DNA Methylation Clusters and Their Relation to Cytogenetic Features in Pediatric AML
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Population
2.2. DNA Methylation Profiling
2.3. Transcriptomic Profiling
2.4. Statistical Analysis
3. Results
3.1. Bootstrap Clustering for Discovery of Methylation and Transcription Subgroups
3.2. Unsupervised Discovery of Methylation Subgroups
3.3. Unsupervised Discovery of Expression Subgroups
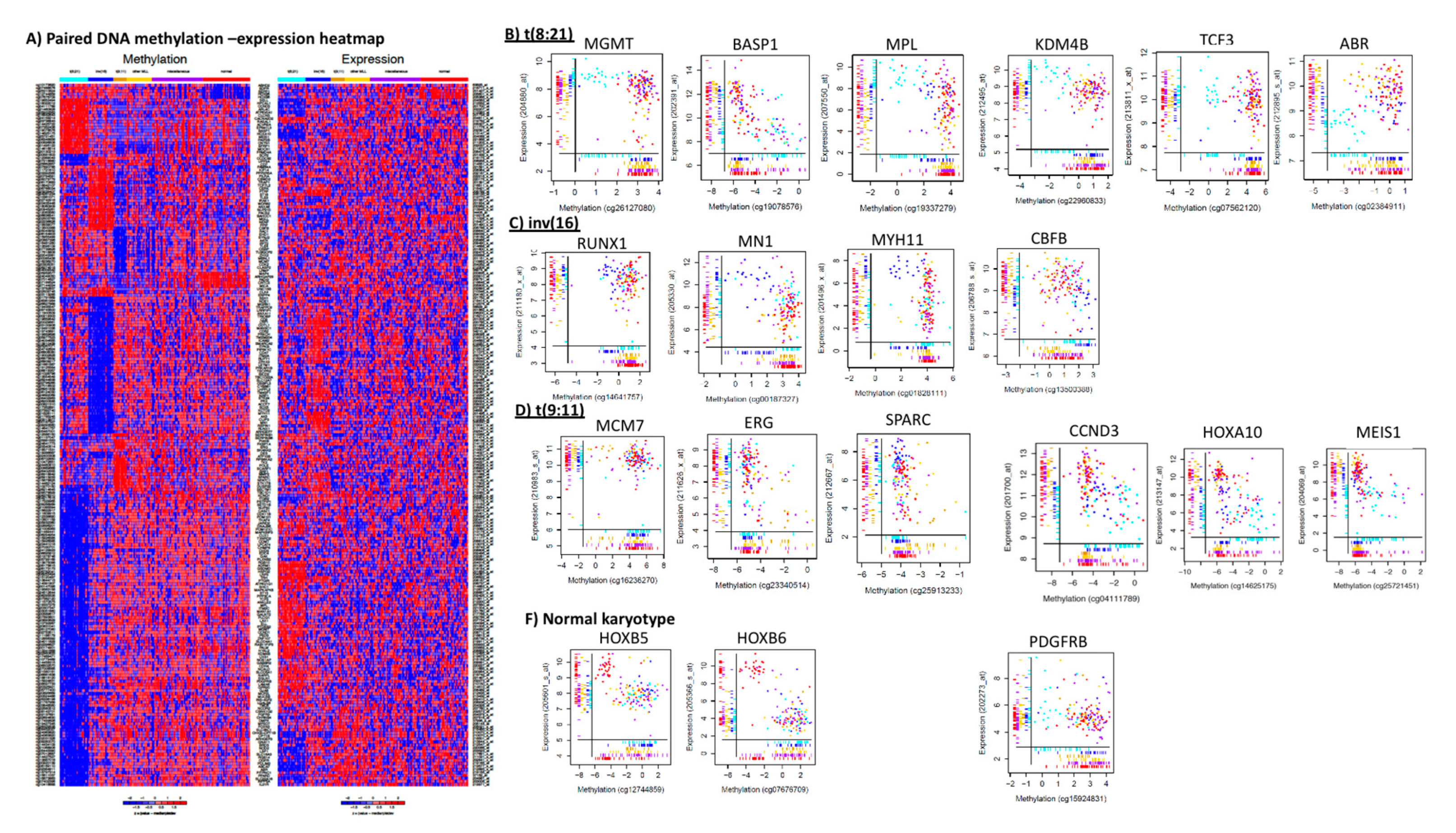
3.4. Supervised Comparisons of Cytogenetic Subgroups and Methylation-Expression Pairs Unique to Cytogenetic Features
3.5. t(8;21) vs. Non-t(8;21) Comparison
3.6. inv(16) vs. Non inv(16)
3.7. t(9;11) vs. Non-t(9;11)
3.8. Other 11q23 MLL vs. Non-Other 11q23
3.9. Other Miscellaneous Abnormalities vs. Other Cytogenetic Features
3.10. FLT3-ITD vs. Non-FLT3-ITD
3.11. Complimentary Epigenetic and Genetics Hits of Significant Relevance
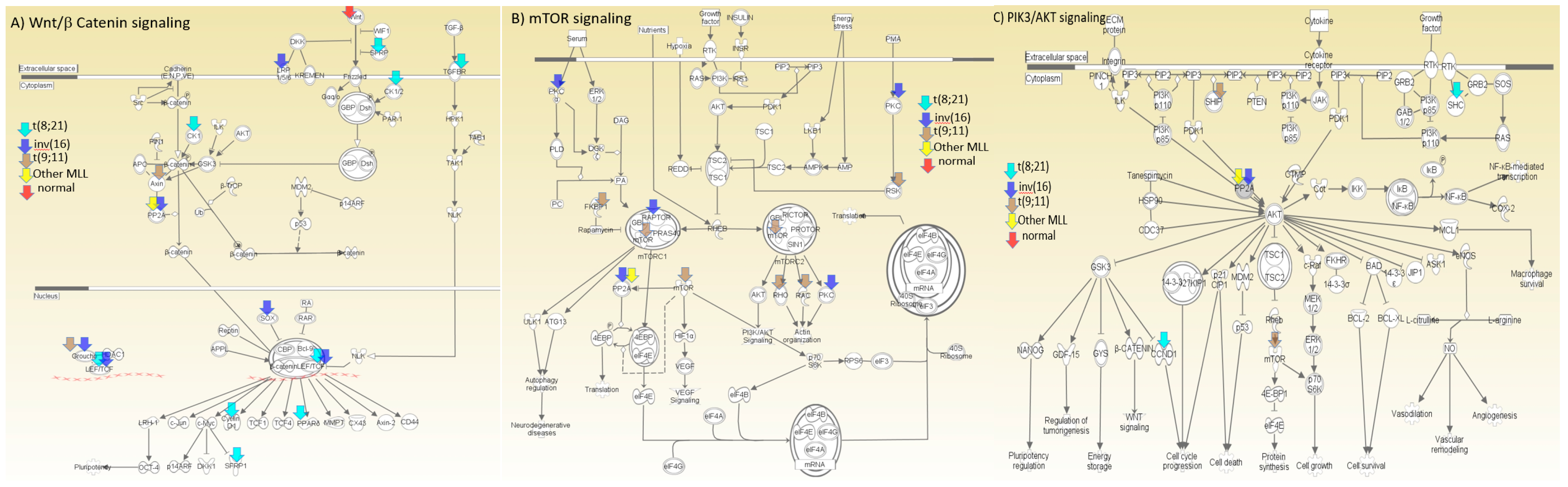
3.12. Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grimwade, D.; Ivey, A.; Huntly, B.J. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood 2016, 127, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Estey, E.H. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am. J. Hematol. 2018, 93, 1267–1291. [Google Scholar] [CrossRef]
- Dohner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Akalin, A.; Garrett-Bakelman, F.E.; Kormaksson, M.; Busuttil, J.; Zhang, L.; Khrebtukova, I.; Milne, T.A.; Huang, Y.; Biswas, D.; Hess, J.L.; et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012, 8, e1002781. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Suela, J.; Valencia, A.; Fernandez, A.; Wunderlich, M.; Agirre, X.; Prosper, F.; Martin-Subero, J.I.; Maiques, A.; Acquadro, F.; et al. DNA methylation profiles and their relationship with cytogenetic status in adult acute myeloid leukemia. PLoS ONE 2010, 5, e12197. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, L.; Gundry, M.C.; Goodell, M.A. DNMT3A in Leukemia. Cold Spring Harb. Perspect. Med. 2017, 7, a030320. [Google Scholar] [CrossRef] [PubMed]
- Claus, R.; Plass, C.; Armstrong, S.A.; Bullinger, L. DNA methylation profiling in acute myeloid leukemia: From recent technological advances to biological and clinical insights. Future Oncol. 2010, 6, 1415–1431. [Google Scholar] [CrossRef]
- Figueroa, M.E.; Lugthart, S.; Li, Y.; Erpelinck-Verschueren, C.; Deng, X.; Christos, P.J.; Schifano, E.; Booth, J.; van Putten, W.; Skrabanek, L.; et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 2010, 17, 13–27. [Google Scholar] [CrossRef]
- Marcucci, G.; Yan, P.; Maharry, K.; Frankhouser, D.; Nicolet, D.; Metzeler, K.H.; Kohlschmidt, J.; Mrozek, K.; Wu, Y.Z.; Bucci, D.; et al. Epigenetics meets genetics in acute myeloid leukemia: Clinical impact of a novel seven-gene score. J. Clin. Oncol. 2014, 32, 548–556. [Google Scholar] [CrossRef]
- Plass, C.; Oakes, C.; Blum, W.; Marcucci, G. Epigenetics in acute myeloid leukemia. Semin. Oncol. 2008, 35, 378–387. [Google Scholar] [CrossRef]
- Prada-Arismendy, J.; Arroyave, J.C.; Rothlisberger, S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017, 31, 63–76. [Google Scholar] [CrossRef]
- Ho, P.A.; Kutny, M.A.; Alonzo, T.A.; Gerbing, R.B.; Joaquin, J.; Raimondi, S.C.; Gamis, A.S.; Meshinchi, S. Leukemic mutations in the methylation-associated genes DNMT3A and IDH2 are rare events in pediatric AML: A report from the Children’s Oncology Group. Pediatr. Blood Cancer 2011, 57, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Hollink, I.H.; Feng, Q.; Danen-van Oorschot, A.A.; Arentsen-Peters, S.T.; Verboon, L.J.; Zhang, P.; de Haas, V.; Reinhardt, D.; Creutzig, U.; Trka, J.; et al. Low frequency of DNMT3A mutations in pediatric AML, and the identification of the OCI-AML3 cell line as an in vitro model. Leukemia 2012, 26, 371–373. [Google Scholar] [CrossRef]
- Liang, D.C.; Liu, H.C.; Yang, C.P.; Jaing, T.H.; Hung, I.J.; Yeh, T.C.; Chen, S.H.; Hou, J.Y.; Huang, Y.J.; Shih, Y.S.; et al. Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood 2013, 121, 2988–2995. [Google Scholar] [CrossRef] [PubMed]
- Juhl-Christensen, C.; Ommen, H.B.; Aggerholm, A.; Lausen, B.; Kjeldsen, E.; Hasle, H.; Hokland, P. Genetic and epigenetic similarities and differences between childhood and adult AML. Pediatr. Blood Cancer 2012, 58, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Bolouri, H.; Farrar, J.E.; Triche, T., Jr.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef]
- Rubnitz, J.E.; Inaba, H.; Dahl, G.; Ribeiro, R.C.; Bowman, W.P.; Taub, J.; Pounds, S.; Razzouk, B.I.; Lacayo, N.J.; Cao, X.; et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: Results of the AML02 multicentre trial. Lancet Oncol. 2010, 11, 543–552. [Google Scholar] [CrossRef]
