elisei, R.; Habra, M.A.; Newbold, K.; Shah, M.H.; Hoff, A.O. Lenvatinib versus placebo in radioiodine-refractory thyroid cancer. New Engl. J. Med. 2015, 372, 621–630. [Google Scholar] [CrossRef]
- Gianoukakis, A.G.; Dutcus, C.E.; Batty, N.; Guo, M.; Baig, M. Prolonged duration of response in lenvatinib responders with thyroid cancer. Endocr. Relat. Cancer 2018, 25, 699–704. [Google Scholar] [CrossRef]
- Wells, S.A., Jr.; Robinson, B.G.; Gagel, R.F.; Dralle, H.; Fagin, J.A.; Santoro, M.; Baudin, E.; Elisei, R.; Jarzab, B.; Vasselli, J.R. Vandetanib in patients with locally advanced or metastatic medullary thyroid cancer: A randomized, double-blind phase III trial. J. Clin. Oncol. 2012, 30, 134. [Google Scholar] [CrossRef]
- Schlumberger, M.; Elisei, R.; Müller, S.; Schöffski, P.; Brose, M.S.; Shah, M.H.; Licitra, L.F.; Jarzab, B.; Medvedev, V.; Kreissl, M. Final overall survival analysis of EXAM, an international, double-blind, randomized, placebo-controlled phase III trial of cabozantinib (Cabo) in medullary thyroid carcinoma (MTC) patients with documented RECIST progression at baseline. Am. Soc. Clin. Oncol. 2015. [Google Scholar] [CrossRef]
- Subbiah, V.; Kreitman, R.J.; Wainberg, Z.A.; Cho, J.Y.; Schellens, J.H.; Soria, J.C.; Wen, P.Y.; Zielinski, C.; Cabanillas, M.E.; Urbanowitz, G. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600–mutant anaplastic thyroid cancer. J. Clin. Oncol. 2018, 36, 7. [Google Scholar] [CrossRef]
- Hong, D.S.; DuBois, S.G.; Kummar, S.; Farago, A.F.; Albert, C.M.; Rohrberg, K.S.; van Tilburg, C.M.; Nagasubramanian, R.; Berlin, J.D.; Federman, N. Larotrectinib in patients with TRK fusion-positive solid tumours: A pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020, 21, 531–540. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: Integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef]
- Robbins, R.J.; Srivastava, S.; Shaha, A.; Ghossein, R.; Larson, S.M.; Fleisher, M.; Tuttle, R.M. Factors influencing the basal and recombinant human thyrotropin-stimulated serum thyroglobulin in patients with metastatic thyroid carcinoma. J. Clin. Endocrinol. Metab. 2004, 89, 6010–6016. [Google Scholar] [CrossRef]
- Schlumberger, M.; Hitzel, A.; Toubert, M.; Corone, C.; Troalen, F.; Schlageter, M.; Claustrat, F.; Koscielny, S.; Taieb, D.; Toubeau, M. Comparison of seven serum thyroglobulin assays in the follow-up of papillary and follicular thyroid cancer patients. J. Clin. Endocrinol. Metab. 2007, 92, 2487–2495. [Google Scholar] [CrossRef]
