Simple Summary
Chemoresistance and metastasis are the main causes of treatment failure in cancers. Autophagy contribute to the survival and metastasis of cancer cells. Competing endogenous RNA (ceRNA), particularly long non-coding RNAs and circular RNA (circRNA), can bridge the interplay between autophagy and chemoresistance or metastasis in cancers via sponging miRNAs. This review aims to discuss on the function of ceRNA-mediated autophagy in the process of metastasis and chemoresistance in cancers. ceRNA network can sequester the targeted miRNA expression to indirectly upregulate the expression of autophagy-related genes, and thereof participate in autophagy-mediated chemoresistance and metastasis. Our clarification of the mechanism of autophagy regulation in metastasis and chemoresistance may greatly improve the efficacy of chemotherapy and survival in cancer patients. The combination of the tissue-specific miRNA delivery and selective autophagy inhibitors, such as hydroxychloroquine, is attractive to treat cancer patients in the future.
Abstract
Chemoresistance and metastasis are the main causes of treatment failure and unfavorable outcome in cancers. There is a pressing need to reveal their mechanisms and to discover novel therapy targets. Autophagy is composed of a cascade of steps controlled by different autophagy-related genes (ATGs). Accumulating evidence suggests that dysregulated autophagy contributes to chemoresistance and metastasis via competing endogenous RNA (ceRNA) networks including lncRNAs and circRNAs. ceRNAs sequester the targeted miRNA expression to indirectly upregulate ATGs expression, and thereof participate in autophagy-mediated chemoresistance and metastasis. Here, we attempt to summarize the roles of ceRNAs in cancer chemoresistance and metastasis through autophagy regulation.
Keywords:
autophagy; ceRNA; lncRNA; circRNAs; miRNA; chemoresistance; metastasis; pre-metastasis niche 1. Introduction
Autophagy is an evolutionarily conserved system to maintain homeostasis in various cells. In general, there are three main types of autophagy: macroautophagy, microautophagy and mitoautophagy [1]. Mitoautophagy can remove damaged mitochondria specifically by autophagosomes for lysosomal degradation while microautophagy, mainly in plants and fungi, may complete the isolation and uptake of cell components by directly enveloping them with the vacuolar/lysosomal membrane. Macroautophagy is the best-characterized process that aims to remove unwanted or damaged organelles and aggregated proteins by lysosome degradation. In the narrow sense, some scientists (including those in this article) have referred to macroautophagy as autophagy.
The dysregulation of autophagy has been reported in many human diseases, such as infection, cardiovascular and neurodegenerative diseases, and cancers [2]. Metastasis and chemoresistance are two major factors that are associated with cancer recurrence and dismal clinical outcomes. Moreover, metastasis and chemoresistance are closely linked in cancers, as metastatic cancer cells have a propensity for chemoresistance [3,4]. The roles of autophagy in the processes of cancer metastasis and chemoresistance are starting to be recognized. Autophagy is critically involved in many aspects of cancer metastasis, including epithelial–mesenchymal transition (EMT), invasion, migration, anoikis resistance, crosstalk between cancer and stromal cells, and immune suppression [5]. Cancer cells may survive chemotherapy through protective autophagy [6,7,8].
Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) are non-coding RNAs (ncRNAs) with respective lengths of more than 200 nucleotides and about 22 nucleotides. Both participate in various physiological and pathological processes involving tumorigenesis, such as cell proliferation, apoptosis, EMT, invasion, migration, chemoresistance, and stemness [9]. lncRNAs can regulate gene expression through repressing chromatin components [10], mediating epigenetic silencing, interacting with Polycomb complex, and modulating transcription activation [11]. Considering that there is a complex interplay among messenger RNA (mRNA) and ncRNAs [12,13], lncRNAs also function as miRNA sponges by competitively binding with miRNAs, thereby releasing the targeted mRNAs [14,15]. These lncRNAs are called competing endogenous RNAs (ceRNAs). They can decrease the activity or expression of miRNAs [16,17]. Circular RNA (circRNA) is another kind of ncRNA comprising covalently closed single-stranded loops. Recent advances in RNA sequencing and bioinformatics tools have led to the discovery and identification of thousands of circRNAs and validated their important roles in the pathogenesis of cancers [18,19,20,21]. They can mainly function as ceRNAs to modulate gene transcription by interacting with miRNAs or lncRNAs, sponging miRNAs, or RNA-binding proteins, and rarely can be translated into proteins [22,23,24,25]. In addition to lncRNA and circRNA, other ceRNAs have also been identified by bioinformatics analysis and experimental evidence, such as pseudogenes, mRNA including those expressing 3′-untranslated regions, virus non-coding RNAs, and genomic viral RNAs [26]. Pseudogenes, the relicts of parental gene that are unable to encode full-length proteins, can regulate the expression of parental genes via ceRNA networks, and participate in tumorigenesis [27]. Some mRNAs can function as ceRNAs and can be associated with human diseases such as cardiovascular diseases and glioblastoma multiforme [28,29]. Accumulative data have indicated that ceRNAs, particularly lncRNAs and circRNAs, can bridge the interplay between autophagy and chemoresistance or metastasis in cancers [30,31,32,33,34]. In this review, we will focus on the function of ceRNA-mediated autophagy in carcinogenesis, specifically regarding metastasis and chemoresistance.
2. The Biological Process of Autophagy
The process of autophagy is composed of several consecutive steps (Figure 1): autophagy initiation (autophagosome formation), autophagosome nucleation regulated by Beclin-1, autophagosome elongation, and autophagosome fusion with lysosome and degradation [5].
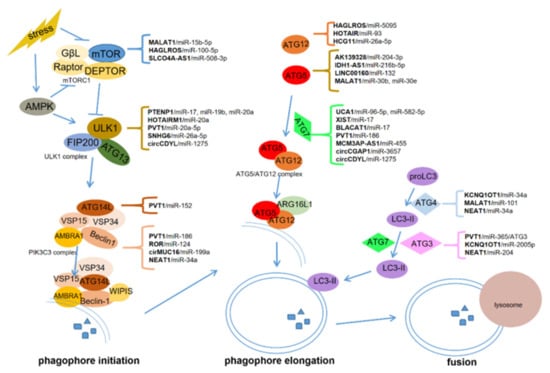
Figure 1.
Competing endogenous RNAs (ceRNAs) regulate autophagy in cancers. ceRNAs regulate autophagosome initiation or phagophore elongation by upregulating mTOR, ULK1, ATG13, ATG14L and Beclin-1, or ATG12, ATG5, ATG7, ATG4, and ATG3, respectively.
Two serine-threonine protein kinases, the mammalian target of rapamycin (mTOR) and the mammalian homologs of yeast ATG1-Unc-51-lilke kinases 1 (ULK1), are critical for autophagosome formation. mTOR can form mTOR complex 1 (mTORC1) with G protein β-subunit-like protein (GβL), Raptor, and Deptor [35,36], whereas ULK1 interacts with the focal adhesion kinase family interacting protein of 200 kDa (FIP200) and ATG13 to assemble the ULK1 complex [37,38]. Activated mTORC1 can disrupt the ULK1 complex by phosphorylating ULK1 and ATG13 [39,40,41]. Upon energy starvation, activated 5′-AMP-activated protein kinase (AMPK) directly phosphorylates ULK1, but inactivates mTORC1 [37,41]. The inactivation of mTORC1 and the intact ULK1 complex subsequently activate the class III phosphatidylinositol 3-kinase (PIK3C3) complex, containing the core components vacuolar protein sorting 15 and 34 (VSP15, VSP34) and Beclin-1 [42]. The PIK3C3 complex then converts phosphatidylinositol (PtdIns) to phosphatidylinositol 3-phosphate (PtdIns3P), which is recognized by WD-repeat protein interacting with phosphoinositides (WIPIS), the key PtdIns3P effector forming the nascent autophagosome [43,44].
Autophagosome elongation is mainly regulated by two ubiquitin-like conjugation systems: the ATG5–ATG12 complex with the help of ATG7, and LC3 with the assistance of ATG3 and ATG7 [45]. The ATG5–ATG12 complex, which interacts with ATG16L1 on the autophagosome membrane, functions as an E3 ubiquitin ligase-like enzyme in the process of autophagosome elongation [45,46]. The proLC3 (unprocessed LC3) can be processed into cytoplasmic soluble LC3-I after the cleavage by ATG4B. Subsequently, LC3-I interacts with the lipid phosphatidylethanolamine (PE), which is named membrane-binding LC3-II. LC3-II recruits p62/SQSTM1 and TBC1D25/OATL1 to provide a base for selective autophagy. Eventually, the fusion of autophagosome and lysosome generates the autophagolysosomes for degradation [45].
3. ceRNAs-Regulated Autophagy in Cancer
ceRNAs, acting as miRNA sponges, participate in many aspects of autophagy, from initiation to maturation. They can modulate autophagy phagophore initiation by upregulating mTOR, ULK1, ATG14L, Beclin-1, and autophagy phagophore elongation by upregulating ATG12, ATG5, ATG7, ATG4, and ATG3. Some ceRNAs, such as HOTAIRM1 and HAGLROS, can be involved in multiple steps in the process of autophagy by targeting different genes via multiple binding sites with miRNAs (Table 1). The interplay between ceRNAs in autophagy has gained particular attention in various cancers, particularly in hepatocellular carcinoma (HCC), colorectal cancer, and pancreatic cancer (Figure 1). Rarely, lncRNAs and circRNAs can also regulate autophagy through RNA–protein or RNA–RNA interactions, independently of the roles of ceRNA. For example, lncRNA–ATB can regulate autophagy by activating Yes-associated protein (YAP) and interacting with ATG5 directly, thereby leading to HCC progression [47]. RNA-binding protein human antigen R (HuR) can increase autophagy in intestinal epithelium by upregulating ATG16L1 expression via binding with ATG16L1 mRNA. circPABPN1 can downregulate ATG16L1 to inhibit autophagy by ejecting HuR [48]

Table 1.
ceRNAs regulate autophagy in cancers.