- Lamba, J.K.; Cao, X.; Raimondi, S.C.; Rafiee, R.; Downing, J.R.; Lei, S.; Gruber, T.; Ribeiro, R.C.; Rubnitz, J.E.; Pounds, S.B. Integrated epigenetic and genetic analysis identifies markers of prognostic significance in pediatric acute myeloid leukemia. Oncotarget 2018, 9, 26711–26723. [Google Scholar] [CrossRef] [PubMed]
- Maksimovic, J.; Gordon, L.; Oshlack, A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012, 13, R44. [Google Scholar] [CrossRef] [PubMed]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef]
- Ross, M.E.; Mahfouz, R.; Onciu, M.; Liu, H.C.; Zhou, X.; Song, G.; Shurtleff, S.A.; Pounds, S.; Cheng, C.; Ma, J.; et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood 2004, 104, 3679–3687. [Google Scholar] [CrossRef]
- Dudoit, S.; Fridlyand, J. A prediction-based resampling method for estimating the number of clusters in a dataset. Genome Biol. 2002, 3, RESEARCH0036. [Google Scholar] [CrossRef] [PubMed]
- Kerr, M.K.; Churchill, G.A. Bootstrapping cluster analysis: Assessing the reliability of conclusions from microarray experiments. Proc. Natl. Acad. Sci. USA 2001, 98, 8961–8965. [Google Scholar] [CrossRef] [PubMed]
- Hartigan, J.A.; Hartigan, P.M. The Dip Test of Unimodality. Ann. Stat. 1985, 13, 70–84. [Google Scholar] [CrossRef]
- Knapp, T.R. Bimodality revisited. J. Mod. Appl. Stat. Methods 2007, 6. [Google Scholar] [CrossRef]
- Royston, J.P. An Extension of Shapiro and Wilk’s W Test for Normality to Large Samples. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1982, 31, 115–124. [Google Scholar] [CrossRef]
- Dunn, J.C. Well-Separated Clusters and Optimal Fuzzy Partitions. J. Cybern. 1974, 4, 95–104. [Google Scholar] [CrossRef]
- Xiao, Q.; Deng, D.; Li, H.; Ye, F.; Huang, L.; Zhang, B.; Ye, B.; Mo, Z.; Yang, X.; Liu, Z. GSTT1 and GSTM1 polymorphisms predict treatment outcome for acute myeloid leukemia: A systematic review and meta-analysis. Ann. Hematol. 2014, 93, 1381–1390. [Google Scholar] [CrossRef]
- Ouerhani, S.; Nefzi, M.A.; Menif, S.; Safra, I.; Douzi, K.; Fouzai, C.; Ben Ghorbel, G.; Ben Bahria, I.; Ben Ammar Elgaaied, A.; Abbes, S. Influence of genetic polymorphisms of xenobiotic metabolizing enzymes on the risk of developing leukemia in a Tunisian population. Bull. Cancer 2011, 98, 95–106. [Google Scholar] [CrossRef]
- Li, N.; Jia, X.; Wang, J.; Li, Y.; Xie, S. Knockdown of homeobox A5 by small hairpin RNA inhibits proliferation and enhances cytarabine chemosensitivity of acute myeloid leukemia cells. Mol. Med. Rep. 2015, 12, 6861–6866. [Google Scholar] [CrossRef]
- Musialik, E.; Bujko, M.; Kober, P.; Grygorowicz, M.A.; Libura, M.; Przestrzelska, M.; Juszczynski, P.; Borg, K.; Florek, I.; Jakobczyk, M.; et al. Promoter DNA methylation and expression levels of HOXA4, HOXA5 and MEIS1 in acute myeloid leukemia. Mol. Med. Rep. 2015, 11, 3948–3954. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.F.; Li, Z.H.; Wang, N.N.; Fang, F.; Xu, L.X.; Pan, J. tp53-dependent G2 arrest mediator candidate gene, Reprimo, is down-regulated by promoter hypermethylation in pediatric acute myeloid leukemia. Leuk. Lymphoma 2015, 56, 2931–2944. [Google Scholar] [CrossRef]
- Ostronoff, F.; Othus, M.; Gerbing, R.B.; Loken, M.R.; Raimondi, S.C.; Hirsch, B.A.; Lange, B.J.; Petersdorf, S.; Radich, J.; Appelbaum, F.R.; et al. NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: A COG and SWOG report. Blood 2014, 124, 2400–2407. [Google Scholar] [CrossRef] [PubMed]
- Shima, Y.; Yumoto, M.; Katsumoto, T.; Kitabayashi, I. MLL is essential for NUP98-HOXA9-induced leukemia. Leukemia 2017, 31, 2200–2210. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Wen, C.L.; Qin, Y.X.; Tang, X.M.; Shi, M.M.; Shen, B.Y.; Fang, Y. Establishment of a human primary pancreatic cancer mouse model to examine and investigate gemcitabine resistance. Oncol. Rep. 2017, 38, 3335–3346. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, E.M.; Abedin, S.M.; Radecki, S.G.; Fischietti, M.; Arslan, A.D.; Blyth, G.T.; Yang, A.; Lantz, C.; Nelson, A.; Goo, Y.A.; et al. Identification and targeting of novel CDK9 complexes in acute myeloid leukemia. Blood 2019, 133, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Fu, L.; Lv, N.; Liu, J.; Li, Y.; Chen, X.; Xu, Q.; Chen, G.; Pang, B.; Wang, L.; et al. Methylation-associated silencing of BASP1 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. Exp. Mol. Med. 2018, 50, 44. [Google Scholar] [CrossRef]
- Hong, Q.; Chen, X.; Ye, H.; Zhou, A.; Gao, Y.; Jiang, D.; Wu, X.; Tian, B.; Chen, Y.; Wang, M.; et al. Association between the methylation status of the MGMT promoter in bone marrow specimens and chemotherapy outcomes of patients with acute myeloid leukemia. Oncol. Lett. 2016, 11, 2851–2856. [Google Scholar] [CrossRef] [PubMed]