- Spencer, C.A. Clinical utility of thyroglobulin antibody (TgAb) measurements for patients with differentiated thyroid cancers (DTC). J. Clin. Endocrinol. Metab. 2011, 96, 3615–3627. [Google Scholar] [CrossRef]
- Giovanella, L.; Suriano, S.; Ceriani, L.; Verburg, F.A. Undetectable Thyroglobulin in Patients with Differentiated Thyroid Carcinoma and Residual Radioiodine Uptake on a Postablation Whole-Body Scan. Clin. Nucl. Med. 2011, 36, 109–112. [Google Scholar] [CrossRef]
- Diesch, J.; Zwick, A.; Garz, A.-K.; Palau, A.; Buschbeck, M.; Götze, K.S. A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers. Clin. Epigenetics 2016, 8, 71. [Google Scholar] [CrossRef]
- Bainschab, A.; Quehenberger, F.; Greinix, H.T.; Krause, R.; Wölfler, A.; Sill, H.; Zebisch, A. Infections in patients with acute myeloid leukemia treated with low-intensity therapeutic regimens: Risk factors and efficacy of antibiotic prophylaxis. Leuk. Res. 2016, 42, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Slaby, O.; Laga, R.; Sedlacek, O. Therapeutic targeting of non-coding RNAs in cancer. Biochem. J. 2017, 474, 4219–4251. [Google Scholar] [CrossRef]
- Wang, W.-T.; Han, C.; Sun, Y.-M.; Chen, T.-Q.; Chen, Y.-Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 55. [Google Scholar] [CrossRef]
- Xu, Y.; Han, Y.F.; Ye, B.; Zhang, Y.L.; Dong, J.D.; Zhu, S.J.; Chen, J. miR-27b-3p is Involved in Doxorubicin Resistance of Human Anaplastic Thyroid Cancer Cells via Targeting Peroxisome Proliferator-Activated Receptor Gamma. Basic Clin. Pharmacol. Toxicol. 2018, 123, 670–677. [Google Scholar] [CrossRef]
- Liu, Y.; Yue, P.; Zhou, T.; Zhang, F.; Wang, H.; Chen, X. LncRNA MEG3 enhances 131I sensitivity in thyroid carcinoma via sponging miR-182. Biomed. Pharmacother. 2018, 105, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Feng, Z.; Chen, T.; Lv, J.; Liu, P.; Jia, L.; Zhu, J.; Chen, F.; Yang, C.; Deng, Z. Downregulation of NEAT1 reverses the radioactive iodine resistance of papillary thyroid carcinoma cell via miR-101-3p/FN1/PI3K-AKT signaling pathway. Cell Cycle 2019, 18, 167–203. [Google Scholar] [CrossRef]
- Xiang, C.; Zhang, M.-L.; Zhao, Q.-Z.; Xie, Q.-P.; Yan, H.-C.; Yu, X.; Wang, P.; Wang, Y. LncRNA-SLC6A9-5: 2: A potent sensitizer in 131I-resistant papillary thyroid carcinoma with PARP-1 induction. Oncotarget 2017, 8, 22954. [Google Scholar] [CrossRef]