3.1. HOTAIRM1
HOTAIRM1 functions as a ceRNA to regulate autophagy initiation and elongation through ULK1, ATG3, ATG7, and ATG12 in cancers. A recent study has shown that HOTAIRM1 can enhance autophagosome formation to degrade oncoprotein PML–RARA by sponging miR-20a to upregulate ULK1 in acute promyelocytic leukemia (APL)-ascites mouse model and APL cell lines [52]. Analogously, HOTAIRM1 can play a tumor-suppressor role in ovarian cancers. HOTAIRM1 downregulation has been found to be associated with advanced International Federation of Gynecology & Obstetrics (FIGO) stage, while overexpression inhibits cell proliferation in vivo [82]. On the contrary, HOTAIR has been identified to be an oncogene in HCC, colorectal cancer, and chondrosarcoma. The elevated level of HOTAIR expression has been found in HCC tissues relevant to the normal tissues and to correlate with large tumor size. HOTAIR can enhance autophagy in HCC cells by upregulating ATG3 and ATG7 [83]. In colorectal cancer, HOTAIR upregulated ATG12 expression by sponging miR-93. Knockdown of HOTAIR and ATG12, or overexpression of miR-93, suppressed autophagy and restored radiosensitivity in colorectal cancer cells [58]. In chondrosarcoma, elevated expression of HOTAIR predicted advanced tumor stage and poor survival. HOTAIR knockdown impeded chondrosarcoma progression in vitro and in vivo through inhibiting autophagosome formation by downregulating ATG12 [76]. The conflicting roles of HOTAIRM1 in autophagy and carcinogenesis are determined by the different target miRNAs which may have a tissue-specific expression pattern.
3.2. HAGLROS
The overexpression of lncRNA HAGLROS has been found in gastric cancer and HCC and correlated with poor clinical outcomes in patients with these cancers [53,54]. In gastric cancer, HAGLROS was upregulated by the transcription factor STAT3. HAGLROS may inhibit autophagy in two conflicting manners. It can competitively decoy miR-100-5p to antagonize its inhibition on mTOR. On the other hand, it can activate the mTORC1 signaling pathway, an important negative signal of autophagy, via direct interaction with mTORC1. More importantly, the inhibition of autophagy by HAGLROS can promote cell proliferation and migration in gastric cancer cells [53]. In HCC, HAGLROS facilitated cell proliferation, inhibited apoptosis, and enhanced autophagy via regulating the miR-5095/ATG12 axis [54].
3.3. PVT1
The lncRNA human plasmacytoma variant translocation 1 (PVT1) has been identified as an oncogene and regarded as an indicator for poor survival in pancreatic ductal adenocarcinoma [32], HCC [56], and osteosarcoma [84], although the target genes were not identical in these cancers. PVT1 expression levels were positively associated with that of ULK1 in pancreatic ductal adenocarcinoma tissues. It induced cyto-protective autophagy by targeting ULK1 in vitro and in vivo. Further study showed that PVT1 functioned as an miRNA sponge of miR-20a-5p and restored the expression of ULK1 in the progression of pancreatic ductal adenocarcinoma [32]. In HCC, PVT1 enhanced autophagy by regulating ATG3 expression via functioning as the sponge of miR-365 [56], whereas in osteosarcoma, PVT1 promoted cell migration and invasion through decoying miR-485 [84].
3.4. PTENP1
The lncRNA PTENP1, a pseudogene of PTEN, was markedly downregulated in HCC specimens and cell lines [49,50]. As a sponge of miR-17, miR-19b, and miR-20a, PTENP1 can indirectly increase the expression of the miRNA targets including ULK1, ATG7, p62, PTEN, and PHLPP (an inhibitor of AKT); therefore, it can promote HCC progression by provoking pro-death autophagy [49]. In addition, PTENP1 overexpression inhibited migration and invasion in HCC cells through decoying miR-193a-3p, which can activate autophagy by targeting PTEN [50,51].
3.5. Other ceRNAs
In non-small-cell lung cancer (NSCLC), KCNQ1OT1 silencing inhibited autophagy and proliferation in vitro and tumor growth in murine xenograft models, but miR-204-5p inhibitor abrogated these inhibitory effects. Both KCNQ1OT1 and ATG3, a direct target of miR-204-5p, were upregulated in NSCLC tissues. KCNQ1OT1 induced autophagy and facilitated NSCLC progression by decoying miR-204-5p to upregulate ATG3 expression [85].
Overexpression of the lncRNA IDH1–AS1 was found in pancreatic cancer tissues and cell lines. IDH1–AS1 promoted autophagy through upregulating ATG5. Bioinformatics analysis indicated that miR-216b-5p shared complementary base pairing with both ATG5 and IDH1-AS1. Therefore, IDH1–AS1 acted as a ceRNA for miR-216b-5p to enhance ATG5 expression and therefore to facilitate autophagy in pancreatic cancers [59]. Similar approaches have been applied to identify that LINC00160 improved autophagy activity in HCC by sequestering miR-132, resulting in the upregulation of PI3K3R, ATG5, and LC3I/II [60].
The expression level of lncRNA SLCO4A–AS1 was upregulated in colorectal cancer tissues, and positively correlated with that of partition-defective 3 (PARD3) [55], a downstream effector of mTOR and AMPK in the initiation of autophagy [86]. Knockdown of SLCO4A–AS1 inhibited autophagy and proliferation in vivo and in vitro by sponging miR-508-3p targeting PARD3 [55].
The lncRNA MALAT1 can inhibit autophagy by decoying miR-15b-5p to regulate MAPK1 expression, thereby activating the MAPK1/mTOR signaling [87]. Consistently, MALAT1 abrogated excessive autophagy in cutaneous squamous cell carcinoma [88].
In acute myeloid leukemia (AML) cells, the lncRNA UCA1 induced autophagy, which was associated with cell proliferation. Mechanistically, UCA1 may function as an miRNA sponge of miR-96-5p that can inhibit ATG7 expression by binding to the 3′-UTR of ATG7 [57].
4. An Overview of Autophagy-Regulated Cancer Metastasis and Chemoresistance
Emerging evidence has supported the roles of autophagy in the processes of cancer metastasis and chemoresistance. Cancer cells may succeed in metastasis through tumor-promoting autophagy [5], and survival against chemotherapy through protective autophagy [6,7,8].
In most cancers, the activation of autophagy promotes chemoresistance and indicates poor survival. Chemotherapeutic agents induce stress, thus activating autophagy as a cellular adaptive response [89,90]. Recently, it has been demonstrated that DNA damage can induce the process of autophagy and upregulate related genes, essentially resulting in chemoresistance [91]. Therefore, the activation of autophagy becomes a novel molecular mechanism for chemoresistance modulated by some lncRNAs in cancer cells, such as GBCDRlnc1 in gallbladder cancer [92] and MALAT1 in gastric cancer [93]. Under some circumstances, autophagy inhibition may restore chemotherapy sensitivity. For instance, autophagy suppression by Atg7 knockdown enhanced chemosensitivity and prolonged overall survival in AML mouse models [89]. Autophagy mediated by OPN/NF-κB signaling is essential for pancreatic cancer stem cells, while pharmacological inhibition of autophagy increased drug sensitivity [94]. These findings suggest a link between autophagy and cancer stem cells—the root cause for chemoresistance and disease relapse. Additionally, autophagy suppression can enhance chemotherapy response in triple-negative breast cancers (TNBCs) [90]. In HCC cells, miR-541 increased their sensitivity to sorafenib by inhibiting autophagy [95]. Moreover, N6-isopentenyladenosine also improved chemotherapy sensitivity by impairing autophagy in melanoma [96].
Cancer cells experience invasion into the circulation, migration into the pre-metastatic niche, and colonization at the new place [97]. A substantial body of evidence has shown that autophagy is crucial for cancer metastasis through regulating many important steps in this process, such as tumor invasion, migration, EMT, the crosstalk between stromal cells and tumor cells, and immune surveillance [5,98,99,100]. The precision function of autophagy in cancer metastasis remains controversial to date, with tumor-promoting roles in most studies and suppressive roles in a few [101]. A high level of ATG5 is required for metastasis in the mouse models of pancreatic ductal adenocarcinoma [102], while elevated LC3B has been indicated to be associated with invasion and metastasis in solid tumors [103]. Ube2v1, a ubiquitin-conjugating E2 enzyme variant, can facilitate metastasis in colorectal cancer by inhibiting autophagy. Importantly, rapamycin and trehalose treatment may reverse Ube2v1-mediated metastasis in mouse models [104]. In ovarian carcinoma, circMUC16-mediated autophagy promotes metastasis by directly binding to ATG13 and enhancing its expression. Moreover, cirMUC16 could also function as a ceRNA for miR-199a-5p and relieve its inhibitory on Beclin-1 and RUNX1 [81]. Nevertheless, in breast cancer, the expression of autophagy-related genes negatively correlates with pre-metastasis signatures, and thereof restricts tumor metastasis [105,106].
5. ceRNA-Mediated Autophagy and Chemoresistance
Chemoresistance is one of the main causes for therapeutic failure in cancers. Dysregulated autophagy [8,107,108], lncRNAs [33,109,110,111], and circRNAs [107,112,113,114,115] have been indicated to be associated with drug resistance in cancers. ceRNAs can play important roles in the regulation of autophagy-mediated chemoresistance in cancers.
