Cancer-Associated Stemness and Epithelial-to-Mesenchymal Transition Signatures Related to Breast Invasive Carcinoma Prognostic
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
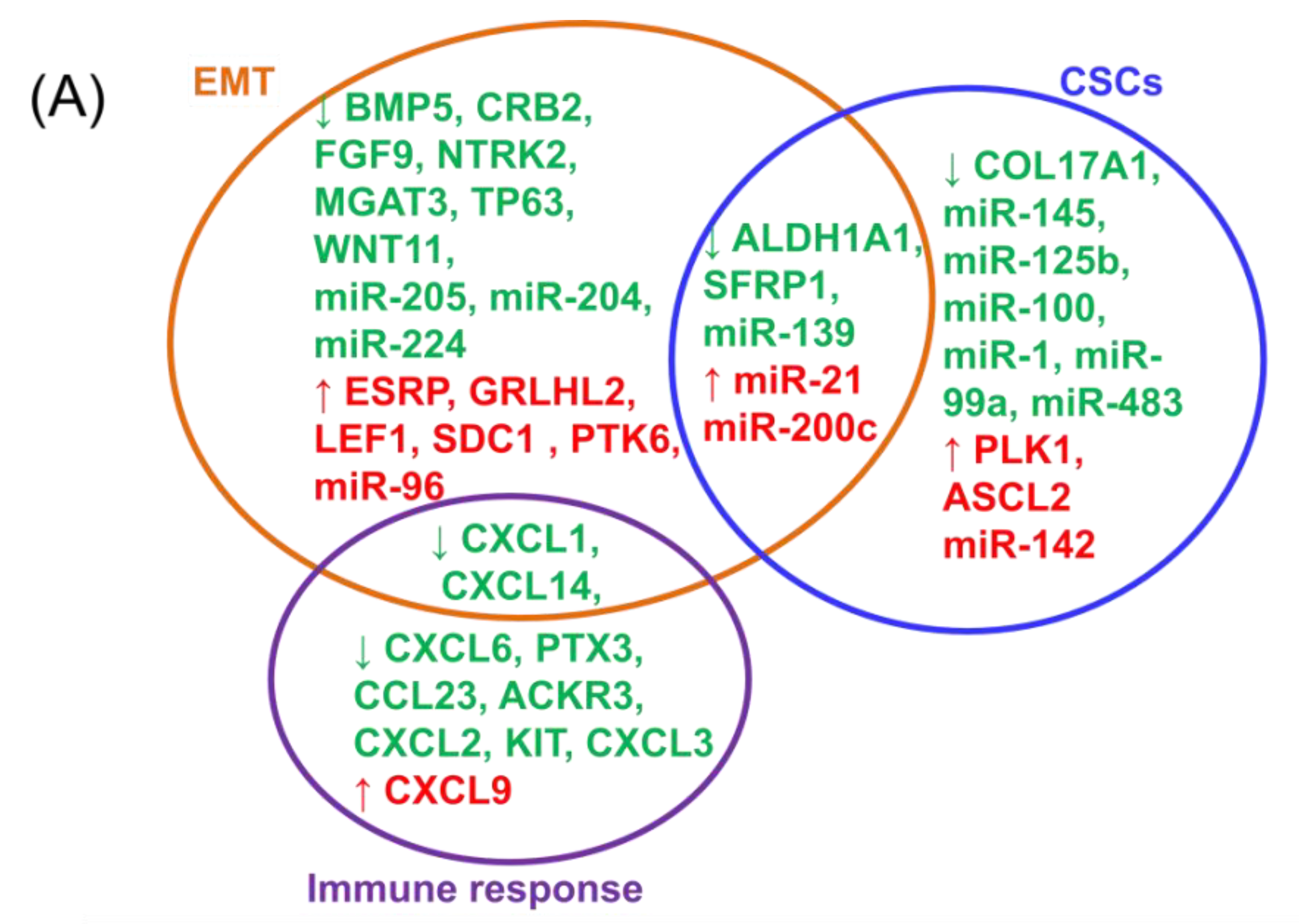
2.1. Breast Cancer Cell Analysis Reveals Distinct Expression Profiles for EMT
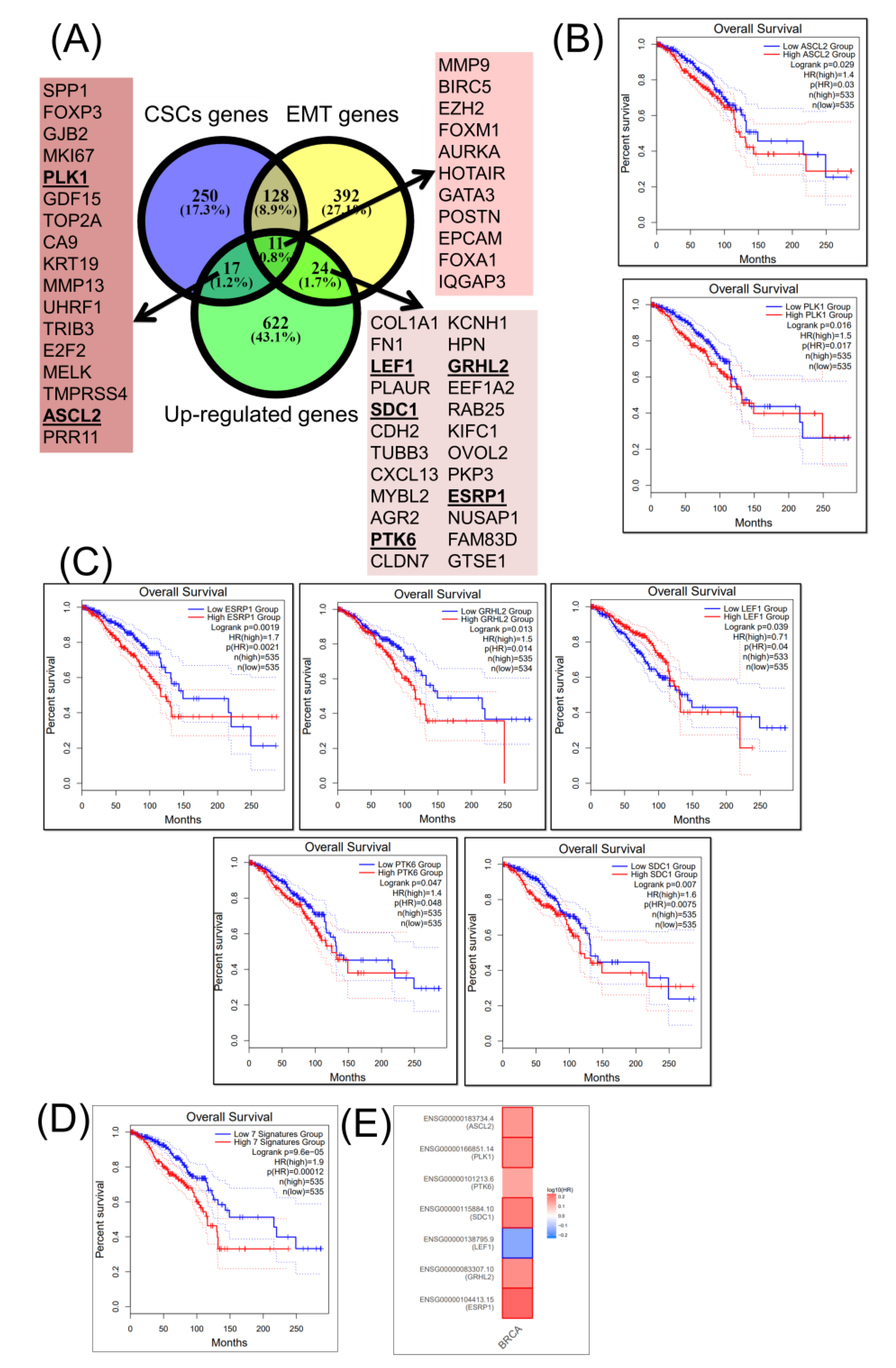
2.2. Integrating Altered Genes to Tumorigenesis and Molecular Pathways
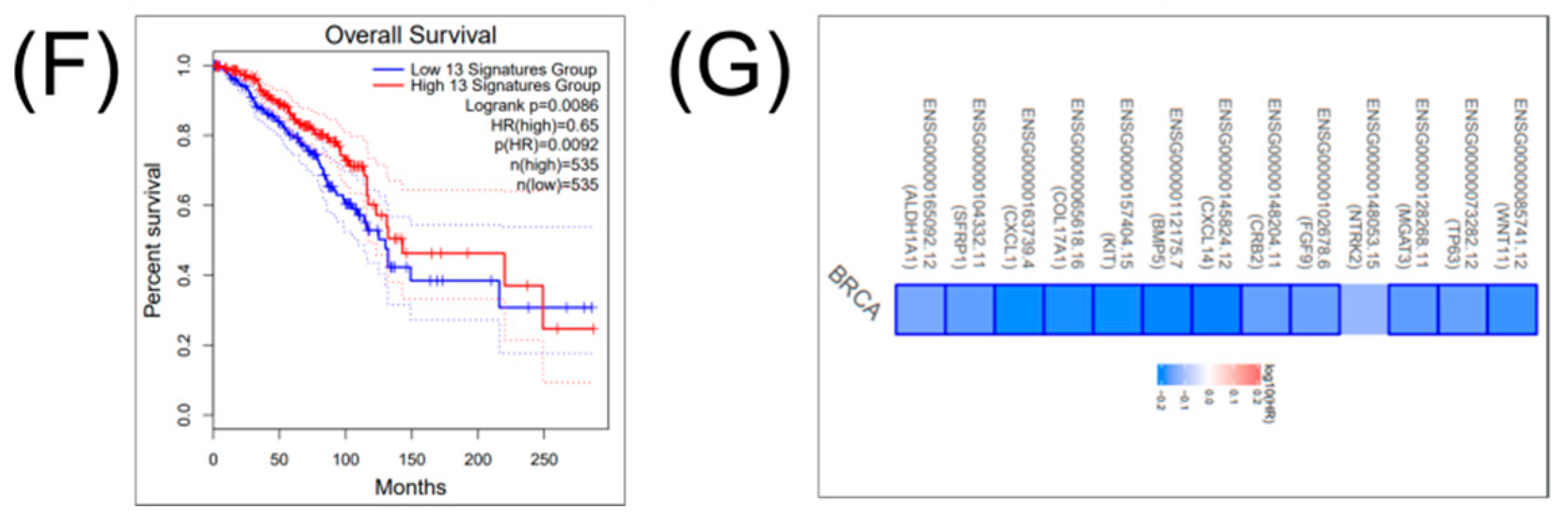
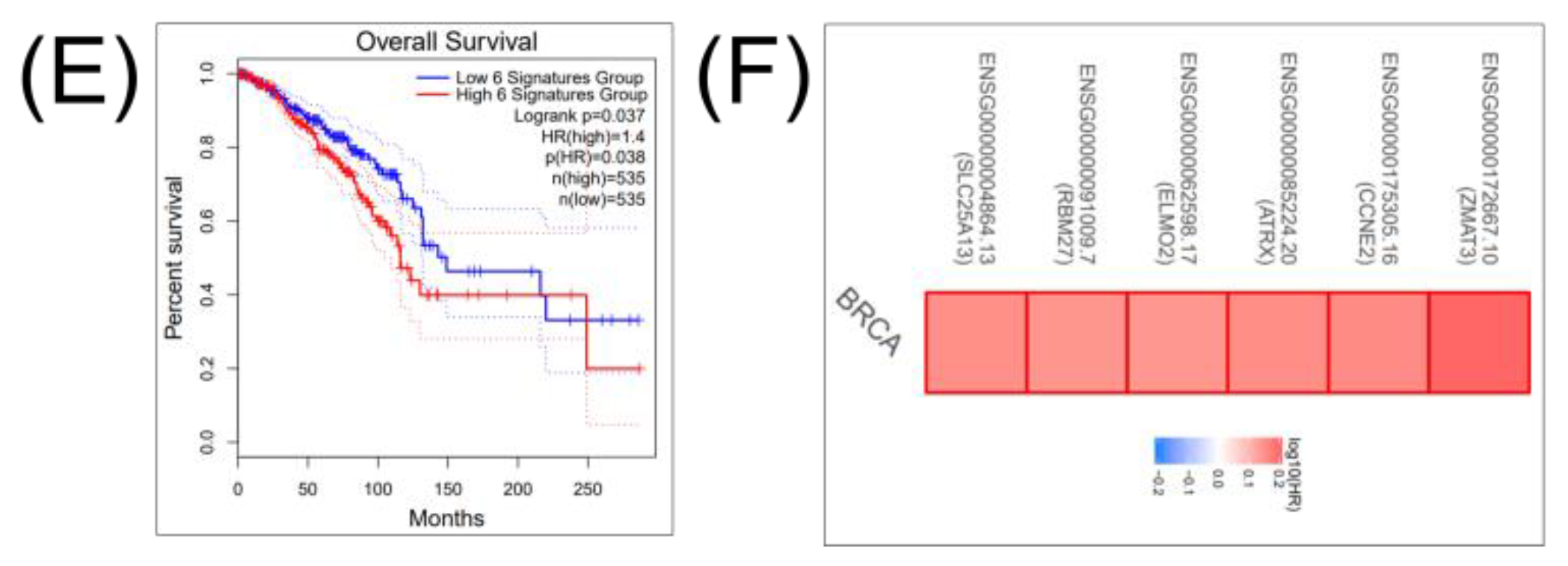
2.3. Construction of a Gene Network Involved in CSCs and EMT for Breast Cancer Genes (BRCA)
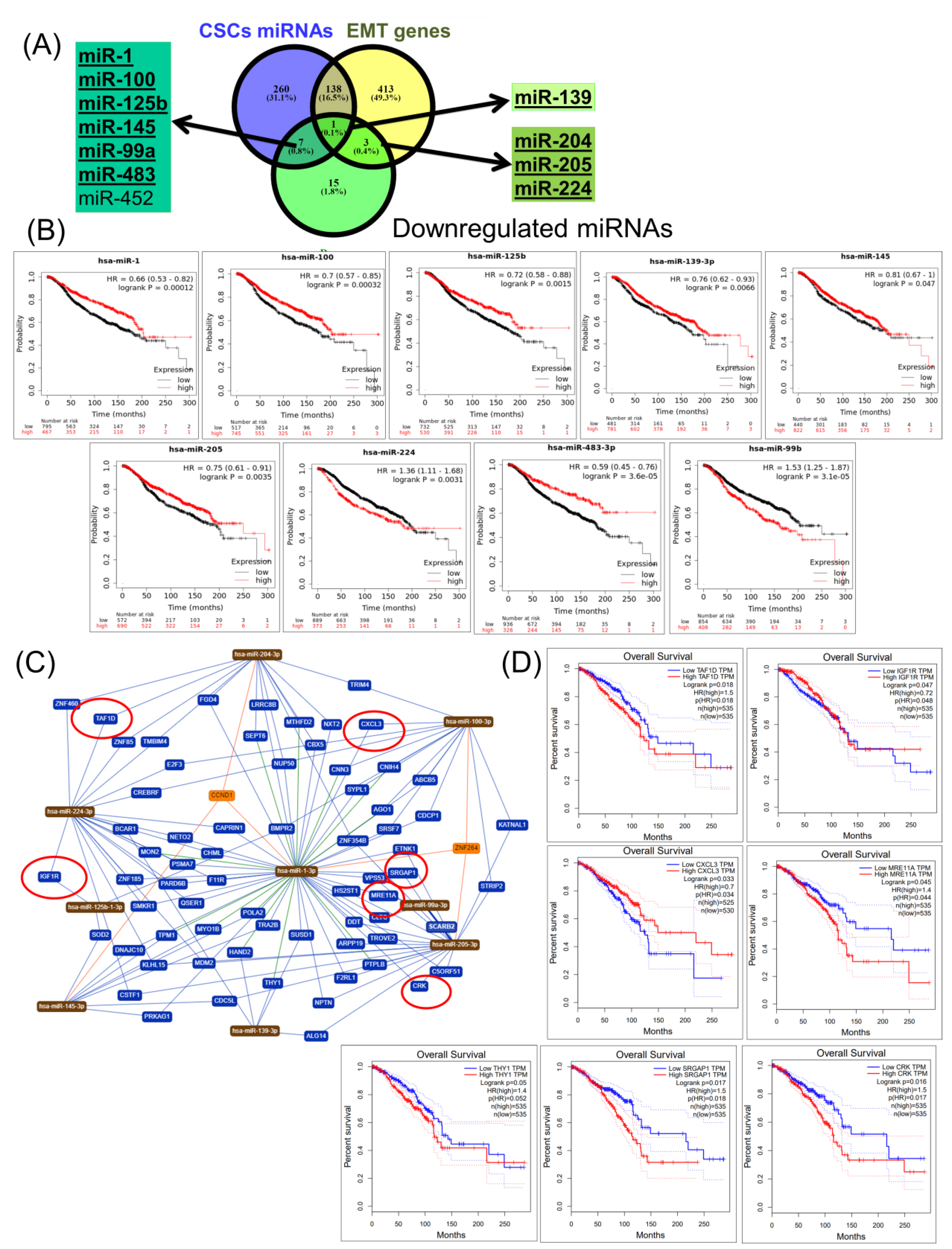
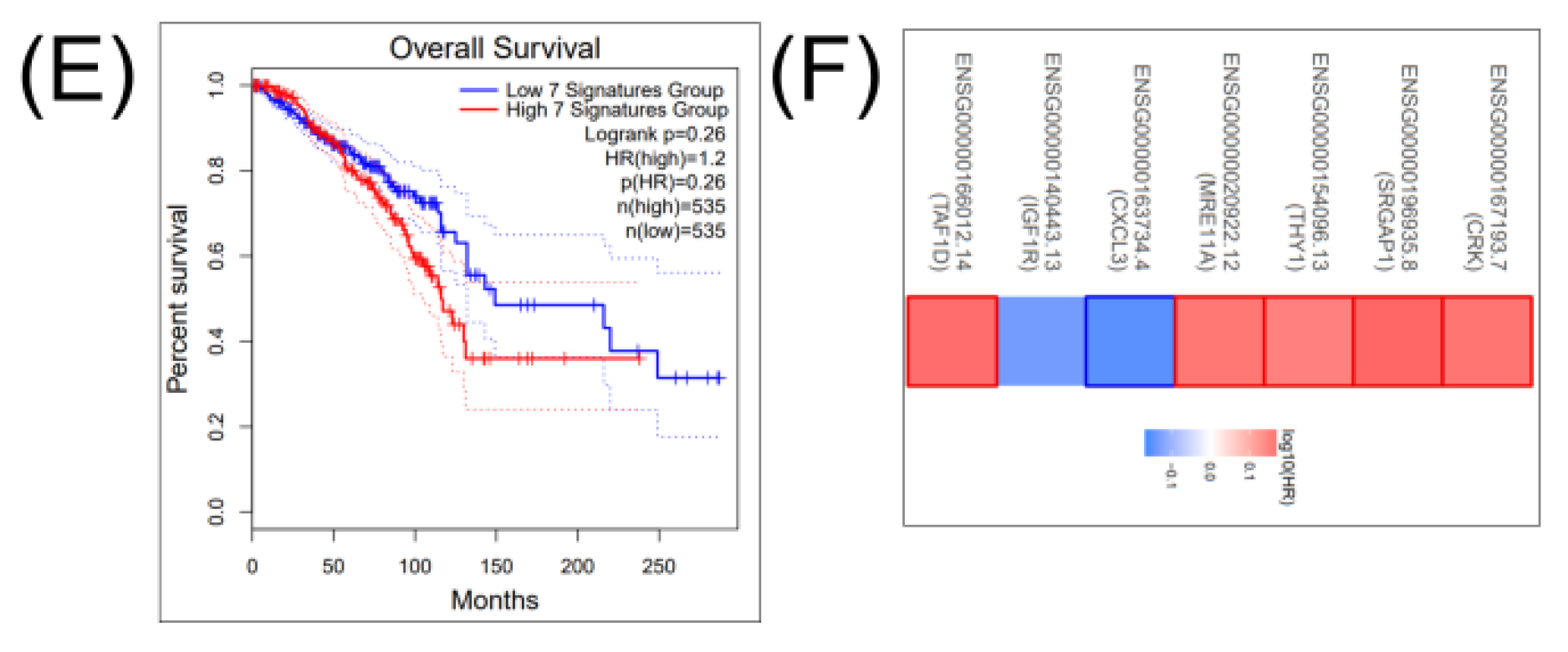
2.4. Construction of a miRNA Network Involved in both CSCs and EMT
2.5. Validation of Altered Transcriptomic Patterns by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
3. Discussion
4. Materials and Methods
4.1. Differential Gene and miRNA Expression Analysis for BRCA
4.2. Network Analysis
4.3. Survival Analysis
4.4. Gene and miRNA Validation Using Early Stage Breast Cancer Tissue Samples
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Toss, M.S.; Abidi, A.; Lesche, D.; Joseph, C.; Mahale, S.; Saunders, H.; Kader, T.; Miligy, I.M.; Green, A.R.; Gorringe, K.L.; et al. The prognostic significance of immune microenvironment in breast ductal carcinoma in situ. Br. J. Cancer 2020, 122, 1496–1506. [Google Scholar] [CrossRef]
- Ciocan-Cȃrtiţă, C.A.; Jurj, A.; Raduly, L.; Cojocneanu, R.; Moldovan, A.; Pileczki, V.; Pop, L.-A.; Budişan, L.; Braicu, C.; Korban, S.S.; et al. New perspectives in triple-negative breast cancer therapy based on treatments with TGFβ1 siRNA and doxorubicin. Mol. Cell. Biochem. 2020. [Google Scholar] [CrossRef]
- Braicu, C.; Chiorean, R.; Irimie, A.; Chira, S.; Tomuleasa, C.; Neagoe, E.; Paradiso, A.; Achimas-Cadariu, P.; Lazar, V.; Berindan-Neagoe, I. Novel insight into triple-negative breast cancers, the emerging role of angiogenesis, and antiangiogenic therapy. Expert Rev. Mol. Med. 2016, 18, e18. [Google Scholar] [CrossRef]
- Braicu, C.; Raduly, L.; Morar-Bolba, G.; Cojocneanu, R.; Jurj, A.; Pop, L.A.; Pileczki, V.; Ciocan, C.; Moldovan, A.; Irimie, A.; et al. Aberrant miRNAs expressed in HER-2 negative breast cancers patient. J. Exp. Clin. Cancer Res. 2018, 37, 257. [Google Scholar] [CrossRef]
- Cojocneanu, R.; Braicu, C.; Raduly, L.; Jurj, A.; Zanoaga, O.; Magdo, L.; Irimie, A.; Muresan, M.-S.; Ionescu, C.; Grigorescu, M.; et al. Plasma and Tissue Specific miRNA Expression Pattern and Functional Analysis Associated to Colorectal Cancer Patients. Cancers 2020, 12, 843. [Google Scholar] [CrossRef]