- Chou, F.S.; Griesinger, A.; Wunderlich, M.; Lin, S.; Link, K.A.; Shrestha, M.; Goyama, S.; Mizukawa, B.; Shen, S.; Marcucci, G.; et al. The thrombopoietin/MPL/Bcl-xL pathway is essential for survival and self-renewal in human preleukemia induced by AML1-ETO. Blood 2012, 120, 709–719. [Google Scholar] [CrossRef]
- Pulikkan, J.A.; Madera, D.; Xue, L.; Bradley, P.; Landrette, S.F.; Kuo, Y.H.; Abbas, S.; Zhu, L.J.; Valk, P.; Castilla, L.H. Thrombopoietin/MPL participates in initiating and maintaining RUNX1-ETO acute myeloid leukemia via PI3K/AKT signaling. Blood 2012, 120, 868–879. [Google Scholar] [CrossRef]
- Agger, K.; Nishimura, K.; Miyagi, S.; Messling, J.E.; Rasmussen, K.D.; Helin, K. The KDM4/JMJD2 histone demethylases are required for hematopoietic stem cell maintenance. Blood 2019, 134, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Karvonen, H.; Perttila, R.; Niininen, W.; Hautanen, V.; Barker, H.; Murumagi, A.; Heckman, C.A.; Ungureanu, D. Wnt5a and ROR1 activate non-canonical Wnt signaling via RhoA in TCF3-PBX1 acute lymphoblastic leukemia and highlight new treatment strategies via Bcl-2 co-targeting. Oncogene 2019, 38, 3288–3300. [Google Scholar] [CrossRef] [PubMed]
- Jastaniah, W.; Elimam, N.; Abdalla, K.; AlAzmi, A.A.; Elgaml, A.M.; Alkassar, A.; Daghistani, M.; Felimban, S. Early vs. late MRD response- and risk-based treatment intensification of childhood acute lymphoblastic leukemia: A prospective pilot study from Saudi Arabia. Exp. Hematol. Oncol. 2018, 7, 29. [Google Scholar] [CrossRef]
- Chen, X.; Wang, F.; Zhang, Y.; Wang, M.; Tian, W.; Teng, W.; Ma, X.; Guo, L.; Fang, J.; Zhang, Y.; et al. Retrospective analysis of 36 fusion genes in 2479 Chinese patients of de novo acute lymphoblastic leukemia. Leuk. Res. 2018, 72, 99–104. [Google Scholar] [CrossRef]
- Lin, A.; Cheng, F.W.T.; Chiang, A.K.S.; Luk, C.W.; Li, R.C.H.; Ling, A.S.C.; Cheuk, D.K.L.; Chang, K.O.; Ku, D.; Lee, V.; et al. Excellent outcome of acute lymphoblastic leukaemia with TCF3-PBX1 rearrangement in Hong Kong. Pediatr. Blood Cancer 2018, 65, e27346. [Google Scholar] [CrossRef] [PubMed]
- Namasu, C.Y.; Katzerke, C.; Brauer-Hartmann, D.; Wurm, A.A.; Gerloff, D.; Hartmann, J.U.; Schwind, S.; Muller-Tidow, C.; Hilger, N.; Fricke, S.; et al. ABR, a novel inducer of transcription factor C/EBPalpha, contributes to myeloid differentiation and is a favorable prognostic factor in acute myeloid leukemia. Oncotarget 2017, 8, 103626–103639. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Fu, Y.; Wu, W.; Xu, J.D.; Chen, T.M.; Qiao, C.; Li, J.Y.; Liu, P. Expression of BAG3 Gene in Acute Myeloid Leukemia and Its Prognostic Value. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2015, 23, 925–929. [Google Scholar]
- Sun, X.; Lu, B.; Hu, B.; Xiao, W.; Li, W.; Huang, Z. Novel function of the chromosome 7 open reading frame 41 gene to promote leukemic megakaryocyte differentiation by modulating TPA-induced signaling. Blood Cancer J. 2014, 4, e198. [Google Scholar] [CrossRef]
- Lin, W.H.; Dai, W.G.; Xu, X.D.; Yu, Q.H.; Zhang, B.; Li, J.; Li, H.P. Downregulation of DPF3 promotes the proliferation and motility of breast cancer cells through activating JAK2/STAT3 signaling. Biochem. Biophys. Res. Commun. 2019, 514, 639–644. [Google Scholar] [CrossRef]
- Alam, A.; Taye, N.; Patel, S.; Thube, M.; Mullick, J.; Shah, V.K.; Pant, R.; Roychowdhury, T.; Banerjee, N.; Chatterjee, S.; et al. SMAR1 favors immunosurveillance of cancer cells by modulating calnexin and MHC I expression. Neoplasia 2019, 21, 945–962. [Google Scholar] [CrossRef]
- Edwards, D.K.t.; Watanabe-Smith, K.; Rofelty, A.; Damnernsawad, A.; Laderas, T.; Lamble, A.; Lind, E.F.; Kaempf, A.; Mori, M.; Rosenberg, M.; et al. CSF1R inhibitors exhibit antitumor activity in acute myeloid leukemia by blocking paracrine signals from support cells. Blood 2019, 133, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Sood, R.; Kamikubo, Y.; Liu, P. Role of RUNX1 in hematological malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [CrossRef]
- Carella, C.; Bonten, J.; Sirma, S.; Kranenburg, T.A.; Terranova, S.; Klein-Geltink, R.; Shurtleff, S.; Downing, J.R.; Zwarthoff, E.C.; Liu, P.P.; et al. MN1 overexpression is an important step in the development of inv(16) AML. Leukemia 2007, 21, 1679–1690. [Google Scholar] [CrossRef]
- Larmonie, N.S.D.; Arentsen-Peters, T.; Obulkasim, A.; Valerio, D.; Sonneveld, E.; Danen-van Oorschot, A.A.; de Haas, V.; Reinhardt, D.; Zimmermann, M.; Trka, J.; et al. MN1 overexpression is driven by loss of DNMT3B methylation activity in inv(16) pediatric AML. Oncogene 2018, 37, 107–115. [Google Scholar] [CrossRef]
- Lai, C.K.; Norddahl, G.L.; Maetzig, T.; Rosten, P.; Lohr, T.; Sanchez Milde, L.; von Krosigk, N.; Docking, T.R.; Heuser, M.; Karsan, A.; et al. Meis2 as a critical player in MN1-induced leukemia. Blood Cancer J. 2017, 7, e613. [Google Scholar] [CrossRef]