Chemoresistance correlates with tumor relapse and contributes to poor clinical outcomes in patients with colorectal cancers. The lncRNA SNHG6 induced 5-fluorouracil (5-FU) resistance in colorectal cancers by regulating an autophagy-related pathway in vitro and in vivo. Bioinformatics analysis and dual-luciferase reporter assay implicated that SNHG6 could competitively bind to miR-26a-5p and upregulate the expression of ULK1, the direct target of miR-26a-5p, thereof leading to autophagy activation and 5-FU resistance in colorectal cancer cells [80]. Consistently, SNHG6 upregulated EZH2 expression and induced EMT, migration, and invasion by binding to miR-26a in colorectal cancers [116]. These studies indicate that SNHG6 may serve as a promising therapeutic target for colorectal cancers.
The lncRNA XIST was overexpressed in NSCLC tissues and cisplatin-resistant A549 cell lines. XIST silencing improved chemotherapy sensitivity by decreasing autophagy. XIST enhanced ATG7 expression by sponging miR-17, thereof contributing to autophagy-mediated drug resistance in NSCLC [63]. Moreover, the silencing of XIST impeded metastasis in NSCLC by sponging other miRNAs [117,118]. It can facilitate TGF-β-induced EMT and metastasis via regulating miR-367 and miR-141 that target ZEB2 [118], or contribute to cell proliferation, migration, and invasion as a ceRNA of miR-374a to modulate LARP1 expression [117]. Similar to XIST, upregulated lnc-ROR expression induced EMT, migration, and invasion in pancreatic cancers [119]. lnc-ROR knockdown restored drug sensitivity by increasing basal autophagy in pancreatic cancer cells via sponging miR-124, which regulates the PTBP1/PKM2 axis [61]. It has been clarified that PKM2 can activate autophagy through phosphorylating Beclin-1 [120]. Given that cancer cells experiencing EMT and metastasis are usually resistant to chemotherapy, we may suggest that XIST and lnc-ROR promote ceRNA-mediated autophagy to facilitate EMT and metastasis, and may be associated with chemoresistance in NSCLC. The lncRNA UCA1 was found to be overexpressed in bladder cancers. UCA1 knockdown suppressed cell proliferation, invasion, migration, and chemoresistance by sponging miR-582-5p to attenuate its inhibition of ATG7 expression [62].
The lncRNA bladder cancer-associated transcript 1 (BLACAT1) regulated chemoresistance in NSCLC via the miR-17/ATG7 axis. Silencing of BLACAT1 alleviated drug resistance in vivo. RIP and RNA pull-down assays confirmed the direct interaction between BLACAT1and miR-17. BLACAT1 promoted ATG7 expression through miR-17 to induce autophagy and enhance chemoresistance in NSCLC cells [64]. Similar to BLACAT1, KCNQ1OT1 overexpression was associated with decreased chemotherapy sensitivity and poor prognosis in colon cancers. The study in vitro further indicated that KCNQ1OT1 induced autophagy and chemotherapy resistance via upregulating ATG4B expression by sponging miR-34a [65].
lncRNA MALAT1 expression was increased in chemoresistant gastric cancer cells and colorectal cancer cells, and was positively related with autophagy activity [66,121,122]. Emerging data have indicated that MALAT1 may regulate autophagosome maturation through a ceRNA mechanism, contributing to chemoresistance in gastric and colorectal cancers. MALAT1 induced CDDP resistance by enhancing autophagy in gastric cancer cells, which could be reversed by miR-30b overexpression. MALAT1 sequestered miR-30e and miR-30b to upregulate ATG5 expression—a direct target of both miRNAs [66,67]. In colorectal cancer, MALAT1 promoted autophagy activity by acting as a ceRNA with miR-101 [121]. Of note, miR-101 is a potent autophagy inhibitor which targets ATG4D, a member of the ATG4 family which is essential for LC3 processing [123]. Further work is required to validate the potential relationship between MALAT1 and ATG4D in colorectal cancers.
In addition, apatinib, a small-molecule inhibitor of vascular endothelial growth factor receptor 2, is helpful in treating gastric cancers. However, in the clinical setting, some patients received a reduced dose of apatinib in terms of their intolerability and severe complications [124]. Proper intervention is a pressing need to help patients achieve great benefits from apatinib treatment. circCGAP1 knockdown increased apatinib sensitivity in gastric cancer cells by autophagy inhibition. Mechanistically, circCGAP1 sponged miR-3657 to upregulate ATG7, the direct target of miR-3657, leading to autophagy activation [125]. Therefore, specific blockage of the circRACGAP1–miR-3657–ATG7 axis is critical for the regulation of apatinib sensitivity in gastric cancers.
Increased expression of lncRNA NEAT1 was found in various cancers, such as HCC [70], anaplastic thyroid carcinoma [71], and colorectal cancer cells [69], and was correlated with poor prognosis in these cancers. NEAT promoted autophagy-mediated chemoresistance as a ceRNA targeting the miR-204/ATG3 axis in HCC [70], miR-9-5p/SPAG9 in anaplastic thyroid carcinoma [71], and miR-34a/HMGB1, ATG9A, and ATG4B in colorectal cancers [69]. Notably, SPAG9 is critical for lysosome localization and autophagy [126], while HMGB1 initiates autophagosomes by binding with Beclin-1 [127]. ATG9A plays essential roles in the delivery of lipids or proteins to the initiating sites of autophagy [128]. Interestingly, the NEAT1/miR-29b/ATG9A axis was modulated by insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) in hepatic stellate cell autophagy [129]. However, the relationship between the NEAT1/miR-29b/ATG9A axis and IGFBPrP1 has not been clarified yet. In addition to NEAT1, circEIF6 is another ceRNA that bridges autophagy and chemoresistance in anaplastic thyroid carcinomas. circEIF6 knockdown can re-sensitize anaplastic thyroid carcinoma cells to cisplatin, and its overexpression increased cisplatin-induced autophagy and inhibited cell apoptosis by targeting the miR-144-3p/TGF-α axis [72].
circCDYL and circABCB10 are two potential biomarkers for chemoresistance in breast cancers. Increased circCDYL level in both tissue and serum samples from breast cancer patients was associated with unfavorable clinical outcomes and drug resistance. circCDYL-induced proliferation in breast cancers depended on autophagy via sponging miR-1275 to upregulate the expression of ATG7 and ULK1 [73]. In an analysis of 30 pairs of paclitaxel-resistant and sensitive breast cancers, circABCB10 expression was positively correlated with paclitaxel resistance. Consistently, silencing of circABCB10 restored paclitaxel sensitivity by inducing apoptosis and inhibiting invasion and autophagy in chemoresistant breast cancer cells through the let-7a-5a/dual specificity phosphatase 7 (DUSP7) axis [74]. Likewise, ceRNA-mediated autophagy also regulated drug resistance in renal cell carcinomas. circ_0035483 overexpression promoted gemcitabine-induced autophagy and drug resistance in renal clear cell carcinoma by upregulating CCNB1 expression by sponging miR-335 [75].
Taken together, the current data demonstrate that ceRNAs can participate in many steps of autophagy by upregulating some key molecules, such as ULK1, ATG3, ATG7, and ATG4, resulting in chemoresistance in various cancers. Most ceRNAs are expressed highly in cancer tissues and enhance autophagy, resulting in chemoresistance. EMT, metastasis, cancer stem cells, proliferation, and apoptosis are the major mechanisms of chemoresistance [130], nevertheless, no evidence has indicated that the ceRNAs-mediated autophagy plays direct roles in chemoresistance and these related processes (Figure 2).
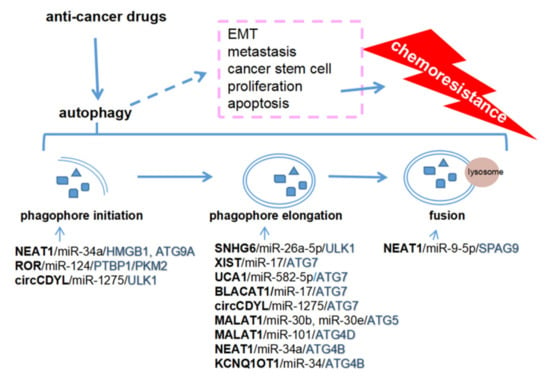
Figure 2.
Competing endogenous RNAs (ceRNAs) regulate autophagy to promote chemoresistance. NEAT1 and ROR are involved in the initiation of phagophore formation, and SNHG6, XIST, UCA1, BLACAT1, KCNQ1OT1, MALAT1, and NEAT1 in the elongation of autophagosome. NEAT1 also regulates lysosome localization by upregulating SPAG9. ceRNAs-mediated autophagy may promote chemoresistance via EMT, metastasis, cancer stem cells, proliferation, and apoptosis.
lncRNA HULC and eosinophil granule ontogeny transcript (EGOT) were found to be associated with chemoresistance through autophagy regulation independently of ceRNA. HULC was markedly downregulated in gastric cancer cells with cisplatin resistance. RNA pull-down and RIP assays identified the interaction between HULC and FoxM1. Silencing of HULC inhibited autophagy and reduced cisplatin resistance through regulating FoxM1 [48]. Intriguingly, HULC indirectly increased USP22 expression via epigenetic or transcriptional modulation of miR-6825-5p, miR-6845-5p, and miR-6886-3p rather than sponging these molecules in HCC cells [131]. USP22 upregulated Sirt1 expression by inhibiting its ubiquitin-mediated degradation. Sirt1 deacetylated ATG5 and ATG7, leading to autophagy activation. EGOT, an antisense intronic lncRNA derived from lncRNA ITPR1, promoted autophagy to sensitize paclitaxel cytotoxicity in breast cancers by upregulating ITPR1 expression through RNA–RNA and RNA–protein interactions [132].