- Jurj, A.; Pop, L.A.; Zanoaga, O.; Ciocan-Cârtiţă, C.A.; Cojocneanu, R.; Moldovan, C.; Raduly, L.; Pop-Bica, C.; Trif, M.; Irimie, A.; et al. New Insights in Gene Expression Alteration as Effect of Paclitaxel Drug Resistance in Triple Negative Breast Cancer Cells. Cell. Physiol. Biochem. 2020, 54, 648–664. [Google Scholar]
- Zimta, A.-A.; Tigu, A.B.; Braicu, C.; Stefan, C.; Ionescu, C.; Berindan-Neagoe, I. An Emerging Class of Long Non-coding RNA With Oncogenic Role Arises From the snoRNA Host Genes. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Jiang, W.; Peng, J.; Zhang, Y.; Sharifi-Rad, J.; Jin, K. The Implications of Cancer Stem Cells for Cancer Therapy. Int. J. Mol. Sci. 2012, 13, 16636–16657. [Google Scholar] [CrossRef]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef]
- Jewer, M.; Lee, L.; Leibovitch, M.; Zhang, G.; Liu, J.; Findlay, S.D.; Vincent, K.M.; Tandoc, K.; Dieters-Castator, D.; Quail, D.F.; et al. Translational control of breast cancer plasticity. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Vila, M.; Etakahashi, R.-U.; Usuba, W.; Kohama, I.; Ochiya, T. Drug Resistance Driven by Cancer Stem Cells and Their Niche. Int. J. Mol. Sci. 2017, 18, 2574. [Google Scholar] [CrossRef] [PubMed]
- Ricardo, S.; Vieira, A.F.; Gerhard, R.; Leitão, D.; Pinto, R.; Cameselle-Teijeiro, J.F.; Milanezi, F.; Schmitt, F.; Paredes, J. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011, 64, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Kabakov, A.E.; Yakimova, A.; Matchuk, O. Molecular Chaperones in Cancer Stem Cells: Determinants of Stemness and Potential Targets for Antitumor Therapy. Cells 2020, 9, 892. [Google Scholar] [CrossRef]
- Pop-Bica, C.; Gulei, D.; Cojocneanu, R.; Braicu, C.; Petrut, B.; Zimta, A. Understanding the Role of Non-Coding RNAs in Bladder Cancer: From Dark Matter to Valuable Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 1514. [Google Scholar] [CrossRef]
- Georgakopoulos-Soares, I.; Chartoumpekis, D.V.; Kyriazopoulou, V.; Zaravinos, A. EMT Factors and Metabolic Pathways in Cancer. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Wilson, M.M.; Weinberg, R.A.; Lees, J.A.; Guen, V.J. Emerging Mechanisms by which EMT Programs Control Stemness. Trends Cancer 2020, 6, 775–780. [Google Scholar] [CrossRef]
- Park, S.-Y.; Choi, J.-H.; Nam, J.-S. Targeting Cancer Stem Cells in Triple-Negative Breast Cancer. Cancers 2019, 11, 965. [Google Scholar] [CrossRef]
- Takebe, N.; Warren, R.Q. Ivy SP: Breast cancer growth and metastasis: Interplay between cancer stem cells, embryonic signaling pathways and epithelial-to-mesenchymal transition. Breast Cancer Res. 2011, 13, 211. [Google Scholar] [CrossRef]