- Kawagoe, H.; Grosveld, G.C. Conditional MN1-TEL knock-in mice develop acute myeloid leukemia in conjunction with overexpression of HOXA9. Blood 2005, 106, 4269–4277. [Google Scholar] [CrossRef] [PubMed]
- Beverly, L.J.; Varmus, H.E. MYC-induced myeloid leukemogenesis is accelerated by all six members of the antiapoptotic BCL family. Oncogene 2009, 28, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; Leenen, P.J.M.; Erkeland, S.J. The interplay between critical transcription factors and microRNAs in the control of normal and malignant myelopoiesis. Cancer Lett. 2018, 427, 28–37. [Google Scholar] [CrossRef]
- Moore, S.D.; Offor, O.; Ferry, J.A.; Amrein, P.C.; Morton, C.C.; Dal Cin, P. ELF4 is fused to ERG in a case of acute myeloid leukemia with a t(X;21)(q25−26;q22). Leuk. Res. 2006, 30, 1037–1042. [Google Scholar] [CrossRef]
- Panagopoulos, I.; Aman, P.; Fioretos, T.; Hoglund, M.; Johansson, B.; Mandahl, N.; Heim, S.; Behrendtz, M.; Mitelman, F. Fusion of the FUS gene with ERG in acute myeloid leukemia with t(16;21)(p11;q22). Genes Chromosomes Cancer 1994, 11, 256–262. [Google Scholar] [CrossRef]
- DiMartino, J.F.; Lacayo, N.J.; Varadi, M.; Li, L.; Saraiya, C.; Ravindranath, Y.; Yu, R.; Sikic, B.I.; Raimondi, S.C.; Dahl, G.V. Low or absent SPARC expression in acute myeloid leukemia with MLL rearrangements is associated with sensitivity to growth inhibition by exogenous SPARC protein. Leukemia 2006, 20, 426–432. [Google Scholar] [CrossRef]
- Ono, R.; Taki, T.; Taketani, T.; Taniwaki, M.; Kobayashi, H.; Hayashi, Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res. 2002, 62, 4075–4080. [Google Scholar]
- Seipel, K.; Marques, M.T.; Bozzini, M.A.; Meinken, C.; Mueller, B.U.; Pabst, T. Inactivation of the p53-KLF4-CEBPA Axis in Acute Myeloid Leukemia. Clin. Cancer Res. 2016, 22, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, J.; Lu, C.; Han, J.; Wang, G.; Song, C.; Zhu, S.; Wang, C.; Li, G.; Kang, J.; et al. HDAC1 and Klf4 interplay critically regulates human myeloid leukemia cell proliferation. Cell Death Dis. 2014, 5, e1491. [Google Scholar] [CrossRef]
- Campregher, P.V.; Halley, N.D.S.; Vieira, G.A.; Fernandes, J.F.; Velloso, E.; Ali, S.; Mughal, T.; Miller, V.; Mangueira, C.L.P.; Odone, V.; et al. Identification of a novel fusion TBL1XR1-PDGFRB in a patient with acute myeloid leukemia harboring the DEK-NUP214 fusion and clinical response to dasatinib. Leuk. Lymphoma 2017, 58, 2969–2972. [Google Scholar] [CrossRef] [PubMed]
- Nagel, S.; Kaufmann, M.; Scherr, M.; Drexler, H.G.; MacLeod, R.A. Activation of HLXB9 by juxtaposition with MYB via formation of t(6;7)(q23;q36) in an AML-M4 cell line (GDM-1). Genes Chromosomes Cancer 2005, 42, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Gross, M.; Mkrtchyan, H.; Glaser, M.; Fricke, H.J.; Hoffken, K.; Heller, A.; Weise, A.; Liehr, T. Delineation of yet unknown cryptic subtelomere aberrations in 50% of acute myeloid leukemia with normal GTG-banding karyotype. Int. J. Oncol. 2009, 34, 417–423. [Google Scholar]
- Steinbach, D.; Schramm, A.; Eggert, A.; Onda, M.; Dawczynski, K.; Rump, A.; Pastan, I.; Wittig, S.; Pfaffendorf, N.; Voigt, A.; et al. Identification of a set of seven genes for the monitoring of minimal residual disease in pediatric acute myeloid leukemia. Clin. Cancer Res. 2006, 12, 2434–2441. [Google Scholar] [CrossRef]
- Jonsson, E.; Ljungkvist, I.; Hamberg, J. Standardized measurement of lateral spinal flexion and its use in evaluation of the effect of treatment of chronic low back pain. Upsala J. Med. Sci. 1990, 95, 75–86. [Google Scholar] [CrossRef]
- He, X.; Li, W.; Liang, X.; Zhu, X.; Zhang, L.; Huang, Y.; Yu, T.; Li, S.; Chen, Z. IGF2BP2 Overexpression Indicates Poor Survival in Patients with Acute Myelocytic Leukemia. Cell. Physiol. Biochem. 2018, 51, 1945–1956. [Google Scholar] [CrossRef]
- Lian, X.Y.; Zhang, W.; Wu, D.H.; Ma, J.C.; Zhou, J.D.; Zhang, Z.H.; Wen, X.M.; Xu, Z.J.; Lin, J.; Qian, J. Methylation-independent ITGA2 overexpression is associated with poor prognosis in de novo acute myeloid leukemia. J. Cell. Physiol. 2018, 233, 9584–9593. [Google Scholar] [CrossRef] [PubMed]
- Lindblad, O.; Chougule, R.A.; Moharram, S.A.; Kabir, N.N.; Sun, J.; Kazi, J.U.; Ronnstrand, L. The role of HOXB2 and HOXB3 in acute myeloid leukemia. Biochem. Biophys. Res. Commun. 2015, 467, 742–747. [Google Scholar] [CrossRef]
- Gambacorta, V.; Gnani, D.; Vago, L.; Di Micco, R. Epigenetic Therapies for Acute Myeloid Leukemia and Their Immune-Related Effects. Front. Cell Dev. Biol. 2019, 7, 207. [Google Scholar] [CrossRef] [PubMed]
- de Rooij, J.D.; Zwaan, C.M.; van den Heuvel-Eibrink, M. Pediatric AML: From Biology to Clinical Management. J. Clin. Med. 2015, 4, 127–149. [Google Scholar] [CrossRef] [PubMed]