6. ceRNA-Mediated Autophagy and Metastasis
The roles of ncRNAs such as lncRNAs [133,134], circRNAs [73,135], and miRNAs [133,136] have been recognized in cancer metastasis and autophagy. Figure 3 summarizes the potential roles of ceRNA-mediated autophagy in modulating metastasis. The microenvironment in the metastatic site is referred to as the pre-metastasis niche, which is prerequisite for cancer cells to colonize. Stromal cells including cancer-associated fibroblasts (CAFs), tumor-associated macrophages (TAMs), and other immunocytes as well as exosomes are indispensable for the formation of the pre-metastasis niche [137,138]. Therefore, ceRNA-mediated autophagy in the microenvironment is potentially crucial for regulating cancer metastasis.
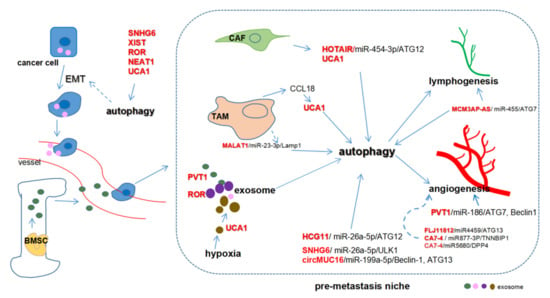
Figure 3.
Competing endogenous RNAs (ceRNAs) regulate metastasis through autophagy. Cancer-associated fibroblasts (CAFs) increase HOTAIR and UCA1 expression while tumor-associated macrophages (TAMs) upregulate UCA1 through CCL18. MALAT1/miR-23-3p regulates autophagy in macrophages. MCM3AP-ASinduces lymphatic vessel formation while PVT1 promotes angiogenesis by sponging miR-186. FLJ11812- and CA7-4-mediated autophagy may be essential for angiogenesis. PVT1, ROR, and UCA1 can be packaged in exosomes to promote metastasis. PVT1 can be encapsulated in exosomes derived from bone marrow mesenchymal stem cells (BMSC) while exosomal UCA1 is induced substantially by hypoxia.
Serval lncRNAs were found to be associated with cancer metastasis and advanced TNM stage, such as HCG11 [78] and MCM3AP-AS [79] in HCC, and SNHG6 [68,139] in osteosarcoma. They functioned as ceRNAs promoting autophagosome maturation by upregulating ATG12 or ATG7. HCG11 knockdown inhibited cell proliferation, metastasis, and autophagy in HCC cells and murine xenografts. Interestingly, miR-26a-5p deletion or ATG12 overexpression could rescue the effects caused by HCG11 silencing. miR-26a-5p can bind to both HCG11 and ATG12. As a collar, the biological functions of HCG11 may be obtained through sponging miR-26a-5p to antagonize its inhibitory effect on ATG12 [78]. MCM3AP-AS1 knockdown inhibited HCC cell invasion and lymphatic vessel formation. Bioinformatics analysis and luciferase reporter assay validated the direct interaction between MCM3AP-AS1 and miR-455. Gain- or loss-of-function studies showed that an miR-455 mimic impaired cell invasion and lymphatic vessel formation, which could be rescued by ATG7 overexpression. Therefore, MCM3AP-AS1 promoted metastasis via regulating the miR-455/ATG7 axis [79]. Silencing of SNHG6 suppressed cell proliferation and invasion, and induced autophagy in osteosarcoma cells, possibly by sponging miR-26a-5p to restore the expression of its target ULK1 [68]. lncRNAs SNHG11 and SNHG15 induced proliferation, apoptosis, migration, and autophagy in HCC cells through sponging miR-184 [140] and miR-141 [141], respectively.
Increased circMUC16 expression has been associated with advanced tumor stage in ovarian carcinomas. circMUC16 knockdown or overexpression showed that circMUC16 promoted autophagy flux to facilitate invasion and metastasis in ovarian carcinomas. Further studies indicated that circMUC16 upregulated Beclin-1, RUNX1 expression via sponging miR-199a-5p, and ATG13 level by direct interaction. RUNX1 facilitated circMUC16 expression in a positive feedback. circMUC16 is a promising prognostic biomarker for metastasis in ovarian carcinomas [81].
CAFs and TAMs are two major components in the microenvironment in the metastatic site. Regarding the crosstalk between autophagy among cancer cells, CAFs and TAMs through a ceRNA mechanism have emerged to be recognized in cancer metastasis. HOTAIR sponged miR-454-3p and indirectly upregulated ATG12 to promote cancer progression in chondrosarcoma cells [76]. In breast cancer cells, CAFs induced metastasis by enhancing the expression of HOTAIR transcriptionally, of which the promoter was bound with SMAD2/3/4. Silencing of HOTAIR impeded breast cancer cell growth and lung metastasis in vivo [142]. CAFs upregulated UCA1 expression in colorectal cancer cells, possibly through mTOR-related pathways [143], which was crucial for the pre-metastatic niche. UCA1 overexpression, induced by TAM-derived CCL18 (C-C motif chemokine ligand 18) [144], was closely associated with pulmonary metastasis and unfavorable clinical outcomes in osteosarcomas [145]. The lncRNA MALAT1 alleviated LAMP1 inhibition and promoted autophagy in macrophages by competitively binding to miR-23-3p [146]. ceRNA-mediated autophagy may be vital for TAM in immune regulation in the metastatic niche, despite the absence of direct evidence at present.
Exosomes, extracellular vesicles containing ncRNAs and proteins, are crucial for cellular communication in tumorigenesis in autocrine, paracrine, and endocrine manners [147]. lncRNAs such as PVT1 and UCA1 can be packaged into exosomes to link the crosstalk between stromal cells and tumor cells partly through autophagy regulation, thereby contributing to pre-metastasis niche formation [148,149]. PVT1 promoted tumor cell metastasis by upregulating autophagy. Moreover, PVT1 was encapsulated into bone marrow mesenchymal stem cells (BMSCs)-derived exosomes and promoted osteosarcoma cell proliferation and migration [148]. The elevated UCA1 upregulated ATG7 in bladder cancers by sponging miR-582-5p [62]. UCA1 expression increased in hypoxic exosomes from bladder cells compared to non-hypoxic exosomes and serum-derived exosomes from bladder cancer patients [150]. The hypoxic exosomes facilitated bladder cancer progression through EMT both in vivo and in vitro [62], which may be caused by UCA1/miR-582-5p/ATG7-mediated autophagy. In pancreatic cancer, lnc-ROR modulated autophagy-related chemoresistance through a ceRNA mechanism and induced EMT by upregulating ZEB1, resulting in cell migration and invasion [61,119]. Interestingly, lnc-ROR was found in the thyroid cancer-stem-like-cell-derived exosomes that regulated EMT and remodeled the tumor microenvironment and pre-metastasis niche [149]. Given that the inhibition of autophagy suppressed cancer metastasis and EMT [151], we postulated that lnc-ROR might regulate EMT and metastasis through a ceRNA-mediated autophagy in cancers.
Hypoxia is a key microenvironmental factor in the pre-metastasis niche formation [152,153]. Hypoxia can promote autophagy through a ceRNA mechanism. lncRNA PVT1 facilitated hypoxia-induced autophagy via PVT1/miR-152-ATG14 pathway in hepatic stellate cells (liver-specific mesenchymal cells that can be transformed to fibroblasts) [154]. Importantly, PVT1-induced autophagy has been involved in the proliferation, migration, and angiogenesis of gliomas. Bioinformatics analysis and dual-luciferase assay revealed that PVT1 restored the expression of ATG7 and Beclin-1 via a ceRNA mechanism targeting miR-186 [77]. However, no direct evidence is available for hypoxia-induced autophagy in cancer metastasis to date.
7. ceRNAs as Potential Therapeutic Targets in Cancers
As shown in Table 1, most of the ceRNAs involving autophagy are oncogenes and are expressed highly in cancer tissues compared to normal adjacent tissues—especially in HCC, colorectal cancer, and pancreatic cancer (Table 2), which are among the most devastating cancers worldwide [155,156,157]. Upregulation of HAGLROS [54], PVT1 [56], LINC00160 [60], NEAT1 [70], HCG11 [78], and MCM3AP-AS [79] in cancers were associated with poor clinical outcomes in patients with HCC. The oncogenic roles of LINC00160 and HCG11 have been confirmed not only in vitro but also in murine xenograft models. Likewise, SLCO4A-AS1 [55] and HOTAIR [58] were found to be overexpressed in colorectal cancers, and acted as oncogenes in in vitro and in vivo studies. PVT1 overexpression indicated poor clinical outcomes in patients with pancreatic cancers [32]. Its oncogenic roles were confirmed through in vitro and in vivo experiments, and its interaction with miRNAs was confirmed by the RIP assay.

Table 2.
ceRNAs regulate autophagy in HCC, colorectal cancer, and pancreatic cancer.
These ceRNAs are suggested to induce protective autophagy and to promote cancer progression, chemoresistance, and metastasis. Therefore, they and their related pathways have a great potential to serve as therapeutic targets. It has been reported that miRNAs could be delivered through monomethoxy-poly-poly nanoparticles [158]. We propose that the miRNAs sponged by those ceRNAs might be delivered to the cancers to correct the interaction among miRNA, ceRNA, and target genes, thereof alleviating the protective autophagy and eventually achieving therapeutic effects such as the restoration of sensitivity to chemotherapy and inhibition of metastasis. Moreover, CRISPR-based gene-editing tools are potential applicable treatment strategies [159]. The CRISPR/Cas9 system may be designed to downregulate the ceRNAs in cancers. One major limitation of these systems is adverse reactions owing to a lack of tissue specificity. The use of tissue-specific promoters may overcome these problems. Chloroquine and hydroxychloroquine were observed to have anti-tumor effects by targeting autophagy [160]. Moreover, multiple groups of data from Phase I/II clinical trials have confirmed that hydroxychloroquine selectively targets autophagy in cancer patients [161,162]. It would be attractive to combine the tissue-specific miRNA delivery or CRISPR/Cas9 system with hydroxychloroquine, the selective autophagy inhibitor, to treat cancer patients in the future.