- Hong, D.; Fritz, A.J.; Zaidi, S.K.; Van Wijnen, A.J.; Nickerson, J.A.; Imbalzano, A.N.; Lian, J.B.; Stein, J.L.; Stein, G.S. Epithelial-to-mesenchymal transition and cancer stem cells contribute to breast cancer heterogeneity. J. Cell. Physiol. 2018, 233, 9136–9144. [Google Scholar] [CrossRef]
- Sun, Q.; Li, M.; Wang, X. The Cancer Omics Atlas: An integrative resource for cancer omics annotations. BMC Med. Genom. 2018, 11, 63. [Google Scholar]
- Gooding, A.J.; Schiemann, W.P. Epithelial-Mesenchymal Transition Programs and Cancer Stem Cell Phenotypes: Mediators of Breast Cancer Therapy Resistance. Mol. Cancer Res. 2020, 18, 1257–1270. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 1–35. [Google Scholar] [CrossRef]
- Bieche, I.; Chavey, C.; Andrieu, C.; Busson, M.; Vacher, S.; Le Corre, L.; Burlinchon, S.; Lidereau, R.; Lazennec, G.; Guinebretière, J.-M. CXC chemokines located in the 4q21 region are up-regulated in breast cancer. Endocr. Relat. Cancer 2007, 14, 1039–1052. [Google Scholar] [CrossRef]
- Xiao, G.; Wang, X.; Wang, J.; Zu, L.; Cheng, G.; Hao, M.; Sun, X.; Xue, Y.; Lu, J.; Wang, J. CXCL16/CXCR6 chemokine signaling mediates breast cancer progression by pERK1/2-dependent mechanisms. Oncotarget 2015, 6, 14165–14178. [Google Scholar] [CrossRef]
- Boussios, S.; Karathanasi, A.; Cooke, D.; Neille, C.; Sadauskaite, A.; Moschetta, M.; Zakynthinakis-Kyriakou, N.; Pavlidis, N. PARP Inhibitors in Ovarian Cancer: The Route to “Ithaca”. Diagnostics 2019, 9, 55. [Google Scholar] [CrossRef]
- Yao, Y.; Liu, R.; Gao, C.; Zhang, T.; Qi, L.; Liu, G.; Zhang, W.; Wang, X.; Li, J.; Li, J.; et al. Identification of prognostic biomarkers for breast cancer based on miRNA and mRNA co-expression network. J. Cell. Biochem. 2019, 120, 15378–15388. [Google Scholar] [CrossRef]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef]
- Scheau, C.; Badarau, I.A.; Costache, R.S.; Caruntu, C.; Mihai, G.L.; Didilescu, A.C.; Constantin, C.; Neagu, M. The Role of Matrix Metalloproteinases in the Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma. Anal. Cell. Pathol. 2019, 2019, 9423907. [Google Scholar] [CrossRef]
- Li, J.; Liu, C.; Chen, Y.; Gao, C.; Wang, M.; Ma, X.; Zhang, W.; Zhuang, J.; Yao, Y.; Sun, C. Tumor Characterization in Breast Cancer Identifies Immune-Relevant Gene Signatures Associated With Prognosis. Front. Genet. 2019, 10, 1119. [Google Scholar] [CrossRef]
- Neves, M.; Fumagalli, A.; Bor, J.V.D.; Marin, P.; Smit, M.J.; Mayor, F. The Role of ACKR3 in Breast, Lung, and Brain Cancer. Mol. Pharmacol. 2019, 96, 819–825. [Google Scholar] [CrossRef]
- Küçükköse, C.; Yalcin-Ozuysal, O. Effects of Notch signalling on the expression of SEMA3C, HMGA2, CXCL14, CXCR7, and CCL20 in breast cancer. Turk. J. Boil. 2019, 43, 70–76. [Google Scholar] [CrossRef]