- Tan, H.-L.; Chan, K.-G.; Pusparajah, P.; Saokaew, S.; Duangjai, A.; Lee, L.-H.; Goh, B.-H. Anti-cancer properties of the naturally occurring aphrodisiacs: Icariin and its derivatives. Front. Pharmacol. 2016, 7, 191. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, K.; Zhao, F. Icariin regulates the proliferation and apoptosis of human ovarian cancer cells through microRNA-21 by targeting PTEN, RECK and Bcl-2. Oncol. Rep. 2015, 33, 2829–2836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, Y.; Zhu, Z.; Gong, W.; Liu, X.; Hou, Q.; Sun, Y.; Chai, J.; Zou, L.; Zhou, T. Icariin enhances radiosensitivity of colorectal cancer cells by suppressing NF-κB activity. Cell Biochem. Biophys. 2014, 69, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Leon, A.; Ezponda, T.; Meydan, C.; Valcárcel, L.V.; Ordoñez, R.; Kulis, M.; Garate, L.; Miranda, E.; Segura, V.; Guruceaga, E. Role of lncRNAs as prognostic factor and potential therapeutic target in Multiple Myeloma. Clin. Lymphoma Myeloma Leuk. 2019, 19, e354–e355. [Google Scholar] [CrossRef]
- Xiang, Z.; Song, S.; Zhu, Z.; Sun, W.; Sun, S.; Li, Q.S.; Yu, Y.; Li, K.K. LncRNAs GIHCG and SPINT1-AS1 are crucial factors for pan-cancer cells sensitivity to lapatinib. Front. Genet. 2019, 10, 25. [Google Scholar] [CrossRef]
- Tsang, S.; Patel, T.; Yustein, J.T. Long non-coding RNAs regulation of therapeutic resistance. Cancer Drug Resist. 2019, 2, 550–567. [Google Scholar] [CrossRef]
- Sheng, S.-R.; Wu, J.-S.; Tang, Y.-L.; Liang, X.-H. Long noncoding RNAs: Emerging regulators of tumor angiogenesis. Future Oncol. 2017, 13, 1551–1562. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, R45–R53. [Google Scholar] [CrossRef]
- Moradi, A.; Naiini, M.R.; Yazdanpanahi, N.; Tabatabaeian, H.; Nabatchian, F.; Baghi, M.; Azadeh, M.; Ghaedi, K. Evaluation of The Expression Levels of Three Long Non-Coding RNAs in Multiple Sclerosis. Cell J. 2020, 22, 165–170. [Google Scholar]
- Shao, F.; Huang, M.; Meng, F.; Huang, Q. Circular RNA signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Front. Pharmacol. 2018, 9, 584. [Google Scholar] [CrossRef]
- Kun-Peng, Z.; Xiao-Long, M.; Lei, Z.; Chun-Lin, Z.; Jian-Ping, H.; Tai-Cheng, Z. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics 2018, 10, 1327–1346. [Google Scholar] [CrossRef]
- Yu, W.; Peng, W.; Sha, H.; Li, J. Hsa_circ_0003998 promotes chemoresistance via modulation of miR-326 in lung adenocarcinoma cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2019, 27, 623–628. [Google Scholar] [CrossRef]
- Shang, J.; Chen, W.-M.; Wang, Z.-H.; Wei, T.-N.; Chen, Z.-Z.; Wu, W.-B. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p–XIAP axis. Exp. Hematol. 2019, 70, 42–54.e43. [Google Scholar] [CrossRef]
- Huang, X.; Li, Z.; Zhang, Q.; Wang, W.; Li, B.; Wang, L.; Xu, Z.; Zeng, A.; Zhang, X.; Zhang, X. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer 2019, 18, 71. [Google Scholar] [CrossRef]