8. Conclusion and Future Perspectives
In conclusion, ceRNAs play important roles in many human cancers. Most ceRNAs positively regulate autophagy unless they sponge miRNAs targeting mTOR. Nearly all ceRNAs function as oncogenes, with rare exceptions (Table 1).
Most metastatic cancer cells are chemoresistant [3,163]. It is prerequisite to explore the underlying molecular link between metastasis and chemotherapy. Many factors significantly contribute to drug resistance, such as cancer cell heterogeneity, drug efflux, strength of DNA repair, cancer cell stemness, and dysregulated proliferation/apoptosis pathways [164,165,166]. Under unfavorable circumstances, autophagy activation can protect cells from stress-induced injury [167,168]. Unequivocally, chemotherapy drugs can activate autophagy in cancer cells [169]. The metastasis of cancer harbors some key events, such as EMT, invasion into vessels, resistance to anoikis in circulation, and travelling to the secondary site [99]. During this process, cancer cells struggle to survive against various stresses. Likewise, autophagy also plays an important part in metastasis against these stresses [170]. Moreover, even in the secondary site, it is important to build up an appropriate microenvironment—that is, the pre-metastasis niche [171,172]. Autophagy is essential for niche formation owing to its biological effects on stromal cells, immune escape, angiogenesis, and lymphangiogenesis [171]. Therefore, further clarification of the mechanism of autophagy regulation in metastasis and chemoresistance may greatly improve the efficacy of chemotherapy and survival in cancer patients.
lncRNAs, circRNAs, and miRNAs are critical in tumorigenesis and may serve as promising therapeutic targets and biomarkers for cancers [9,173,174]. ceRNAs communicate with different types of RNAs in cancer progression, forming the network among lncRNAs, circRNAs, miRNAs, and mRNAs [15,131]. In this manuscript, we summarized the role of ceRNA-regulated autophagy in chemoresistance and metastasis. ceRNAs can regulate the expression of autophagy-related genes via binding with some miRNAs. These genes include mTOR, ULK1, ATG3, ATG4, ATG5, ATG7, ATG12, ATG13, and ATG14, which are critical for autophagosome initiation, elongation, and fusion with lysosomes (Figure 1).
To date, ceRNA-mediated autophagy is mostly known as a positive regulator of chemoresistance and metastasis. These ceRNAs can promote chemoresistance through proliferation inhibition and apoptosis induction, and facilitate metastasis by inducing EMT, invasion, migration, angiogenesis, and modulating the pre-metastasis niche (Table 1; Figure 2 and Figure 3). Some ceRNAs can be packaged in exosomes which are derived from cancer cells or bone marrow mesenchymal stem cells. The exosomal ceRNAs contribute to optimizing the microenvironment for cancer metastasis. However, more work is required to uncover how ceRNA-mediated autophagy regulates chemoresistance and metastasis in cancers. Particular issues might be focused on the potential role of ceRNAs in the crosstalk among cancer cells and the tumor microenvironment composed of stromal cells, endothelial cells (angiogenesis), and immunocytes. Nevertheless, several critical issues should first be addressed.
The ceRNA theory is supported by a great body of experimental data on lncRNAs and circRNAs. The studies are invariably associated with ectopic vectors which produce much higher than physiological levels in cancer cells, and unavoidably present non-physiological phenotypes [175]. The stoichiometry required for a ceRNA, a competitor for miRNAs, includes a substantially higher endogenous content and affinity with linear transcripts over miRNAs; therefore, it is very unlikely that a low-level ceRNA or physiological changes in the expression of an individual ceRNA would effectively regulate a far more abundant miRNA target. In fact, the abundance of ceRNAs such as lncRNA and circRNA in cancers has always been a major concern. The absolute levels of functional ceRNAs in cancer tissues have not been well addressed to date, owing to the difficulty in accurately quantifying ceRNAs and due to some technical problems in tissue preparation. The unification of accurate quantification becomes a prerequisite for ceRNA studies in cancers before we can find a functional ceRNA with considerably high contents.
A ceRNA may target multiple miRNAs, and the potential target miRNAs may also have several target genes (Table 1). It is reasonable to reckon that a subtle alteration of the miRNA by its upstream ceRNA may have a significant effect on the target genes in the process of autophagy. The development of a ceRNA network is crucial for better understanding the molecular mechanisms and potential utilities underlying gene expression regulated by ceRNAs. However, ceRNA networks have so far predominantly been constructed by bioinformatic prediction, with very limited experimental data. Another important issue with regard to the interaction between ceRNA and miRNA is the problem of “degradation” or “inhibition”, which is crucial for ceRNA function [175]. There is still conflicting evidence describing the effect of ceRNA–protein complexes on the degradation of miRNA after binding. It may be determined by the status of the binding sites between circRNAs and miRNAs: “degradation” in the case of completely complementary, or “inhibition” when partially matched [176].
Bioinformatic analysis, dual-luciferase assay, and methods measuring gene expression (e.g., reverse transcription-PCR (qRT-PCR) and Western blot) are the most commonly applied approaches in ceRNA investigations [26]. Results from bioinformatic prediction should be validated by experimental data. Dual-luciferase assay and gene expression analysis are not direct and sound methods to establish the interaction among ceRNA, miRNA, and mRNA. The reliable methods are currently RIP and RNA pull-down. However, as listed in Table 2, both methods have only been applied in a small number of studies. Importantly, the experimental results are obtained from gain- or loss-of-function assays in vitro and, uncommonly, in vivo. The technical limitations and defects should be appreciated in the assessment of the current studies. From the perspective of clinical relevance, future elaborate work in primary-cancer-derived organoids which repeat the pathological features of cancer [177], and studies in vivo will consolidate the interaction among ceRNA, miRNA, and RNA; therefore, they will ultimately provide great insights into carcinogenesis including autophagy-related cancer chemoresistance and metastasis.
Funding
This study is supported by grants from the Natural Science Foundation of China (no. 81872112) and National Key Research and Development Program of China (2016YFC1302900).
Conflicts of Interest
There are no competing financial interests in relation to the work described.
References
- Zhang, H.; Duan, C.; Yang, H. Defective autophagy in Parkinson’s disease: Lessons from genetics. Mol. Neurobiol. 2015, 51, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Mizushima, N. Autophagy and human diseases. Cell Res. 2014, 24, 69–79. [Google Scholar] [CrossRef]
- Acharyya, S.; Oskarsson, T.; Vanharanta, S.; Malladi, S.; Kim, J.; Morris, P.G.; Manova-Todorova, K.; Leversha, M.; Hogg, N.; Seshan, V.E.; et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012, 150, 165–178. [Google Scholar] [CrossRef]
- Ren, H.; Du, P.; Ge, Z.; Jin, Y.; Ding, D.; Liu, X.; Zou, Q. TWIST1 and BMI1 in cancer metastasis and chemoresistance. J. Cancer 2016, 7, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Dower, C.M.; Wills, C.A.; Frisch, S.M.; Wang, H.-G. Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy 2018, 14, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Chen, R.; Wang, Z.; Huang, Z.; Kong, N.; Zhang, M.; Han, W.; Lou, F.; Yang, J.; Zhang, Q.; et al. Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis. 2013, 4, e838. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef]
- Pérez-Hernández, M.; Arias, A.; Martínez-García, D.; Pérez-Tomás, R.; Quesada, R.; Soto-Cerrato, V. Targeting autophagy for cancer treatment and tumor chemosensitization. Cancers 2019, 11, 1599. [Google Scholar] [CrossRef]
- Wei, L.; Sun, J.; Zhang, N.; Zheng, Y.; Wang, X.; Lv, L.; Liu, J.; Xu, Y.; Shen, Y.; Yang, M. Noncoding RNAs in gastric cancer: Implications for drug resistance. Mol. Cancer 2020, 19, 62. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Guttman, M. Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014, 24, 651–663. [Google Scholar] [CrossRef]