- Chen, W.-Y.; Wu, F.; You, Z.-Y.; Zhang, Z.-M.; Guo, Y.-L.; Zhong, L.-X. Analyzing the differentially expressed genes and pathway cross-talk in aggressive breast cancer. J. Obstet. Gynaecol. Res. 2014, 41, 132–140. [Google Scholar] [CrossRef]
- Thomas, C.; Henry, W.; Cuiffo, B.G.; Collmann, A.Y.; Marangoni, E.; Benhamo, V.; Bhasin, M.K.; Fan, C.; Fuhrmann, L.; Baldwin, A.S.; et al. Pentraxin-3 is a PI3K signaling target that promotes stem cell–like traits in basal-like breast cancers. Sci. Signal. 2017, 10, eaah4674. [Google Scholar] [CrossRef]
- Scimeca, M.; Antonacci, C.; Colombo, D.; Bonfiglio, R.; Buonomo, O.C.; Bonanno, E. Emerging prognostic markers related to mesenchymal characteristics of poorly differentiated breast cancers. Tumor Biol. 2016, 37, 5427–5435. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP–dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilicstress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef]
- Althobiti, M.; El Ansari, R.; Aleskandarany, M.; Joseph, C.; Toss, M.S.; Green, A.R.; Rakha, E.A. The prognostic significance of ALDH1A1 expression in early invasive breast cancer. Histopathology 2020. [Google Scholar] [CrossRef]
- Wanandi, S.I.; Syahrani, R.A.; Arumsari, S.; Wideani, G.; Hardiany, N.S. Profiling of Gene Expression Associated with Stemness and Aggressiveness of ALDH1A1-Expressing Human Breast Cancer Cells. Malays. J. Med. Sci. 2019, 26, 38–52. [Google Scholar] [CrossRef]
- Yaghjyan, L.; Esnakula, A.K.; Scott, C.G.; Wijayabahu, A.T.; Jensen, M.R.; Vachon, C.M. Associations of mammographic breast density with breast stem cell marker-defined breast cancer subtypes. Cancer Causes Control. 2019, 30, 1103–1111. [Google Scholar] [CrossRef]
- Clemenceau, A.; Diorio, C.; Durocher, F. Role of Secreted Frizzled-Related Protein 1 in Early Mammary Gland Tumorigenesis and Its Regulation in Breast Microenvironment. Cells 2020, 9, 208. [Google Scholar] [CrossRef]
- Yodsurang, V.; Tanikawa, C.; Miyamoto, T.; Lo, P.H.Y.; Hirata, M.; Matsuda, K. Identification of a novel p53 target, COL17A1, that inhibits breast cancer cell migration and invasion. Oncotarget 2017, 8, 55790–55803. [Google Scholar] [CrossRef]
- Farré, P.L.; Scalise, G.D.; Duca, R.B.; Dalton, G.N.; Massillo, C.; Porretti, J.; Graña, K.; Gardner, K.; De Luca, P.; De Siervi, A. CTBP1 and metabolic syndrome induce an mRNA and miRNA expression profile critical for breast cancer progression and metastasis. Oncotarget 2018, 9, 13848–13858. [Google Scholar] [CrossRef]
- Sundqvist, A.; Vasilaki, E.; Voytyuk, O.; Bai, Y.; Morikawa, M.; Moustakas, A.; Miyazono, K.; Heldin, C.-H.; Dijke, P.T.; Van Dam, H. TGFβ and EGF signaling orchestrates the AP-1- and p63 transcriptional regulation of breast cancer invasiveness. Oncogene 2020, 39, 4436–4449. [Google Scholar] [CrossRef]