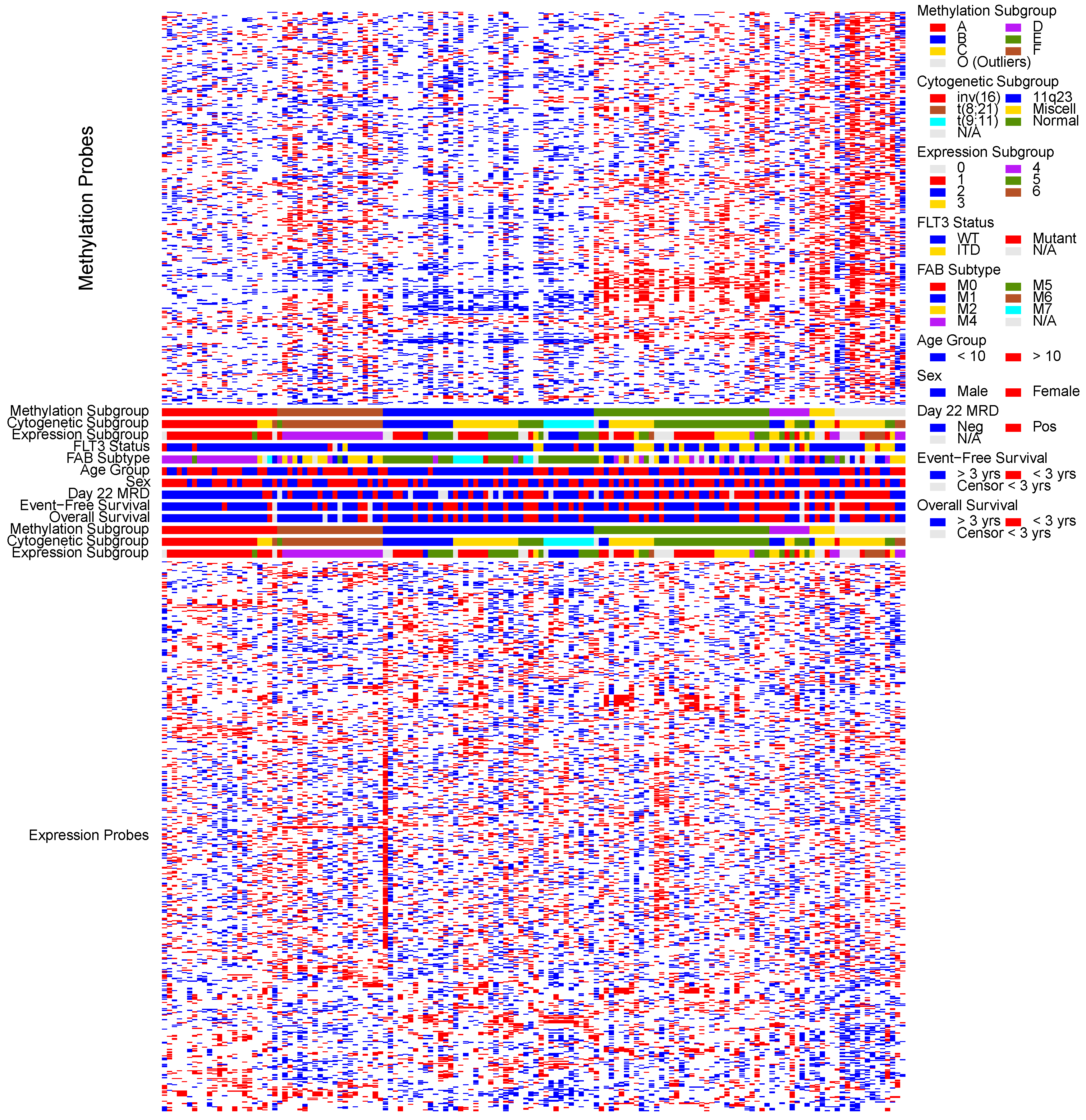
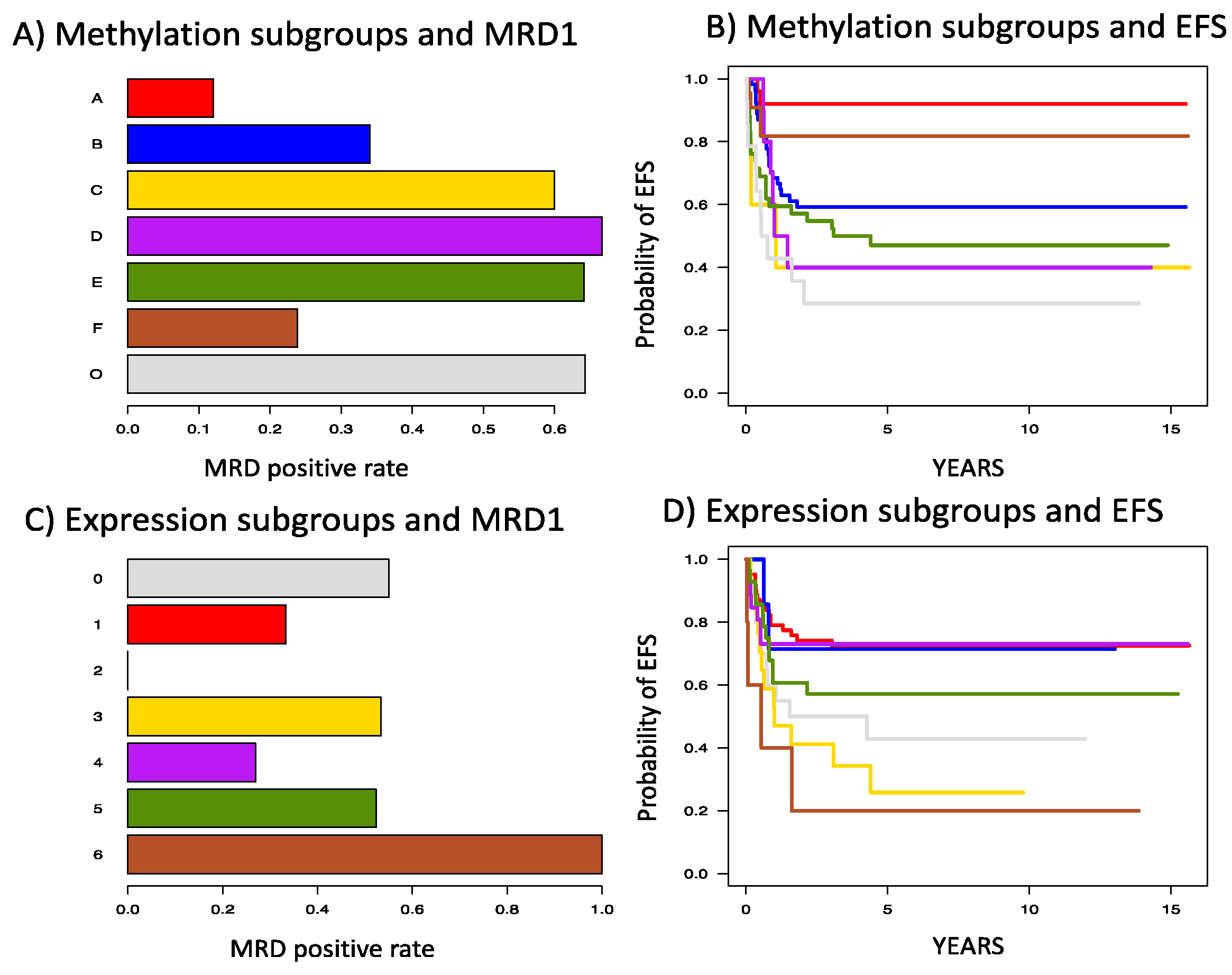

| Methylation Subgroup | 11q23 (n = 23) | Insuff (n = 3) | inv(16) (n = 22) | Miscell (n = 47) | Normal (N = 39) | t(8;21) (n = 24) | t(9;11) (n = 14) | Total | MRD-Inevaluable (n = 12) | MRD-Negative n (%) | MRD-Positive n (%) |
| A | 0 | 0 | 20 | 3 | 1 | 1 | 0 | 25 | 0 | 22 (88%) | 3 (12%) |
| B | 16 | 1 | 0 | 17 | 7 | 0 | 13 | 54 | 7 (13%) | 31 (57.4%) | 16 (29.6%) |
| C | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 5 | 0 | 2 (40%) | 3 (60%) |
| D | 3 | 1 | 0 | 2 | 3 | 0 | 1 | 10 | 1 (10%) | 3 (30%) | 6 (60%) |
| E | 3 | 1 | 1 | 11 | 26 | 0 | 0 | 42 | 3 (7.15%) | 14 (33.3%) | 25 (59.5%) |
| F | 0 | 0 | 0 | 0 | 1 | 21 | 0 | 22 | 1 (4.5%) | 16 (72.72%) | 5 (22.7%) |
| O | 0 | 0 | 1 | 10 | 1 | 2 | 0 | 14 | 0 | 5 (35.71%) | 9 (64.3%) |
| Expression Subgroup | 11q23 (n = 21) | Insuff (n = 1) | inv(16) (n = 21) | Miscell (n = 45) | Normal (N = 42) | t(8;21) (n = 24) | t(9;11) (n = 11) | Total | MRD-Inevaluable (n = 12) | MRD-Negative n (%) | MRD-Positive n (%) |
| 0 | 2 | 0 | 1 | 9 | 6 | 1 | 1 | 20 | 0 | 9 (45%) | 11 (55%) |
| 1 | 8 | 0 | 18 | 19 | 16 | 0 | 1 | 62 | 2 (3.2%) | 40 (64.5%) | 20 (32.3%) |
| 2 | 1 | 0 | 0 | 0 | 0 | 0 | 6 | 7 | 1 (14.3%) | 6 (85.7%) | 0 (0%) |
| 3 | 3 | 0 | 0 | 2 | 12 | 0 | 0 | 17 | 2 (11.7%) | 7 (41.2%) | 8 (47.1%) |
| 4 | 0 | 0 | 1 | 1 | 1 | 23 | 0 | 26 | 0 | 19 (73.1%) | 7 (26.9%) |
| 5 | 7 | 1 | 1 | 9 | 7 | 0 | 3 | 28 | 7 (38.8%) | 10 (35.7%) | 11 (39.3%) |
| 6 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | 0 (0%) | 5 (100%) |
| inv(16) vs. Non inv(16) | t(8;21) vs. Non t(8;21) | t(9;11) vs Non t(9;11) | Other 11q23 vs Non 11q23 | Normal Cytogenetics vs. Non Normal Cytogentics | Other Abnormalities vs. Not | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| illumina_ID | CHR | Gene Name | inv(16)vs. Non inv 16 p Value | illumina_ID | CHR | Gene Name | t(8;21) vs. Non t(8;21) p Value | illumina_ID | CHR | Gene Name | t(9;11) vs. Non t(9;11) p Value | illumina_ID | CHR | Gene Name | 11q23 vs. Non 11q23 p Value | illumina_ID | CHR | Gene Name | Normal vs. non Normal p Value | illumina_ID | CHR | Gene Name | Others vs. Non Others p Value |
| cg11757040 | 14 | DPF3 | 1.08 × 10−19 | cg06640020 | 6 | BX649158 | 1.10 × 10−29 | cg27486692 | 12 | PLBD1 | 1.07 × 10−13 | cg22364219 | 1 | AL832937 | 1.16 × 10−10 | cg04498190 | 1 | TRIM46 | 1.21 × 10−10 | cg04738464 | 2 | TNS1 | 6.66 × 10−10 |
| cg13588403 | 14 | DPF3 | 1.38 × 10−19 | cg00948274 | 16 | SOLH | 1.07 × 10−27 | cg25670451 | 12 | SCNN1A | 2.55 × 10−13 | cg05817709 | 11 | RARRES3 | 2.22 × 10−9 | cg13372766 | 1 | OBSCN | 1.42 × 10−10 | cg02147126 | 19 | AZU1 | 8.94 × 10−10 |
| cg01138706 | 2 | TNS1 | 1.65 × 10−17 | cg04194341 | 13 | DZIP1 | 2.15 × 10−27 | cg20373894 | 2 | BC071802 | 7.20 × 10−13 | cg18061433 | 10 | C10orf113 | 6.98 × 10−9 | cg13199615 | 1 | TRIM46 | 2.43 × 10−9 | cg26677004 | 1 | TOR1AIP2 | 1.04 × 10−9 |
| cg26953469 | 10 | JA682651 | 2.88 × 10−17 | cg09057885 | 21 | COL18A1 | 2.15 × 10−27 | cg15605307 | 11 | MIR1237 | 9.99 × 10−13 | cg07568841 | 7 | MIR550B1 | 8.02 × 10−9 | cg18832152 | 1 | EFNA3 | 3.79 × 10−9 | cg23678154 | 2 | MIR3126 | 3.74 × 10−9 |
| cg15256743 | 12 | PLXNC1 | 7.04 × 10−17 | cg24378253 | 1 | RNF223 | 3.00 × 10−27 | cg01219000 | 4 | CXCL5 | 1.70 × 10−12 | cg04753583 | 21 | PCBP3 | 8.02 × 10−9 | cg15474859 | 2 | KLF7 | 1.80 × 10−9 | cg25361961 | 19 | ZNF550 | 3.83 × 10−9 |
| cg26909217 | 16 | ZFHX3 | 1.31 × 10−15 | cg00502209 | 15 | LRRK1 | 4.12 × 10−27 | cg19954448 | 17 | CYTH1 | 1.88 × 10−12 | cg13933080 | 10 | SPAG6 | 5.27 × 10−8 | cg06500792 | 2 | AK097952 | 2.56 × 10−9 | cg01877450 | 7 | BRI3 | 7.62 × 10−9 |
| cg09221269 | 21 | RUNX1 | 1.31 × 10−15 | cg07088935 | 2 | TPO | 1.01 × 10−26 | cg00722188 | 14 | CCDC88C | 2.55 × 10−12 | cg03124514 | 5 | AK056817 | 6.16 × 10−8 | cg02913882 | 3 | TBL1XR1 | 1.80 × 10−9 | cg15134583 | 1 | TP53BP2 | 1.07 × 10−8 |
| cg15796340 | 17 | PRCD | 1.42 × 10−15 | cg24216893 | 2 | TPO | 1.01 × 10−26 | cg00704819 | 6 | ANKRD6 | 2.82 × 10−12 | cg01067139 | 5 | AK056817 | 1.11 × 10−7 | cg05429527 | 6 | FOXC1 | 5.86 × 10−10 | cg15487251 | 3 | IGF2BP2 | 1.12 × 10−8 |
| cg09599228 | 4 | RXFP1 | 1.91 × 10−15 | cg02914962 | 6 | BX649158 | 1.34 × 10−26 | cg19690404 | 7 | CPA4 | 4.18 × 10−12 | cg13494087 | 9 | KLF4 | 2.33 × 10−7 | cg25583983 | 6 | MICA | 9.57 × 10−10 | cg18780288 | 10 | XPNPEP1 | 1.25 × 10−8 |
| cg06343673 | 17 | RPTOR | 3.43 × 10−15 | cg04734276 | 5 | AX747985 | 1.76 × 10−26 | cg03969651 | 17 | TBX4 | 4.60 × 10−12 | cg01146808 | 6 | PRDM1 | 4.77 × 10−7 | cg27144224 | 6 | MICA | 1.06 × 10−9 | cg02119938 | 15 | ACSBG1 | 1.34 × 10−8 |