- Bonasio, R.; Shiekhattar, R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014, 48, 433–455. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Fu, P. Pseudogene-derived lncRNAs and their miRNA sponging mechanism in human cancer. Front. Cell Dev. Biol. 2020, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wang, J.; Guo, Z.; Li, M.; Li, M.; Liu, S.; Liu, H.; Li, W.; Yin, X.; Tao, J.; et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol. Cancer 2018, 17, 123. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nat. Cell Biol. 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Karreth, F.A.; Pandolfi, P.P. ceRNA cross-talk in cancer: When ce-bling rivalries go awry. Cancer Discov. 2013, 3, 1113–1121. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar] [CrossRef]
- Li, S.-P.; Xu, H.-X.; Yu, Y.; He, J.-D.; Wang, Z.; Xu, Y.-J.; Wang, C.-Y.; Zhang, H.-M.; Zhang, R.-X.; Zhang, J.-J.; et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget 2016, 7, 42431–42446. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, W.; Zhou, Q.; Chen, C.; Yuan, W.; Liu, J.; Li, X.; Sun, Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer 2020, 19, 14. [Google Scholar] [CrossRef]
- Shan, C.; Zhang, Y.; Hao, X.; Gao, J.; Chen, X.; Wang, K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol. Cancer 2019, 18, 136. [Google Scholar] [CrossRef]
- Lei, M.; Zheng, G.; Ning, Q.; Zheng, J.; Dong, D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 2020, 19, 30. [Google Scholar] [CrossRef]
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 90. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, B.; Shu, C.; Ma, Q.; Wang, J. Functions and clinical significance of circular RNAs in glioma. Mol. Cancer 2020, 19, 34. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Li, X.; Nan, A.; Zhang, N.; Chen, L.; Zhou, H.; Zhang, H.; Qiu, M.; Zhu, J.; Ling, Y.; et al. Circular RNA 406961 interacts with ILF2 to regulate PM2.5-induced inflammatory responses in human bronchial epithelial cells via activation of STAT3/JNK pathways. Environ. Int. 2020, 141, 105755. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Wang, Y.; Wang, Y.; Dai, X.; Zhou, T.; Chen, J.; Tao, B.; Zhang, J.; Cao, F. The tumor-suppressive human circular RNA CircITCH sponges miR-330-5p to ameliorate doxorubicin-induced cardiotoxicity through upregulating SIRT6, Survivin, and SERCA2a. Circ. Res. 2020, 127. [Google Scholar] [CrossRef]
- Chen, L.-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- An, Y.; Furber, K.L.; Ji, S. Pseudogenes regulate parental gene expression via ceRNA network. J. Cell. Mol. Med. 2017, 21, 185–192. [Google Scholar] [CrossRef]
- Song, C.; Zhang, J.; Qi, H.; Feng, C.; Chen, Y.; Cao, Y.; Ba, L.; Ai, B.; Wang, Q.; Huang, W.; et al. The global view of mRNA-related ceRNA cross-talks across cardiovascular diseases. Sci. Rep. 2017, 7, 10185. [Google Scholar] [CrossRef]
- Zhu, X.; Jiang, L.; Yang, H.; Chen, T.; Wu, X.; Lv, K. Analyzing the lncRNA, miRNA, and mRNA-associated ceRNA networks to reveal potential prognostic biomarkers for glioblastoma multiforme. Cancer Cell Int. 2020, 20, 393. [Google Scholar] [CrossRef]
- Sun, T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol. Res. 2018, 129, 151–155. [Google Scholar] [CrossRef]
- Kulkarni, B.; Kirave, P.; Gondaliya, P.; Jash, K.; Jain, A.; Tekade, R.K.; Kalia, K. Exosomal miRNA in chemoresistance, immune evasion, metastasis and progression of cancer. Drug Discov. Today 2019, 24, 2058–2067. [Google Scholar] [CrossRef]
- Huang, F.; Chen, W.; Peng, J.; Li, Y.; Zhuang, Y.; Zhu, Z.; Shao, C.-K.; Yang, W.; Yao, H.; Zhang, S. LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol. Cancer 2018, 17, 98. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez, M.; Aguilar-Medina, M.; Lizárraga-Verdugo, E.; Avendaño-Félix, M.; Silva-Benítez, E.; López-Camarillo, C.; Ramos-Payán, R. LncRNAs as regulators of autophagy and drug resistance in colorectal Cancer. Front. Oncol. 2019, 9, 1008. [Google Scholar] [CrossRef] [PubMed]
- Cristóbal, I.; Caramés, C.; Rubio, J.; Sanz-Álvarez, M.; Luque, M.; Madoz-Gúrpide, J.; Rojo, F.; García-Foncillas, J. Functional and clinical impact of CircRNAs in oral cancer. Cancers 2020, 12, 1041. [Google Scholar] [CrossRef] [PubMed]
- Foster, K.G.; Acosta-Jaquez, H.A.; Romeo, Y.; Ekim, B.; Soliman, G.A.; Carriere, A.; Roux, P.P.; Ballif, B.A.; Fingar, D.C. Regulation of mTOR Complex 1 (mTORC1) by raptor Ser863and multisite phosphorylation. J. Biol. Chem. 2010, 285, 80–94. [Google Scholar] [CrossRef]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; Latek, R.R.; Guntur, K.V.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 2003, 11, 895–904. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Ganley, I.G.; Lam, D.H.; Wang, J.; Ding, X.; Chen, S.; Jiang, X. ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.-I.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef]
- Chan, E.Y. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci. Signal. 2009, 2, pe51. [Google Scholar] [CrossRef]
- Paquette, M.; El-Houjeiri, L.; Pause, A. mTOR Pathways in Cancer and Autophagy. Cancers 2018, 10, 18. [Google Scholar] [CrossRef]
- Reidick, C.; El Magraoui, F.; Meyer, H.E.; Stenmark, H.; Platta, H.W. Regulation of the Tumor-Suppressor Function of the Class III Phosphatidylinositol 3-Kinase Complex by Ubiquitin and SUMO. Cancers 2014, 7, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Proikas-Cezanne, T.; Takacs, Z.; Dönnes, P.; Kohlbacher, O. WIPI proteins: Essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 2015, 128, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Baskaran, S.; Ragusa, M.J.; Hurley, J.H. How Atg18 and the WIPIs sense phosphatidylinositol 3-phosphate. Autophagy 2012, 8, 1851–1852. [Google Scholar] [CrossRef]
- Cao, Y.; Luo, Y.; Zou, J.; Ouyang, J.; Cai, Z.; Zeng, X.; Ling, H.; Zeng, T. Autophagy and its role in gastric cancer. Clin. Chim. Acta 2019, 489, 10–20. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Schulman, B.A. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 2014, 21, 336–345. [Google Scholar] [CrossRef]
- Wang, C.-Z.; Yan, G.-X.; Dong, D.-S.; Xin, H.; Liu, Z. LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J. Gastroenterol. 2019, 25, 5310–5322. [Google Scholar] [CrossRef]
- Li, X.-X.; Xiao, L.; Chung, H.K.; Ma, X.-X.; Liu, X.; Song, J.-L.; Jin, C.Z.; Rao, J.N.; Gorospe, M.; Wang, J.-Y. Interaction between HuR and circPABPN1 Modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol. Cell. Biol. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-L.; Tseng, Y.-W.; Wu, J.-C.; Chen, G.-Y.; Lin, K.-C.; Hwang, S.-M.; Hu, Y.-C. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomater. 2015, 44, 71–81. [Google Scholar] [CrossRef]
- Qian, Y.-Y.; Li, K.; Liu, Q.-Y.; Liu, Z.-S. Long non-coding RNA PTENP1 interacts with miR-193a-3p to suppress cell migration and invasion through the PTEN pathway in hepatocellular carcinoma. Oncotarget 2017, 8, 107859–107869. [Google Scholar] [CrossRef]
- Errafiy, R.; Aguado, C.; Ghislat, G.; Esteve, J.M.; Gil, A.; Loutfi, M.; Knecht, E. PTEN increases autophagy and inhibits the ubiquitin-proteasome pathway in glioma cells independently of its lipid phosphatase activity. PLoS ONE 2013, 8, e83318. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-H.; Wang, W.-T.; Huang, W.; Fang, K.; Sun, Y.-M.; Liu, S.-R.; Luo, X.-Q.; Chen, Y.-Q. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017, 24, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-F.; Wu, P.; Xia, R.; Yang, J.; Huo, X.-Y.; Gu, D.-Y.; Tang, C.-J.; De, W.; Yang, F. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol. Cancer 2018, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Hu, J.; Pu, J.; Tang, Q.; Li, W.; Ma, R.; Xu, Z.; Tan, C.; Yao, T.; Wu, X.; et al. Long noncoding RNA HAGLROS promotes cell proliferation, inhibits apoptosis and enhances autophagy via regulating miR-5095/ATG12 axis in hepatocellular carcinoma cells. Int. Immunopharmacol. 2019, 73, 72–80. [Google Scholar] [CrossRef]
- Wang, Z.; Jin, J. LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging 2019, 11, 4876–4889. [Google Scholar] [CrossRef]
- Yang, L.; Peng, X.; Jin, H.; Liu, J. Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene 2019, 697, 94–102. [Google Scholar] [CrossRef]
- Li, J.J.; Chen, X.F.; Wang, M.; Zhang, P.P.; Zhang, F.; Zhang, J.J. Long non-coding RNA UCA1 promotes autophagy by targeting miR-96-5p in acute myeloid leukaemia. Clin. Exp. Pharmacol. Physiol. 2020, 47, 877–885. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Chen, X.; Liu, J.; Gu, H.; Fan, R.; Ge, H. Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis. 2020, 11, 175. [Google Scholar] [CrossRef]
- Zhang, N.; Li, Z.; Bai, F.; Zhang, S. PAX5-induced upregulation of IDH1-AS1 promotes tumor growth in prostate cancer by regulating ATG5-mediated autophagy. Cell Death Dis. 2019, 10, 734. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Fu, Y.; Han, W.; Xu, H.; Wen, L.; Deng, Y.; Liu, K. Long non-coding RNA LINC00160 functions as a decoy of microRNA-132 to mediate autophagy and drug resistance in hepatocellular carcinoma via inhibition of PIK3R3. Cancer Lett. 2020, 478, 22–33. [Google Scholar] [CrossRef]