- Coates, P.J.; Nenutil, R.; Holcakova, J.; Nekulova, M.; Podhorec, J.; Svoboda, M.; Vojtesek, B. p63 isoforms in triple-negative breast cancer: ΔNp63 associates with the basal phenotype whereas TAp63 associates with androgen receptor, lack of BRCA mutation, PTEN and improved survival. Virchows Arch. 2018, 472, 351–359. [Google Scholar] [CrossRef]
- Mendoza-Rodríguez, M.G.; Ayala-Sumuano, J.T.; García-Morales, L.; Zamudio-Meza, H.; Pérez-Yepez, E.A.; Meza, I. IL-1β Inflammatory Cytokine-Induced TP63 Isoform ∆NP63α Signaling Cascade Contributes to Cisplatin Resistance in Human Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 270. [Google Scholar] [CrossRef]
- Romagnoli, M.; Belguise, K.; Yu, Z.; Wang, X.; Landesman-Bollag, E.; Seldin, D.C.; Chalbos, D.; Barillé-Nion, S.; Jézéquel, P.; Seldin, M.L.; et al. Epithelial-to-mesenchymal transition induced by TGF-β1 is mediated by Blimp-1-dependent repression of BMP-5. Cancer Res. 2012, 72, 6268–6278. [Google Scholar] [CrossRef]
- Liao, S.; Liu, C.; Zhu, G.; Wang, K.; Yang, Y.; Wang, C. Relationship between SDC1 and cadherin signalling activation in cancer. Pathol. Res. Pract. 2020, 216, 152756. [Google Scholar] [CrossRef]
- Qiao, W.; Liu, H.; Guo, W.; Li, P.; Deng, M. Prognostic and clinical significance of syndecan-1 expression in breast cancer: A systematic review and meta-analysis. Eur. J. Surg. Oncol. 2019, 45, 1132–1137. [Google Scholar] [CrossRef]
- Sayyad, M.R.; Puchalapalli, M.; Vergara, N.G.; Wangensteen, S.M.; Moore, M.; Mu, L.; Edwards, C.; Anderson, A.; Kall, S.; Sullivan, M.; et al. Syndecan-1 facilitates breast cancer metastasis to the brain. Breast Cancer Res. Treat. 2019, 178, 35–49. [Google Scholar] [CrossRef]
- Gao, X.-H.; Zhang, Y.-L.; Zhang, Z.-Y.; Guo, S.-S.; Chen, X.-B.; Guo, Y.-Z. MicroRNA-96-5p represses breast cancer proliferation and invasion through Wnt/β-catenin signaling via targeting CTNND1. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Wu, X. Expressions of miR-21 and miR-210 in Breast Cancer and Their Predictive Values for Prognosis. Iran. J. Public Health 2020, 49. [Google Scholar] [CrossRef]
- Peng, Q.; Zhang, X.; Min, M.; Zou, L.; Shen, P.; Zhu, Y. The clinical role of microRNA-21 as a promising biomarker in the diagnosis and prognosis of colorectal cancer: A systematic review and meta-analysis. Oncotarget 2017, 8, 44893–44909. [Google Scholar] [CrossRef]