| cg19755069 | 1 | CSF1 | 4.26 × 10−15 | cg05408203 | 16 | CACNA1H | 3.87 × 10−26 | cg07877629 | 16 | CMIP | 6.12 × 10−12 | cg19744908 | 1 | LPHN2 | 6.85 × 10−7 | cg20028974 | 6 | SLC2A12 | 1.27 × 10−9 | cg10752406 | 19 | AZU1 | 1.53 × 10−8 |
| cg20536921 | 15 | MEF2A | 5.65 × 10−15 | cg05019221 | 2 | OTOS | 8.10 × 10−26 | cg22042908 | 20 | RASSF2 | 7.38 × 10−12 | cg08004620 | 8 | AX747124 | 1.31 × 10−6 | cg12837744 | 6 | SYNGAP1 | 1.48 × 10−9 | cg14870461 | 3 | C3orf64 | 1.94 × 10−8 |
| cg20331980 | 6 | TAB2 | 6.98 × 10−15 | cg04011470 | 8 | PHYHIP | 8.10 × 10−26 | cg10100652 | 10 | LOC643529 | 8.88 × 10−12 | cg19743406 | 6 | LHFPL5 | 1.54 × 10−6 | cg24899846 | 6 | MICA | 3.61 × 10−9 | cg16535752 | 17 | MIR4729 | 2.03 × 10−8 |
| cg23013977 | 19 | CLIP3 | 9.20 × 10−15 | cg13967936 | 1 | GJA4 | 1.29 × 10−25 | cg15063726 | 11 | HPX | 1.06 × 10−11 | cg08390877 | 2 | EN1 | 1.67 × 10−6 | cg05852760 | 7 | IGF2BP3 | 4.51 × 10−10 | cg04087271 | 1 | CDA | 2.36 × 10−8 |
| cg27587645 | 19 | BTBD2 | 1.39 × 10−14 | cg12946705 | 6 | FAM120B | 1.29 × 10−25 | cg11726288 | 1 | NPPA-AS1 | 1.16 × 10−11 | cg01369082 | 5 | BC045578 | 2.28 × 10−6 | cg19507725 | 7 | HIP1 | 5.28 × 10−10 | cg16643542 | 19 | AZU1 | 2.42 × 10−8 |
| cg08124030 | 3 | TM4SF1 | 2.36 × 10−14 | cg13751417 | 9 | VAV2 | 2.54 × 10−25 | cg13412283 | 7 | AK024243 | 1.27 × 10−11 | cg25714774 | 2 | TRAPPC12 | 2.34 × 10−6 | cg10902549 | 10 | MTG1 | 3.35 × 10−9 | cg07018107 | 8 | MIR1208 | 2.94 × 10−8 |
| cg20299572 | 11 | MOB2 | 2.69 × 10−14 | cg08178815 | 20 | ADRM1 | 3.15 × 10−25 | cg04241282 | 3 | LARS2 | 1.39 × 10−11 | cg07217350 | 5 | MAT2B | 2.34 × 10−6 | cg18571531 | 11 | AK097446 | 6.17 × 10−10 | cg03082779 | 20 | LOC100131496 | 2.94 × 10−8 |
| cg22549879 | 2 | PKI55 | 3.72 × 10−14 | cg17712092 | 4 | LARP1B | 3.91 × 10−25 | cg11235411 | 10 | FW312330 | 1.39 × 10−11 | cg11702085 | 6 | TRNA_Leu | 2.34 × 10−6 | cg00667026 | 14 | ZFP36L1 | 3.20 × 10−10 | cg23606421 | 9 | ENTPD8 | 3.07 × 10−8 |
| cg04596071 | 7 | MAD1L1 | 4.51 × 10−14 | cg05716567 | 8 | ZC3H3 | 3.91 × 10−25 | cg09107053 | 1 | C1orf198 | 1.66 × 10−11 | cg01826444 | 1 | FNDC7 | 2.40 × 10−6 | cg14876117 | 14 | PYGL | 2.62 × 10−9 | cg14663914 | 19 | AZU1 | 3.20 × 10−8 |
| cg02065790 | 12 | PPFIBP1 | 6.19 × 10−14 | cg13593887 | 9 | C9orf62 | 3.91 × 10−25 | cg15889057 | 12 | ITGB7 | 1.66 × 10−11 | cg06626359 | 14 | PRIMA1 | 2.46 × 10−6 | cg15698066 | 14 | EVL | 3.19 × 10−9 | cg13551117 | 2 | Mir_562 | 3.27 × 10−8 |
| cg11944797 | 13 | STK24 | 1.22 × 10−13 | cg07912453 | 15 | PGPEP1L | 4.82 × 10−25 | cg05000598 | 1 | TTC24 | 1.81 × 10−11 | cg18281743 | 5 | STC2 | 2.73 × 10−6 | cg07199524 | 15 | CRTC3 | 3.37 × 10−10 | cg10221596 | 2 | ALS2CR12 | 3.34 × 10−8 |
| cg10839684 | 17 | LOC100499466 | 1.55 × 10−13 | cg27111634 | 9 | DQ593554 | 5.93 × 10−25 | cg09579869 | 1 | Mir_584 | 1.97 × 10−11 | cg20312179 | 1 | EPS15 | 3.10 × 10−6 | cg18942110 | 15 | CRTC3 | 9.82 × 10−10 | cg00153306 | 3 | TMIE | 3.34 × 10−8 |
| cg03451731 | 17 | GP1BA | 1.65 × 10−13 | cg23731501 | 1 | MIR429 | 7.28 × 10−25 | cg21642076 | 22 | TMEM191A | 1.97 × 10−11 | cg03847796 | 1 | MAB21L3 | 3.10 × 10−6 | cg19134665 | 15 | CRTC3 | 1.76 × 10−9 | cg03216580 | 10 | KLF6 | 3.34 × 10−8 |
| cg05247914 | 19 | FXYD1 | 1.65 × 10−13 | cg08166720 | 17 | 5S_rRNA | 1.09 × 10−24 | cg17262492 | 20 | PCIF1 | 2.34 × 10−11 | cg01296593 | 16 | FOXC2 | 3.51 × 10−6 | cg27326306 | 16 | AK056683 | 1.99 × 10−9 | cg16808455 | 18 | TUBB6 | 4.05 × 10−8 |
| cg16747164 | 4 | TLR10 | 3.16 × 10−13 | cg13028635 | 11 | LSP1 | 1.32 × 10−24 | cg12167489 | 8 | RBPMS | 2.55 × 10−11 | cg12354192 | 3 | Y_RNA | 3.60 × 10−6 | cg18200326 | 16 | FHOD1 | 2.43 × 10−9 | cg24534743 | 1 | AHDC1 | 4.14 × 10−8 |
| cg17384769 | 6 | LOC100128176 | 3.76 × 10−13 | cg17820878 | 1 | SLC9A1 | 1.61 × 10−24 | cg09518969 | 11 | PLA2G16 | 2.55 × 10−11 | cg06626126 | 3 | MECOM | 3.79 × 10−6 | cg24210813 | 17 | HOXB5 | 5.71 × 10−14 | cg16727006 | 16 | ZCCHC14 | 4.14 × 10−8 |
| cg18908677 | 10 | BAG3 | 3.98 × 10−13 | cg17696563 | 2 | NEU4 | 1.61 × 10−24 | cg14949065 | 2 | TCF23 | 2.78 × 10−11 | cg20961007 | 1 | AX747534 | 3.88 × 10−6 | cg05487507 | 17 | HOXB5 | 1.55 × 10−12 | cg09053680 | 10 | UTF1 | 4.80 × 10−8 |
| cg08715862 | 16 | FHOD1 | 4.21 × 10−13 | cg26786253 | 17 | 5S_rRNA | 1.94 × 10−24 | cg05498785 | 16 | IRF8 | 2.78 × 10−11 | cg12610471 | 10 | SPAG6 | 3.88 × 10−6 | cg12744859 | 17 | HOXB5 | 3.08 × 10−12 | cg13521941 | 9 | ENTPD8 | 4.90 × 10−8 |
| cg03440386 | 7 | TCRBV9S1A1T | 5.60 × 10−13 | cg18851522 | 14 | KIF26A | 2.35 × 10−24 | cg22718696 | 1 | MTOR | 3.02 × 10−11 | cg07777156 | 17 | TRNA | 3.88 × 10−6 | cg07676709 | 17 | HOXB6 | 4.06 × 10−12 | cg23098529 | 19 | PPAN-P2RY11 | 5.12 × 10−8 |
| cg10368834 | 16 | RUNDC2C | 6.63 × 10−13 | cg01074955 | 1 | NPHP4 | 3.40 × 10−24 | cg05483534 | 2 | BC048424 | 3.02 × 10−11 | cg23361764 | 11 | ODZ4 | 4.08 × 10−6 | cg18127922 | 17 | HOXB6 | 8.14 × 10−12 | cg04848343 | 19 | PLIN5 | 5.22 × 10−8 |