- Selvam, C.; Mutisya, D.; Prakash, S.; Ranganna, K.; Thilagavathi, R. Therapeutic potential of chemically modified si RNA: Recent trends. Chem. Biol. Drug Des. 2017, 90, 665–678. [Google Scholar] [CrossRef]
- Steichen, S.D.; Caldorera-Moore, M.; Peppas, N.A. A review of current nanoparticle and targeting moieties for the delivery of cancer therapeutics. Eur. J. Pharm. Sci. 2013, 48, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Kramps, T.; Probst, J. Messenger RNA-based vaccines: Progress, challenges, applications. Wiley Interdiscip. Rev. RNA 2013, 4, 737–749. [Google Scholar] [CrossRef]
- Tatiparti, K.; Sau, S.; Kashaw, S.K.; Iyer, A.K. siRNA delivery strategies: A comprehensive review of recent developments. Nanomaterials 2017, 7, 77. [Google Scholar] [CrossRef]
- Dammes, N.; Peer, D. Paving the road for RNA therapeutics. Trends Pharmacol. Sci. 2020, 41, 755–775. [Google Scholar] [CrossRef]
- Sugawa, H.; Smith, E.; Imura, H.; Mori, T. A thyroid cancer specific monoclonal antibody which recognizes cryptic epitope (s) of human thyroglobulin. Mol. Cell. Endocrinol. 1993, 93, 207–211. [Google Scholar] [CrossRef]
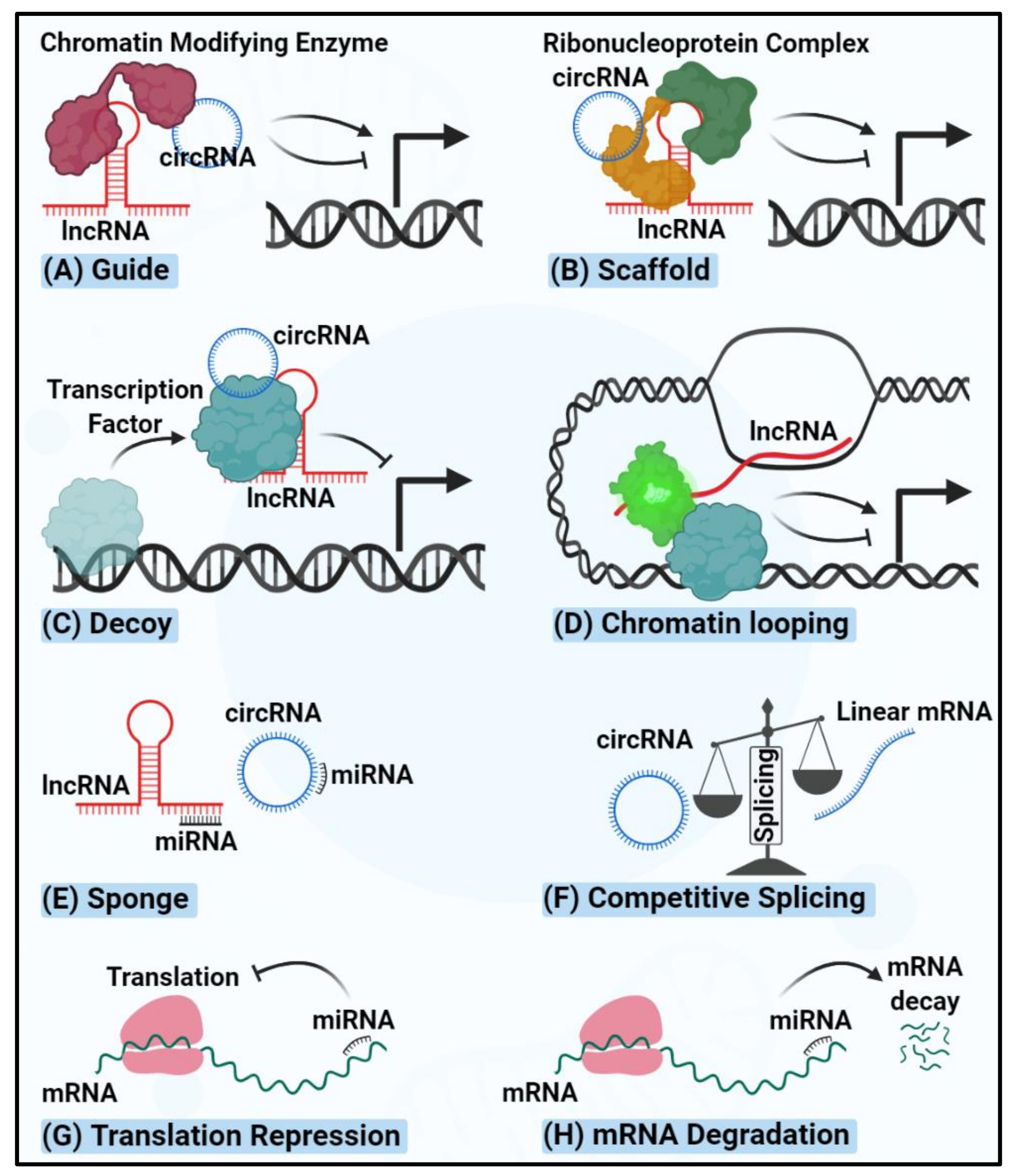
| miRNA | Alteration | Mechanism | Thyroid Cancer Type | Sample | Ref. |
|---|---|---|---|---|---|
| miR-146a/b-5p | ↑ | ↓ RAR-β | PTC | Cell line and tissue | [52] |
| miR-155 | ↓ SOCS1 | ATC | [53] | ||
| miR-21 | ↓ VHL → ↑ N-cadherin and vimentin | PTC | [54] | ||
| miR-574-5p | ↓ SCAI → ↑ β-catenin | [55] | |||
| miR-625-3p | ↑ Bcl-2, ↓ Bax, and cleaved caspase 3/9 | [56] | |||
| miR-96 | ↓ FOXO1 → ↓ Bim | Cell line | [57] | ||
| Let-7a | ↓ | Not known | Not known | Tissue | [58] |
| miR-125a-5p | ↑ CD147 → ↓ glucose metabolism | PTC, FTC, MTC and ATC | Cell line and tissue | [59] | |
| miR-1266 | ↑ FGFR2 | PTC | [60] | ||
| miR-141-3p | ↑ YY1 | [61] | |||
| miR-144 | ↑ TGF-α | ATC | [62] | ||
| miR-148a | ↑ INO80 | ATC cancer stem cells | [63] | ||
| miR-150 | ↑ROCK1 | PTC | Cell line and tissue | [64] | |
| miR-195 | ↑ CCND1 and FGF2 → ↑ β-catenin, c-Myc, cyclin D1 and MMP-13 | [65] | |||
| miR-199b-5p | ↑ STON2 → ↑ N-cadherin and fibronectin | [66] | |||
| miR-375 | ↑ ERBB2 | [57] | |||
| miR-497 | ↑ AKT3 | [67] | |||
| miR-524-5p | ↑ FOXE1 and ITGA3 | [68] | |||
| miR-7 | ↑ PAK1 | [69] | |||
| miR-873-5p | ↑ CXCL16 → ↑ p-p65 and p-Rel-B, MMP1, MMP9 and MMP13 | [70] | |||
| miR-9-5p | ↑ BRAF | [67] |
| LncRNA | Alteration | Mechanism | Thyroid Cancer Type | Sample | Ref. |
|---|---|---|---|---|---|
| CASC9 | ↑ | ↓ miR-488-3p → ↑ ADAM9 → ↑ EGFR/PI3K/Akt pathway activation. | PTC | Cell line and tissue | [114] |