- Kjaer-Frifeldt, S.; Hansen, T.F.; Nielsen, B.S.; Joergensen, S.; Lindebjerg, J.; Soerensen, F.B.; dePont Christensen, R.; Jakobsen, A. Danish Colorectal Cancer G: The prognostic importance of miR-21 in stage II colon cancer: A population-based study. Br. J. Cancer 2012, 107, 1169–1174. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Bottai, G.; Nuciforo, P.G.; Di Tommaso, L.; Giovannetti, E.; Peg, V.; Losurdo, A.; Pérez-Garcia, J.; Masci, G.; Corsi, F.; et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget 2015, 6, 37269–37280. [Google Scholar] [CrossRef]
- Yan-Hui, L.; Liu, Y.-H.; Xiang, J.-W.; Wu, Q.-N.; Xu, L.-B.; Luo, X.-L.; Zhu, X.-L.; Liu, C.; Xu, F.-P.; Luo, D.-L.; et al. PIK3R1 targeting by miR-21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT, and predicts clinical outcome of breast cancer. Int. J. Oncol. 2015, 48, 471–484. [Google Scholar] [CrossRef]
- Chang, C.-J.; Chao, C.-H.; Xia, W.; Yang, J.-Y.; Xiong, Y.; Li, C.-W.; Yu, W.-H.; Rehman, S.K.; Hsu, J.L.; Lee, H.-H.; et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 2011, 13, 317–323. [Google Scholar] [CrossRef]
- Alam, F.; Mezhal, F.; El Hasasna, H.; Nair, V.A.; Aravind, S.R.; Ayad, M.S.; El-Serafi, A.; Abdel-Rahman, W.M. The role of p53-microRNA 200-Moesin axis in invasion and drug resistance of breast cancer cells. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef]
- Chang, B.P.; Wang, D.S.; Xing, J.W.; Yang, S.H.; Chu, Q.; Yu, S.Y. miR-200c inhibits metastasis of breast cancer cells by targeting HMGB1. J. Huazhong Univ. Sci. Technol. Med. Sci. 2014, 34, 201–206. [Google Scholar] [CrossRef]
- Liu, B.; Du, R.; Zhou, L.; Xu, J.; Chen, S.; Chen, J.; Yang, X.; Liu, D.X.; Shao, Z.M.; Zhang, L.; et al. miR-200c/141 Regulates Breast Cancer Stem Cell Heterogeneity via Targeting HIPK1/β-Catenin Axis. Theranostics 2018, 8, 5801–5813. [Google Scholar] [CrossRef]
- Vieira, A.F.; Schmitt, F. An Update on Breast Cancer Multigene Prognostic Tests-Emergent Clinical Biomarkers. Front. Med. (Lausanne) 2018, 5, 248. [Google Scholar] [CrossRef]
- Mi, H.; Thomas, P. PANTHER pathway: An ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 2009, 563, 123–140. [Google Scholar]
- Lánczky, A.; Nagy, Á.; Bottai, G.; Munkácsy, G.; Szabó, A.; Santarpia, L.; Győrffy, B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016, 160, 439–446. [Google Scholar] [CrossRef]





| Gene Expression | Gene | p-Value | Log2 (Fold Change) | FDR (p-Value) | miRNA Expression Level | Symbol | FDR (p-Value) | Log2 (Fold Change) | FDR (p-Value) |
|---|---|---|---|---|---|---|---|---|---|
| Upregulated genes | COL10A1 | 2.40 × 10−176 | 6.76 | 5.47 × 10−173 | Upregulated miRNAs | hsa-mir-183 | 4.01 × 10−92 | 2.95 | 1.40 × 10−89 |
| COL11A1 | 3.75 × 10−101 | 5.88 | 2.81 × 10−99 | hsa-mir-96 | 3.59 × 10−90 | 2.87 | 9.38 × 10−88 | ||
| MMP11 | 1.27 × 10−176 | 5.82 | 3.27 × 10−173 | hsa-mir-141 | 2.12 × 10−76 | 2.71 | 3.16 × 10−74 | ||
| MMP13 | 6.76 × 10−94 | 5.79 | 3.94 × 10−92 | hsa-mir-429 | 4.08 × 10−63 | 2.68 | 3.88 × 10−61 | ||