| cg14381313 | 16 | AK126852 | 7.42 × 10−13 | cg14141549 | 7 | DQ583756 | 3.40 × 10−24 | cg01391022 | 12 | WDR66 | 3.28 × 10−11 | cg23739746 | 13 | RASA3 | 4.62 × 10−6 | cg00711072 | 17 | HOXB6 | 9.73 × 10−12 | cg08498533 | 10 | TTC40 | 5.57 × 10−8 |
| cg20436086 | 10 | COL13A1 | 7.84 × 10−13 | cg06782041 | 1 | LDLRAD2 | 4.08 × 10−24 | cg06099459 | 12 | C12orf77 | 3.28 × 10−11 | cg02273477 | 17 | NXN | 4.62 × 10−6 | cg00072689 | 17 | HOXB6 | 1.00 × 10−11 | cg16672770 | 16 | PIEZO1 | 5.93 × 10−8 |
| cg02035018 | 4 | MXD4 | 8.29 × 10−13 | cg08447200 | 22 | TRABD | 4.89 × 10−24 | cg00979704 | 6 | TNXB | 3.56 × 10−11 | cg07038400 | 3 | PPP2R3A | 5.49 × 10−6 | cg24173049 | 17 | SLC16A13 | 3.32 × 10−11 | cg16378063 | 17 | FLJ40194 | 6.05 × 10−8 |
| cg17873456 | 13 | EFNB2 | 9.78 × 10−13 | cg23841288 | 7 | AX746826 | 5.84 × 10−24 | cg02746781 | 16 | DPEP2 | 3.56 × 10−11 | cg22429199 | 3 | CRYBG3 | 5.77 × 10−6 | cg26916621 | 17 | MIR10A | 4.41 × 10−11 | cg17645677 | 5 | PPARGC1B | 6.58 × 10−8 |
| cg00303450 | 2 | MFSD6 | 1.03 × 10−12 | cg22141235 | 11 | MRGPRE | 5.84 × 10−24 | cg03543120 | 5 | MIR143 | 3.87 × 10−11 | cg00446123 | 20 | LIME1 | 5.77 × 10−6 | cg01405107 | 17 | HOXB5 | 5.54 × 10−11 | cg13984746 | 17 | HEXDC | 6.58 × 10−8 |
| cg01541443 | 7 | C7orf41 | 1.09 × 10−12 | cg21876283 | 1 | LDLRAD2 | 6.97 × 10−24 | cg00468395 | 16 | XPO6 | 3.87 × 10−11 | cg07989568 | 17 | TEKT1 | 5.91 × 10−6 | cg10308785 | 17 | HOXB6 | 6.37 × 10−11 | cg01980222 | 6 | TREM2 | 6.86 × 10−8 |
| cg24400630 | 1 | GBP5 | 1.15 × 10−12 | cg13411506 | 13 | MCF2L | 8.30 × 10−24 | cg03721994 | 19 | MED16 | 3.87 × 10−11 | cg23570923 | 4 | HTRA3 | 6.21 × 10−6 | cg00690402 | 17 | HOXB5 | 8.19 × 10−11 | cg07170045 | 8 | FGF17 | 7.94 × 10−8 |
| cg00346376 | 5 | SLC12A7 | 1.15 × 10−12 | cg01403811 | 22 | ELFN2 | 8.30 × 10−24 | cg14839919 | 7 | C7orf45 | 4.20 × 10−11 | cg16419354 | 1 | FAM163A | 6.36 × 10−6 | cg04196862 | 17 | LOC404266 | 1.87 × 10−10 | cg07714276 | 6 | RREB1 | 8.81 × 10−8 |
| cg00059737 | 1 | VPS13D | 1.22 × 10−12 | cg00450617 | 2 | TPO | 9.86 × 10−24 | cg26164773 | 10 | TIAL1 | 4.56 × 10−11 | cg06179127 | 7 | AK024243 | 6.36 × 10−6 | cg01572694 | 17 | MIR10A | 2.38 × 10−10 | cg06495615 | 19 | ARID3A | 8.81 × 10−8 |
| cg11216632 | 1 | ARHGEF2 | 1.29 × 10−12 | cg16520049 | 17 | FBF1 | 9.86 × 10−24 | cg01826337 | 1 | CALML6 | 4.94 × 10−11 | cg09785391 | 18 | ZFP161 | 6.36 × 10−6 | cg21864868 | 17 | LOC404266 | 2.87 × 10−10 | cg04891053 | 1 | PVRL4 | 8.99 × 10−8 |
| cg21042919 | 17 | ARHGAP23 | 1.43 × 10−12 | cg02664177 | 7 | PDGFA | 1.17 × 10−23 | cg11493223 | 17 | TMC6 | 5.35 × 10−11 | cg04392469 | 1 | MOSC2 | 6.68 × 10−6 | cg15435170 | 17 | HOXB6 | 2.95 × 10−10 | cg08190450 | 17 | NXN | 8.99 × 10−8 |
| cg15031685 | 7 | ATP6V1F | 1.51 × 10−12 | cg02113067 | 11 | FERMT3 | 1.17 × 10−23 | cg05165250 | 18 | RAB31 | 5.35 × 10−11 | cg22874893 | 6 | RBM24 | 7.35 × 10−6 | cg21816532 | 17 | HOXB6 | 5.01 × 10−10 | cg10973881 | 1 | TMOD4 | 9.18 × 10−8 |
| cg21950720 | 1 | EXTL1 | 1.68 × 10−12 | cg00155679 | 7 | CNPY1 | 1.64 × 10−23 | cg24166628 | 3 | TNNC1 | 5.80 × 10−11 | cg03930153 | 3 | TBL1XR1 | 7.53 × 10−6 | cg14232289 | 17 | SSTR2 | 1.14 × 10−9 | cg23327851 | 14 | RNASE2 | 9.97 × 10−8 |
| cg19939077 | 10 | PPIF | 1.68 × 10−12 | cg15769475 | 17 | 1.64 × 10−23 | cg05480883 | 10 | ITIH5 | 5.80 × 10−11 | cg16536330 | 8 | LEPROTL1 | 7.53 × 10−6 | cg01986016 | 17 | HOXB6 | 1.99 × 10−9 | cg12136387 | 12 | MIR4472-2 | 1.04 × 10−7 | |
| cg15482928 | 1 | PTPRF | 1.98 × 10−12 | cg05345823 | 11 | MUC6 | 1.93 × 10−23 | cg12146447 | 17 | TAC4 | 5.80 × 10−11 | cg16949914 | 10 | IL2RA | 8.09 × 10−6 | cg01723934 | 17 | HOXB6 | 2.26 × 10−9 | cg17125472 | 11 | AK094674 | 1.06 × 10−7 |
| cg04155862 | 3 | MGLL | 1.98 × 10−12 | cg05322931 | 17 | NXN | 2.27 × 10−23 | cg15064964 | 2 | ASB1 | 7.35 × 10−11 | cg18075627 | 12 | MIR1178 | 8.29 × 10−6 | cg22660299 | 17 | LOC404266 | 2.89 × 10−9 | cg06383124 | 16 | LOC400548 | 1.15 × 10−7 |
| cg11667400 | 13 | UBAC2 | 2.09 × 10−12 | cg21268658 | 1 | PTPRF | 2.67 × 10−23 | cg22014983 | 2 | U6 | 7.35 × 10−11 | cg15871647 | 4 | PARM1 | 8.49 × 10−6 | cg10749413 | 19 | ABCA7 | 2.42 × 10−11 | cg23230830 | 1 | LOC646627 | 1.17 × 10−7 |
| cg03790192 | 16 | BANP | 2.09 × 10−12 | cg02960418 | 7 | PDGFA | 3.14 × 10−23 | cg22870280 | 15 | BCL2L10 | 7.94 × 10−11 | cg02603128 | 9 | FRMD3 | 8.49 × 10−6 | cg01614597 | 19 | LOC113230 | 3.35 × 10−9 | cg08493063 | 16 | RMI2 | 1.22 × 10−7 |
| cg05497253 | 5 | LHFPL2 | 2.32 × 10−12 | cg07503662 | 16 | KLHDC4 | 3.14 × 10−23 | cg05923736 | 10 | ECHS1 | 9.27 × 10−11 | cg24891133 | 13 | C13orf33 | 8.91 × 10−6 | cg21053323 | 21 | SUMO3 | 3.11 × 10−10 | cg19573490 | 17 | PCYT2 | 1.30 × 10−7 |