| DLX6-AS1 | Negatively correlated with UPF1 | Not known | [115] | ||
| ENST00000539653.1 (ENS-653) | Not known | PTC | Tissue | [116] | |
| H19 | Not known | ATC | [117] | ||
| Not known | Not known | [58] | |||
| ↓ miR-3126-5p → ↑ ER-β | PTCSCs and PTC tissue | [118] | |||
| HCP5 | ↓ miR-22-3p, miR-186-5p and miR-216a-5p → ↑ ST6GAL2 | FTC | Cell line and tissue | [119] | |
| LINC00152 | ↓ miR-497 → ↑ BDNF | PTC | [120] | ||
| LINC00514 | ↓ miR-204-3p → ↑ CDC23 | [121] | |||
| LINC00941 | ↓ CDH6 | [122] | |||
| MALAT1 | No mechanism | [123] | |||
| n340790 | ↓ miR-1254 | Not known | [119] | ||
| NEAT1 | ↓ miR-9-5p ↑ SPAG9 | ATC | [67] | ||
| UNC5B-AS1 | Not known | PTC | Tissue | [124] | |
| XIST | ↓ miR-34a → ↑ MET → PI3K/Akt activation | Not known | Cell line and tissue | [125] | |
| H19 | ↓ | Not known | FTC | Tissue | [126] |
| H19 | Not known | PTC | [127] | ||
| LINC003121 | ↑ PI3K and p-Akt | Not known | Cell line and tissue | [128] | |
| PAR5 | ↑ EZH2 → ↓ E-cadherin | ATC | [129] | ||
| PTCSC3 | ↑ STAT3 → ↑ INO80 | [130] | |||
| ↑ miR-574-5p → ↓ SCAI → ↑ β-catenin → ↑ Wnt pathway activation | PTC | [55] | |||
| SNHG3 | ↑ PI3K/Akt/mTOR pathway | [131] |
| circRNA | Alteration | Mechanism | Sample | Thyroid Cancer Type | Ref. |
|---|---|---|---|---|---|
| circ_0008274 | ↑ | ↓ AMPK/mTOR signaling pathway | Cell line and tissue | PTC | [141] |
| circEIF6 | ↑ | ↓ miR-144-3p → ↑ TGF-α | Cell line and tissue | PTC & ATC | [142] |
| circFOXM1 | ↑ | ↓ miR-1179 → ↑ HMGB1 | Cell line and tissue | PTC | [143] |
| ncRNA | ncRNA Type | Source | Finding | Thyroid Samples | Ref. |
|---|---|---|---|---|---|
| miR-138-1-3p miR-139-5p miR-146b-5p miR-155miR-204-5p miR-222-3pmiR-29b-1-5p miR-31-5p miR-375 miR-551b-3p | miRNA | FNA | miRNA testing, recently commercialized as ThyraMIR, identified 64% of malignant cases and 98% of benign cases correctly. | Not known | [170] |
| miR-146b | Higher expression in PTC FNA samples | PTC | [171] | ||
| miR-132-3p miR-146a-5pmiR-17-5p miR-183-3p miR-222-3p miR-451a | Serum | miR-222-3p and miR-17-5p can accurately discriminate MTC from the benign nodule and healthy control groups | PTC, MTC, benign nodules and controls | [172] | |
| miR-146a-5p miR-221-3p miR-222-3p | High pre-surgical expressionLow post-surgical expression | PTC | [173] | ||
| miR-146b miR-21 miR-221 miR-222 | Higher expression in PTM serum samples | [174] | |||
| miR-146a-5p miR-199b-3p | Lower expression in PTC serum as compared to benign serum samples | [175] | |||