| CST1 | 1.22 × 10−65 | 5.64 | 2.52 × 10−64 | hsa-mir-200a | 2.21 × 10−60 | 2.58 | 1.93 × 10−58 | ||
| MMP1 | 1.43 × 10−61 | 4.95 | 2.58 × 10−60 | hsa-mir-196a-1 | 2.43 × 10−19 | 2.41 | 2.86 × 10−18 | ||
| PPAPDC1A | 9.77 × 10−121 | 4.87 | 1.50 × 10−118 | hsa-mir-182 | 4.55 × 10−72 | 2.36 | 5.29 × 10−70 | ||
| COMP | 2.45 × 10−90 | 4.76 | 1.27 × 10−88 | hsa-mir-210 | 2.78 × 10−22 | 2.36 | 3.87 × 10−21 | ||
| NEK2 | 8.02 × 10−196 | 4.59 | 8.23 × 10−192 | hsa-mir-21 | 9.49 × 10−139 | 2.32 | 9.93 × 10−136 | ||
| PKMYT1 | 9.28 × 10−174 | 4.56 | 1.73 × 10−170 | hsa-mir-190b | 1.14 × 10−31 | 2.31 | 2.34 × 10−30 | ||
| Downregulated genes | ITSN1 | 5.46 × 10−89 | −1.5 | 2.61 × 10−87 | Downregulated miRNAs | hsa-mir-378c | 2.06 × 10 -47 | −1.51 | 9.00 × 10−46 |
| C3orf64 | 2.19 × 10−88 | −1.5 | 1.02 × 10−86 | hsa-mir-675 | 2.29 × 10−17 | −1.52 | 2.40 × 10−16 | ||
| C2CD2 | 1.62 × 10−60 | −1.5 | 2.85 × 10−59 | hsa-mir-1258 | 1.54 × 10−58 | −1.56 | 1.24 × 10−56 | ||
| CAB39L | 4.35 × 10−49 | −1.5 | 5.34 × 10−48 | hsa-mir-1-2 | 2.60 × 10−32 | −1.60 | 5.44 × 10−31 | ||
| SLC5A4 | 1.08 × 10−44 | −1.5 | 1.13 × 10−43 | hsa-mir-205 | 8.64 × 10−10 | −1.71 | 4.97 × 10−09 | ||
| KALRN | 3.50 × 10−40 | −1.5 | 3.09 × 10−39 | hsa-mir-584 | 2.52 × 10−33 | −1.71 | 5.49 × 10−32 | ||
| FZD7 | 2.30 × 10−37 | −1.5 | 1.85 × 10−36 | hsa-mir-511-1 | 1.39 × 10−40 | −1.75 | 4.55 × 10−39 | ||
| TMIE | 2.38 × 10−37 | −1.5 | 1.92 × 10−36 | hsa-mir-483 | 2.22 × 10−32 | −1.78 | 4.74 × 10−31 | ||
| NCOA7 | 3.07 × 10−37 | −1.5 | 2.46 × 10−36 | hsa-mir-125b-2 | 5.03 × 10−44 | −1.79 | 1.95 × 10−42 | ||
| HAPLN4 | 4.90 × 10−29 | −1.5 | 2.87 × 10−28 | hsa-mir-511-2 | 7.51 × 10−45 | −1.82 | 3.02 × 10−43 |
| No. | Age | Diagnostic | T | N | M | Stage | ER | PR | HER2/New |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 57 | invasive ductal carcinomas | 2 | 0 | X | IIB | + | + | − |
| 2 | 68 | invasive ductal carcinomas | 1C | 1 | X | IIA | + | + | − |
| 3 | 42 | invasive ductal carcinomas | 1a | 1a | X | I | + | + | + |
| 4 | 56 | invasive ductal carcinomas | 2 | 0 | 0 | IIB | + | + | − |
| 5 | 70 | invasive ductal carcinomas | 2 | 0 | 0 | IIa | + | + | − |
| 6 | 43 | invasive ductal carcinomas | 2 | 1 | 0 | IIB | + | + | + |
| 7 | 73 | invasive ductal carcinomas | 2 | 0 | 0 | IIA | + | + | + |
| 8 | 76 | invasive ductal carcinomas | 2 | 1 | X | IIB | + | + | − |
| 9 | 66 | invasive ductal carcinomas | 2 | 1 | 0 | IIB | + | + | − |
| 10 | 67 | invasive ductal carcinomas | 2 | 0 | 0 | IIA | + | + | − |
| 11 | 62 | invasive ductal carcinomas | 2 | 1 | 0 | IIB | − | − | + |
| 12 | 33 | invasive ductal carcinomas | 1C | 0 | 0 | IA | + | + | − |