- Li, C.; Zhao, Z.; Zhou, Z.; Liu, R. Linc-ROR confers gemcitabine resistance to pancreatic cancer cells via inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis. Cancer Chemother. Pharmacol. 2016, 78, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, W.; Ning, J.; Yu, W.; Rao, T.; Cheng, F. Long noncoding RNA UCA1 targets miR-582-5p and contributes to the progression and drug resistance of bladder cancer cells through ATG7-mediated autophagy inhibition. OncoTargets Ther. 2019, 12, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zu, Y.; Fu, X.; Deng, Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol. Rep. 2017, 38, 3347–3354. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Chen, H.; Zheng, F.; Gao, Z.; Sun, P.; Peng, Q.; Liu, Y.; Deng, X.; Huang, Y.; Zhao, C.; et al. LncRNA BLACAT1 is involved in chemoresistance of non-small cell lung cancer cells by regulating autophagy. Int. J. Oncol. 2019, 54, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, C.; Li, D.; Yang, L.; Jin, J.; Zhang, B. lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. OncoTargets Ther. 2019, 12, 2649–2660. [Google Scholar] [CrossRef]
- Zhang, Y.-F.; Li, C.-S.; Zhou, Y.; Lu, X. Propofol facilitates cisplatin sensitivity via lncRNA MALAT1/miR-30e/ATG5 axis through suppressing autophagy in gastric cancer. Life Sci. 2020, 244, 117280. [Google Scholar] [CrossRef]
- Xi, Z.; Si, J.; Nan, J. LncRNA MALAT1 potentiates autophagy-associated cisplatin resistance by regulating the microRNA-30b/autophagy-related gene 5 axis in gastric cancer. Int. J. Oncol. 2018, 54, 239–248. [Google Scholar] [CrossRef]
- Zhu, X.; Yang, G.; Xu, J.; Zhang, C. Silencing of SNHG6 induced cell autophagy by targeting miR-26a-5p/ULK1 signaling pathway in human osteosarcoma. Cancer Cell Int. 2019, 19, 82. [Google Scholar] [CrossRef]
- Liu, F.; Ai, F.; Zhang, D.; Tian, L.; Yang, Z.; Liu, S. LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 2019, 9, 1079–1091. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Y.; Yang, L.; Ma, Y.; Peng, X.; Yang, S.; Li, H.; Liu, J. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J. Cell. Physiol. 2019, 235, 3402–3413. [Google Scholar] [CrossRef]
- Yan, P.; Su, Z.; Zhang, Z.; Gao, T. LncRNA NEAT1 enhances the resistance of anaplastic thyroid carcinoma cells to cisplatin by sponging miR-9-5p and regulating SPAG9 expression. Int. J. Oncol. 2019, 55, 988–1002. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, J.; Qin, L.; Yang, Z.; Xiong, J.; Zhang, Y.; Li, R.; Li, S.; Wang, H.; Yu, B.; et al. Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging 2018, 10, 3806–3820. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Ling, Y.; Mehrpour, M.; Saw, P.E.; Liu, Z.; Tan, W.; Tian, Z.; Zhong, W.; Lin, W.; Luo, Q.; et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol. Cancer 2020, 19, 65. [Google Scholar] [CrossRef]
- Yang, W.; Gong, P.; Yang, Y.; Yang, C.; Yang, B.; Ren, L. Circ-ABCB10 contributes to paclitaxel resistance in breast cancer through Let-7a-5p/DUSP7 axis. Cancer Manag. Res. 2020, 12, 2327–2337. [Google Scholar] [CrossRef]
- Yan, L.; Liu, G.; Cao, H.; Zhang, H.; Shao, F. Hsa_circ_0035483 sponges hsa-miR-335 to promote the gemcitabine-resistance of human renal cancer cells by autophagy regulation. Biochem. Biophys. Res. Commun. 2019, 519, 172–178. [Google Scholar] [CrossRef]
- Bao, X.; Ren, T.; Huang, Y.; Sun, K.; Wang, S.; Liu, K.; Zheng, B.; Guo, W. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 2017, 8, e2605. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, P.; Xue, Y.; Qu, C.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y. PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef]
- Li, M.-L.; Zhang, Y.; Ma, L.-T. LncRNA HCG11 accelerates the progression of hepatocellular carcinoma via miR-26a-5p/ATG12 axis. Eur Rev Med Pharmacol Sci 2019, 23, 10708–10720. [Google Scholar]
- Zhang, H.; Luo, C.; Zhang, G. LncRNA MCM3AP-AS1 regulates epidermal growth factor receptor and autophagy to promote hepatocellular carcinoma metastasis by interacting with miR-455. DNA Cell Biol. 2019, 38, 857–864. [Google Scholar] [CrossRef]
- Wang, X.; Lan, Z.; He, J.; Lai, Q.; Yao, X.; Li, Q.; Liu, Y.; Lai, H.; Gu, C.; Yan, Q.; et al. LncRNA SNHG6 promotes chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in colorectal cancer cells. Cancer Cell Int. 2019, 19, 234. [Google Scholar] [CrossRef]
- Gan, X.; Zhu, H.; Jiang, X.; Obiegbusi, S.C.; Yong, M.; Long, X.; Hu, J. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. Cancer 2020, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.; Zhang, M.; Hou, H.; Zhang, Z.; Li, N. HOTAIRM1 suppresses cell proliferation and invasion in ovarian cancer through facilitating ARHGAP24 expression by sponging miR-106a-5p. Life Sci. 2020, 243, 117296. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, X.; Li, H.; Liu, J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol. BioSyst. 2016, 12, 2605–2612. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Pan, X.-F.; Liu, Y.; Zhao, S.; Gong, W.-Q.; Liu, W. Long noncoding RNA PVT1 promotes metastasis via miR-484 sponging in osteosarcoma cells. Eur Rev Med Pharmacol Sci 2020, 24, 2229–2238. [Google Scholar] [PubMed]
- Kang, Y.; Jia, Y.; Wang, Q.; Zhao, Q.; Song, M.; Ni, R.; Wang, J. Long noncoding RNA KCNQ1OT1 promotes the progression of non-small cell lung cancer via regulating miR-204-5p/ATG3 Axis. OncoTargets Ther. 2019, 12, 10787–10797. [Google Scholar] [CrossRef]
- Choi, E.J.; Lee, J.H.; Kim, M.S.; Song, S.Y.; Yoo, N.J.; Lee, S.H. Intratumoral heterogeneity of somatic mutations for NRIP1, DOK1, ULK1, ULK2, DLGAP3, PARD3 and PRKCI in colon cancers. Pathol. Oncol. Res. 2018, 24, 827–832. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, T.; Duan, J.; Mu, N.; Zhang, T. MALAT1/miR-15b-5p/MAPK1 mediates endothelial progenitor cells autophagy and affects coronary atherosclerotic heart disease via mTOR signaling pathway. Aging 2019, 11, 1089–1109. [Google Scholar] [CrossRef]
- Tan, M.; Jiang, B.; Wang, H.; Ouyang, W.; Chen, X.; Wang, T.; Dong, D.; Yi, S.; Yi, J.; Huang, Y.; et al. Dihydromyricetin induced lncRNA MALAT1-TFEB-dependent autophagic cell death in cutaneous squamous cell carcinoma. J. Cancer 2019, 10, 4245–4255. [Google Scholar] [CrossRef]
- Piya, S.; Kornblau, S.M.; Ruvolo, V.; Mu, H.; Ruvolo, P.P.; McQueen, T.; Davis, E.; Hail, N.; Kantarjian, H.; Andreeff, M.; et al. Atg7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood 2016, 128, 1260–1269. [Google Scholar] [CrossRef]
- Chittaranjan, S.; Bortnik, S.; Dragowska, W.H.; Xu, J.; Abeysundara, N.; Leung, A.; Go, N.E.; DeVorkin, L.; Weppler, S.A.; Gelmon, K.; et al. Autophagy inhibition augments the anticancer effects of epirubicin treatment in anthracycline-sensitive and -resistant triple-negative breast cancer. Clin. Cancer Res. 2014, 20, 3159–3173. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, J.; Liang, G.; Geng, G.; Zhao, F.; Yin, P.; Nowsheen, S.; Wu, C.; Li, Y.; Li, L.; et al. CHK2-FOXK axis promotes transcriptional control of autophagy programs. Sci. Adv. 2020, 6, eaax5819. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Wang, S.; Jin, L.; Weng, M.; Zhou, D.; Wang, J.; Tang, Z.; Quan, Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer 2019, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- Yiren, H.; Yingcong, Y.; Sunwu, Y.; Keqin, L.; XiaoChun, T.; Senrui, C.; Ende, C.; Xizhou, L.; Yanfan, C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer 2017, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.-C.; Wang, H.-C.; Hou, Y.-C.; Tung, H.-L.; Chiu, T.-J.; Shan, Y.-S. Blockade of autophagy reduces pancreatic cancer stem cell activity and potentiates the tumoricidal effect of gemcitabine. Mol. Cancer 2015, 14, 179. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.-P.; Liu, J.-P.; Feng, J.-F.; Zhu, C.-P.; Yang, Y.; Zhou, W.-P.; Ding, J.; Huang, C.-K.; Cui, Y.-L.; Ding, C.-H.; et al. miR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut 2019, 69, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, R.; Ciaglia, E.; Amodio, G.; Picardi, P.; Proto, M.C.; Gazzerro, P.; Laezza, C.; Remondelli, P.; Bifulco, M.; Pisanti, S. N6-isopentenyladenosine dual targeting of AMPK and Rab7 prenylation inhibits melanoma growth through the impairment of autophagic flux. Cell Death Differ. 2018, 25, 353–367. [Google Scholar] [CrossRef]
- Su, Z.; Yang, Z.; Xu, Y.; Chen, Y.; Yu, Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer 2015, 14, 48. [Google Scholar] [CrossRef]
- Vera-Ramirez, L. Cell-intrinsic survival signals. The role of autophagy in metastatic dissemination and tumor cell dormancy. Semin. Cancer Biol. 2020, 60, 28–40. [Google Scholar] [CrossRef]
- E Mowers, E.; Sharifi, M.N.; MacLeod, K.F. Autophagy in cancer metastasis. Oncogene 2017, 36, 1619–1630. [Google Scholar] [CrossRef]