| let-7b-5pmiR-10a-5p | Higher expression in PTC serum as compared to benign serum samples | ||||
| miR-423-5p | Higher expression in PTC serum samples | [176] | |||
| let-7a | Tissue | Lower expression in thyroid tumor samples | Not known | [58] | |
| H19 | lncRNA | Higher expression in thyroid tumor samples | |||
| H19 | Lower expression in thyroid tumor samples as compared to benign samples | PTC | [127] | ||
| MALAT1 | Higher expression in thyroid tumor samples | [123] | |||
| n340790 | Not known | [119] | |||
| UNC5B-AS1 | PTC | [124] |
| ncRNA | ncRNA Type | Prognostic Significance | Thyroid Cancer Type | Ref. |
|---|---|---|---|---|
| Let-7a | miRNA | Negative correlation with higher TNM stages lymph node metastasis and lower 5-year survival | Not known | [58] |
| miR-141-3p | Negative association with TNM stage and lymph node metastasis | PTC | [61] | |
| miR-146b miR-21 miR-222 | Poor prognosis | [174] | ||
| miR-150 | Negative association with TNM stage and lymph node metastasis | [64] | ||
| miR-199b-5p | Negative association with stage | [66] | ||
| miR-21 | Poor prognosis | [54] | ||
| miR-21 miR-9 | Independent prognostic factors of PTC recurrence | [201] | ||
| miR-221 | Independent prognostic factors of PTC recurrence | [202] | ||
| miR-654-3p | Down-regulation upon a long-term PTC progression in BRAFV600E-transgenic mice | [203] | ||
| miR-7 | Negative association with stage | [69] | ||
| CASC9 | lncRNA | Positive association with large tumor size, advanced stage, or lymph node metastasis. | [114] | |
| ENST00000539653.1 (ENS-653) | Positive association with larger tumor size, more advanced clinical stage and poorer disease-free survival | [116] | ||
| H19 | Positive correlation with higher TNM stages lymph node metastasis and lower 5-year survival | Not known | [58] | |
| Positive correlation with poor overall survival | PTCSCs and PTC tissue | [118] | ||
| Negative correlation with extrathyroid extension, tumor size, histological aggressive type, pathological lateral node metastasis and poorer disease-free survivalIndependent risk factor for extrathyroidal extension and lymph node metastasis. | PTC | [127] | ||
| Negative association with tumor size, distant metastasis and vascular invasion | FTC | [126] | ||
| MALAT1 | Positive correlation with tumor size and lymph node metastases | PTC | [123] | |
| n340790 | Positive correlation with primary clinicopathological characteristics (good prognostic factor) | Not known | [119] | |
| SNHG3 | Negative association with stage and poor prognosis | PTC | [131] | |
| UNC5B-AS1 | Positive correlation with lymph node metastasis, tumor size and histological type | [124] |