| 13 | 62 | invasive ductal carcinomas | 2 | 0 | 0 | IIA | + | + | − |
| 14 | 67 | invasive ductal carcinomas | 1C | 1 | 0 | IIB | + | + | − |
| 15 | 39 | invasive ductal carcinomas | 2 | 0 | X | IIA | + | + | + |
| 16 | 53 | invasive ductal carcinomas | 1c | 0 | 0 | I | + | + | − |
| 17 | 59 | invasive ductal carcinomas | 2 | 0 | 0 | IIA | + | + | − |
| 18 | 75 | invasive ductal carcinomas | 2 | 1 | 0 | IIB | + | + | − |
| 19 | 64 | invasive ductal carcinomas | 4 | 1 | 0 | IIIA | + | − | − |
| 20 | 41 | invasive ductal carcinomas | 1c | 1a | 0 | IIA | + | + | − |
| 21 | 33 | invasive ductal carcinomas | 2 | 1 | 0 | IIB | + | + | + |
| 22 | 73 | invasive ductal carcinomas | 1c | 1 | X | IIB | + | + | + |
| 23 | 56 | invasive ductal carcinomas | 2 | 0 | 0 | IB | + | + | − |
| 24 | 51 | invasive ductal carcinomas | 2 | 0 | 0 | IB | + | + | − |
| 25 | 64 | invasive ductal carcinomas | 2 | 0 | 0 | IB | + | + | − |
| 26 | 71 | invasive ductal carcinomas | 1c | 0 | 0 | IA | + | + | + |
| 27 | 53 | invasive ductal carcinomas | 2 | 0 | 0 | IB | + | + | + |
| 28 | 38 | invasive ductal carcinomas | 1c | 0 | X | IIB | + | + | − |
| 29 | 42 | invasive ductal carcinomas | 1c | 1a | X | IA | + | + | − |
| 30 | 35 | invasive ductal carcinomas | 2 | 0 | 0 | IC | + | + | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Groza, I.-M.; Braicu, C.; Jurj, A.; Zanoaga, O.; Lajos, R.; Chiroi, P.; Cojocneanu, R.; Paun, D.; Irimie, A.; Korban, S.S.; et al. Cancer-Associated Stemness and Epithelial-to-Mesenchymal Transition Signatures Related to Breast Invasive Carcinoma Prognostic. Cancers 2020, 12, 3053. https://doi.org/10.3390/cancers12103053
Groza I-M, Braicu C, Jurj A, Zanoaga O, Lajos R, Chiroi P, Cojocneanu R, Paun D, Irimie A, Korban SS, et al. Cancer-Associated Stemness and Epithelial-to-Mesenchymal Transition Signatures Related to Breast Invasive Carcinoma Prognostic. Cancers. 2020; 12(10):3053. https://doi.org/10.3390/cancers12103053
Chicago/Turabian StyleGroza, Iulia-Monica, Cornelia Braicu, Ancuta Jurj, Oana Zanoaga, Raduly Lajos, Paul Chiroi, Roxana Cojocneanu, Diana Paun, Alexandru Irimie, Schuyler S. Korban, and et al. 2020. "Cancer-Associated Stemness and Epithelial-to-Mesenchymal Transition Signatures Related to Breast Invasive Carcinoma Prognostic" Cancers 12, no. 10: 3053. https://doi.org/10.3390/cancers12103053
APA StyleGroza, I.-M., Braicu, C., Jurj, A., Zanoaga, O., Lajos, R., Chiroi, P., Cojocneanu, R., Paun, D., Irimie, A., Korban, S. S., Achimas-Cadariu, P., & Berindan-Neagoe, I. (2020). Cancer-Associated Stemness and Epithelial-to-Mesenchymal Transition Signatures Related to Breast Invasive Carcinoma Prognostic. Cancers, 12(10), 3053. https://doi.org/10.3390/cancers12103053