| cg14749678 | 3 | CCRL2 | 2.72 × 10−12 | cg03614513 | 1 | TSPAN2 | 4.31 × 10−23 | cg26616258 | 16 | PDIA2 | 1.00 × 10−10 | cg09335658 | 7 | RBM28 | 9.12 × 10−6 | cg16932065 | 22 | LIMK2 | 2.82 × 10−9 | cg18734433 | 7 | ZNF775 | 1.33 × 10−7 |
| Cytogenetic Subgroup | Chr | Gene | GroupMedian: Presence of Cytogenetic Feature | GroupMedian: Absence of Cytogenetic Feature | p Value | FDR Adjusted p | Complementary Cytogenetic Lesion |
|---|---|---|---|---|---|---|---|
| inv(16) [CBFB-MYH11] | 16 | CBFB | 2.456 | 1.012 | 3.97 × 10−14 | 3.08 × 10−10 | Cis-cytogenetic lesion |
| inv(16) [CBFB-MYH11] | 16 | MYH11 | 2.448 | 4.063 | 3.18 × 10−12 | 7.26 × 10−9 | Cis-cytogenetic lesion |
| inv(16) [CBFB-MYH11] | 21 | RUNX1 | 2.512 | 4.213 | 1.31 × 10−15 | 2.91 × 10−11 | Trans Cytogenetic lesion-Fusion gene for t(8;21) |
| inv(16) [CBFB-MYH11] | 01 | RUNX3 | −0.6088 | 0.7442 | 6.27 × 10−13 | 2.30 × 10−9 | |
| inv(16) [CBFB-MYH11] | 21 | RUNX1 | 0.703 | 2.305 | 2.01 × 10−11 | 2.89 × 10−8 | Trans Cytogenetic lesion-Fusion gene for t(8;21) |
| inv(16) [CBFB-MYH11] | 21 | RUNX1 | −0.6719 | 0.9548 | 2.10 × 10−11 | 2.94 × 10−8 | Trans Cytogenetic lesion-Fusion gene for t(8;21) |
| inv(16) [CBFB-MYH11] | 1 | RUNX3 | −3.508 | −0.8562 | 1.09 × 10−10 | 1.00 × 10−7 | |
| inv(16) [CBFB-MYH11] | 19 | FLT3LG | −2.486 | −0.4993 | 4.45 × 10−11 | 5.19 × 10−8 | |
| inv(16) [CBFB-MYH11] | 1 | GLIS1 | 0.096 | 2.2 | 4.33 × 10−12 | 9.24 × 10−9 | |
| Normal | 17 | MLLT6 | 0.348 | −1.732 | 9.67 × 10−9 | 8.17 × 10−6 | |
| O11q23 [MLL-X] | 16 | CBFA2T3 | 1.023 | −1.555 | 4.58 × 10−9 | 4.92 × 10−5 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| O11q23 [MLL-X] | 16 | CBFA2T3 | 2.398 | 0.9998 | 7.49 × 10−9 | 6.22 × 10−5 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| O11q23 [MLL-X] | 16 | CBFA2T3 | −0.2261 | −2.417 | 3.71 × 10−8 | 0.00014808 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| O11q23 [MLL-X] | 16 | CBFA2T3 | 1.76 | −0.2214 | 6.36 × 10−8 | 0.00019022 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| O11q23 [MLL-X] | 16 | CBFA2T3 | −0.4395 | −2.155 | 3.03 × 10−7 | 0.00039603 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| O11q23 [MLL-X] | 09 | MLLT3 | −0.6595 | −1.311 | 3.59 × 10−8 | 0.00014478 | |
| t(8;21) [RUNX1-RUNX1T1] | 16 | CBFA2T3 | −0.5581 | 3.259 | 1.93 × 10−23 | 6.22 × 10−20 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| t(8;21) [RUNX1-RUNX1T1] | 16 | CBFA2T3 | 0.1236 | 3.28 | 2.97 × 10−22 | 5.79 × 10−19 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| t(8;21) [RUNX1-RUNX1T1] | 16 | CBFA2T3 | 0.0312 | −3.232 | 5.93 × 10−22 | 1.04 × 10−18 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| t(8;21) [RUNX1-RUNX1T1] | 16 | CBFA2T3 | 0.1551 | 2.453 | 2.96 × 10−20 | 2.57 × 10−17 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| t(8;21) [RUNX1-RUNX1T1] | 16 | CBFA2T3 | 0.1614 | 3.657 | 6.32 × 10−20 | 4.85 × 10−17 | Trans Cytogenetic lesion-Fusion gene inv16-CBFA2T3-GLIS2) |
| t(8;21) [RUNX1-RUNX1T1] | 06 | MLLT4 | −0.1954 | 3.923 | 1.10 × 10−29 | 1.30 × 10−24 | Trans Cytogenetic lesion MLL fusion partner (11q23) |
| t(8;21) [RUNX1-RUNX1T1] | 19 | MLLT1 | −1.907 | 2.869 | 7.39 × 10−28 | 3.74 × 10−23 | Trans Cytogenetic lesion MLL fusion partner (11q23) |
| t(8;21) [RUNX1-RUNX1T1] | 6 | MLLT4 | 0.4282 | 5.277 | 1.34 × 10−26 | 2.15 × 10−22 | Trans Cytogenetic lesion MLL fusion partner (11q23) |
| t(8;21) [RUNX1-RUNX1T1] | 6 | MLLT4 | −2.402 | 2.485 | 6.86 × 10−23 | 1.80 × 10−19 | Trans Cytogenetic lesion MLL fusion partner (11q23) |
| t(9;11) | 22 | MYH9 | 1.131 | 2.282 | 1.17 × 10−10 | 9.12 × 10−8 | Trans Cytogenetic lesion MLL fusion partner (11q23) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamba, J.K.; Cao, X.; Raimondi, S.; Downing, J.; Ribeiro, R.; Gruber, T.A.; Rubnitz, J.; Pounds, S. DNA Methylation Clusters and Their Relation to Cytogenetic Features in Pediatric AML. Cancers 2020, 12, 3024. https://doi.org/10.3390/cancers12103024
Lamba JK, Cao X, Raimondi S, Downing J, Ribeiro R, Gruber TA, Rubnitz J, Pounds S. DNA Methylation Clusters and Their Relation to Cytogenetic Features in Pediatric AML. Cancers. 2020; 12(10):3024. https://doi.org/10.3390/cancers12103024
Chicago/Turabian StyleLamba, Jatinder K., Xueyuan Cao, Susana Raimondi, James Downing, Raul Ribeiro, Tanja A. Gruber, Jeffrey Rubnitz, and Stanley Pounds. 2020. "DNA Methylation Clusters and Their Relation to Cytogenetic Features in Pediatric AML" Cancers 12, no. 10: 3024. https://doi.org/10.3390/cancers12103024
APA StyleLamba, J. K., Cao, X., Raimondi, S., Downing, J., Ribeiro, R., Gruber, T. A., Rubnitz, J., & Pounds, S. (2020). DNA Methylation Clusters and Their Relation to Cytogenetic Features in Pediatric AML. Cancers, 12(10), 3024. https://doi.org/10.3390/cancers12103024