| ncRNA | ncRNA Type | Therapeutic Significance | Thyroid Cancer Type | Ref. |
|---|---|---|---|---|
| miR-144 | miRNA | ↑ Sensitivity to cisplatin | ATC | [62] |
| miR-146b | ↓ Radioiodine uptake | FTC | [215] | |
| miR-27b-3p | ↑ Resistance to doxorubicin | ATC | [240] | |
| miR-625-3p | Target of icariin anti-tumor substance | PTC | [56] | |
| MEG3a | lncRNA | ↑ Resistance to radioactive iodine | FTC and PTC | [241] |
| NEAT1 | ↑ Resistance to cisplatin | ATC | [67] | |
| NEAT1 | ↑ Resistance to radioactive iodine | PTC | [242] | |
| PTCSC3 | ↑ Resistance to doxorubicin | ATC | [130] | |
| SLC6A9-5:2 | ↑ Resistance to radioactive iodine | PTC | [243] | |
| circEIF6 | circRNA | ↑ Resistance to cisplatin | PTC and ATC | [142] |
| Identifier | ncRNA Type | Type of Sample | Study Type | Observational Model | Clinical Significance | Status |
|---|---|---|---|---|---|---|
| NCT03469544 | HOTAIR | Peripheral blood samples | Observational | Case-Control | Diagnostic biomarker | Not yet recruiting |
| NCT01964508 | miRNAs | FNA samples | Observational | Cohort | Diagnostic biomarker | Not yet recruiting |
| NCT04594720 | lncRNAs | Peripheral blood samples | Observational | Case-Control | Diagnostic biomarker | Recruiting completed |
| NCT01240590 | miRNAs | ATC tumor samples | Interventional | Parallel Assignment | Therapeutic biomarker for Crolibulin and cisplatin | Recruiting completed |
| NCT04285476 | miRNAs | Thyroid carcinoma | Interventional | Single Group Assignment | Diagnostic biomarker | Not yet recruiting |
| NCT00689065 | siRNA | Variety of solid tumors including Thyroid carcinoma | Interventional | Single Group Assignment | RNA-based therapy (CALAA-01) | Recruitment terminated |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabatabaeian, H.; Peiling Yang, S.; Tay, Y. Non-Coding RNAs: Uncharted Mediators of Thyroid Cancer Pathogenesis. Cancers 2020, 12, 3264. https://doi.org/10.3390/cancers12113264
Tabatabaeian H, Peiling Yang S, Tay Y. Non-Coding RNAs: Uncharted Mediators of Thyroid Cancer Pathogenesis. Cancers. 2020; 12(11):3264. https://doi.org/10.3390/cancers12113264
Chicago/Turabian StyleTabatabaeian, Hossein, Samantha Peiling Yang, and Yvonne Tay. 2020. "Non-Coding RNAs: Uncharted Mediators of Thyroid Cancer Pathogenesis" Cancers 12, no. 11: 3264. https://doi.org/10.3390/cancers12113264
APA StyleTabatabaeian, H., Peiling Yang, S., & Tay, Y. (2020). Non-Coding RNAs: Uncharted Mediators of Thyroid Cancer Pathogenesis. Cancers, 12(11), 3264. https://doi.org/10.3390/cancers12113264