- Chen, H.-T.; Liu, H.; Mao, M.-J.; Tan, Y.; Mo, X.-Q.; Meng, X.-J.; Cao, M.-T.; Zhong, C.-Y.; Liu, Y.; Shan, H.; et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Cancer 2019, 18, 101. [Google Scholar] [CrossRef]
- Li, X.; He, S.-K.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Görgülü, K.; Diakopoulos, K.N.; Ai, J.; Schoeps, B.; Kabacaoglu, D.; Karpathaki, A.-F.; Ciecielski, K.J.; Kaya-Aksoy, E.; Ruess, D.A.; Berninger, A.; et al. Levels of the autophagy-related 5 protein affect progression and metastasis of pancreatic tumors in mice. Gastroenterology 2019, 156, 203–217.e20. [Google Scholar]
- Lazova, R.; Camp, R.L.; Klump, V.; Siddiqui, S.F.; Amaravadi, R.K.; Pawelek, J.M. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin. Cancer Res. 2011, 18, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Cai, L.-D.; Liu, Y.-H.; Li, S.; Gan, W.-J.; Li, X.-M.; Wang, J.-R.; Guo, P.-D.; Zhou, Q.; Lu, X.-X.; et al. Ube2v1-mediated ubiquitination and degradation of Sirt1 promotes metastasis of colorectal cancer by epigenetically suppressing autophagy. J. Hematol. Oncol. 2018, 11, 95. [Google Scholar] [CrossRef]
- Marsh, T.; Kenific, C.M.; Suresh, D.; Gonzalez, H.; Shamir, E.R.; Mei, W.; Tankka, A.; Leidal, A.M.; Kalavacherla, S.; Woo, K.; et al. Autophagic degradation of NBR1 restricts metastatic outgrowth during mammary tumor progression. Dev. Cell 2020, 52, 591–604.e6. [Google Scholar] [CrossRef]
- Mao, L.; Zhan, Y.-Y.; Wu, B.; Yu, Q.; Xu, L.; Hong, X.; Zhong, L.; Mi, P.; Xiao, L.; Wang, X.; et al. ULK1 phosphorylates Exo70 to suppress breast cancer metastasis. Nat. Commun. 2020, 11, 117. [Google Scholar] [CrossRef]
- Hombach-Klonisch, S.; Mehrpour, M.; Shojaei, S.; Harlos, C.; Pitz, M.; Hamaï, A.; Siemianowicz, K.; Likus, W.; Wiechec, E.; Toyota, B.D.; et al. Glioblastoma and chemoresistance to alkylating agents: Involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol. Ther. 2018, 184, 13–41. [Google Scholar] [CrossRef]
- Skarkova, V.; Králová, V.; Vitovcova, B.; Rudolf, E. Selected aspects of chemoresistance mechanisms in colorectal carcinoma—A focus on epithelial-to-mesenchymal transition, autophagy, and apoptosis. Cells 2019, 8, 234. [Google Scholar] [CrossRef]
- Wang, W.-T.; Han, C.; Sun, Y.-M.; Chen, T.-Q.; Chen, Y.-Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 55. [Google Scholar] [CrossRef]
- Jiang, W.; Xia, J.; Xie, S.; Zou, R.; Pan, S.; Wang, Z.-W.; Assaraf, Y.G.; Zhu, X. Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist. Updat. 2020, 50, 100683. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, K.; Zhou, H.; Wu, Y.; Li, C.; Liu, Y.; Liu, Z.; Xu, Q.; Liu, S.; Xiao, D.; et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol. Cancer 2020, 19, 1–26, 47. [Google Scholar] [CrossRef]
- Hong, X.; Liu, N.; Liang, Y.; He, Q.; Yang, X.; Lei, Y.; Zhang, P.; Zhao, Y.; He, S.; Wang, Y.; et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol. Cancer 2020, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Yi, X.; Wu, X.; Bu, X.; Wang, D.; Wu, Z.; Zhang, G.; Gu, J.; Kang, D. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020, 479, 1–12. [Google Scholar] [CrossRef]
- Cui, C.; Yang, J.; Li, X.; Liu, D.; Fu, L.-W.; Wang, X. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol. Cancer 2020, 19, 58. [Google Scholar] [CrossRef]
- Jian, X.; He, H.; Zhu, J.; Zhang, Q.; Zheng, Z.; Liang, X.; Chen, L.; Yang, M.; Peng, K.; Zhang, Z.; et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol. Cancer 2020, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Duan, W.; Sun, W. LncRNA SNHG6 promotes the migration, invasion, and epithelial-mesenchymal transition of colorectal cancer cells by miR-26a/EZH2 axis. OncoTargets Ther. 2019, 12, 3349–3360. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Xu, J.; Lu, H.; Lin, B.; Cai, S.; Guo, J.; Zang, F.; Chen, R. LARP1 is regulated by the XIST/miR-374a axis and functions as an oncogene in non-small cell lung carcinoma. Oncol. Rep. 2017, 38, 3659–3667. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wan, L.; Liu, Z.; Xu, G.; Wang, S.; Su, Z.; Zhang, Y.; Zhang, C.; Liu, X.; Lei, Z.; et al. Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018, 418, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.-X.; Wang, Y.; Li, C.; Xu, J.-W.; Zhou, B.; Zhu, J.-K.; Han, H.-F.; Wang, L.; Wang, Y.; Hu, S. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016, 374, 261–271. [Google Scholar] [CrossRef]
- Wang, L.; Yang, L.; Yang, Z.; Tang, Y.; Tao, Y.; Zhan, Q.; Lei, L.; Jing, Y.; Jiang, X.; Jin, H.; et al. Glycolytic Enzyme PKM2 Mediates Autophagic Activation to Promote Cell Survival in NPM1-Mutated Leukemia. Int. J. Biol. Sci. 2019, 15, 882–894. [Google Scholar] [CrossRef]
- Si, Y.; Yang, Z.; Ge, Q.; Yu, L.; Yao, M.; Sun, X.; Ren, Z.; Ding, C. Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell. Mol. Biol. Lett. 2019, 24, 50. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, X.; Wang, H.; Wang, L.; Liu, T.; Du, L.; Yang, Y.; Wang, C. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol. Cancer Ther. 2017, 16, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Frankel, L.B.; Wen, J.; Lees, M.; Høyer-Hansen, M.; Farkas, T.; Krogh, A.; Jäättelä, M.; Lund, A.H. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011, 30, 4628–4641. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qin, S.; Xu, J.; Xiong, J.; Wu, C.; Bai, Y.; Liu, W.; Tong, J.; Liu, Y.; Xu, R.; et al. Randomized, double-blind, placebo-controlled phase iii trial of apatinib in patients with chemotherapy-refractory advanced or metastatic adenocarcinoma of the stomach or gastroesophageal junction. J. Clin. Oncol. 2016, 34, 1448–1454. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, Z.; Xie, M.; Quan, Y.; Zhu, W.; Yang, F.; Zhao, C.; Fan, Y.; Fang, N.; Jiang, H.; et al. Silencing of circRACGAP1 sensitizes gastric cancer cells to apatinib via modulating autophagy by targeting miR-3657 and ATG7. Cell Death Dis. 2020, 11, 169. [Google Scholar] [CrossRef]
- Suzuki, R.; Gunarta, I.K.; Boldbaatar, J.; Erdenebaatar, P.; Odongoo, R.; Yoshioka, K. Functional role of c-Jun NH2-terminal kinase-associated leucine zipper protein (JLP) in lysosome localization and autophagy. Drug Discov. Ther. 2020, 14, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Livesey, K.M.; Kang, R.; Vernon, P.; Buchser, W.J.; Loughran, P.; Watkins, S.C.; Zhang, L.; Manfredi, J.J.; Zeh, H.J.; Li, L.; et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res. 2012, 72, 1996–2005. [Google Scholar] [CrossRef]
- Judith, D.; A Tooze, S. ATG9A supplies PtdIns4P to the autophagosome initiation site. Autophagy 2019, 15, 1660–1661. [Google Scholar] [CrossRef]
- Kong, Y.; Huang, T.; Zhang, H.; Zhang, Q.; Ren, J.; Guo, X.; Fan, H.; Liu, L. The lncRNA NEAT1/miR-29b/Atg9a axis regulates IGFBPrP1-induced autophagy and activation of mouse hepatic stellate cells. Life Sci. 2019, 237, 116902. [Google Scholar] [CrossRef]
- Zhang, H.; Lü, B. microRNAs as biomarkers of ovarian cancer. Expert Rev. Anticancer. Ther. 2020, 20, 373–385. [Google Scholar] [CrossRef]
- Guo, L.-L.; Song, C.-H.; Wang, P.; Dai, L.-P.; Zhang, J.-Y.; Wang, K. Competing endogenous RNA networks and gastric cancer. World J. Gastroenterol. 2015, 21, 11680–11687. [Google Scholar] [CrossRef]
- Xu, S.; Wang, P.; Zhang, J.; Wu, H.; Sui, S.; Zhang, J.; Wang, Q.; Qiao, K.; Yang, W.; Xu, H.; et al. Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol. Cancer 2019, 18, 89. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-A.; Lee, C.-T.; Lee, J.-C.; Wang, Y.-W.; Huang, C.-T.; Lan, S.-H.; Lin, P.-C.; Lin, B.-W.; Tian, Y.-F.; Liu, H.-S.; et al. MiR-338-5p promotes metastasis of colorectal cancer by inhibition of phosphatidylinositol 3-kinase, catalytic subunit type 3-mediated autophagy pathway. EBioMedicine 2019, 43, 270–281. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, D.; Liu, H.; Yang, K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Huang, M.; Xing, L.; Yang, R.; Wang, X.; Jiang, R.; Zhang, L.; Chen, J. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol. Cancer 2020, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Tang, W.; Lin, Z.; Xu, L.; Dong, R.; Li, Y.; Li, J.; Zhang, Z.; Li, X.; et al. miR-130a upregulates mTOR pathway by targeting TSC1 and is transactivated by NF-κB in high-grade serous ovarian carcinoma. Cell Death Differ. 2017, 24, 2089–2100. [Google Scholar] [CrossRef]
- Houg, D.S.; Bijlsma, M.F. The hepatic pre-metastatic niche in pancreatic ductal adenocarcinoma. Mol. Cancer 2018, 17, 95. [Google Scholar] [CrossRef]
- Guo, Y.; Ji, X.; Liu, J.; Fan, D.; Zhou, Q.; Chen, C.; Wang, W.; Wang, G.; Wang, H.; Yuan, W.; et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol. Cancer 2019, 18, 39. [Google Scholar] [CrossRef]
- Wang, K.; Yang, C.; Shi, J.; Gao, T. Ox-LDL-induced lncRNA MALAT1 promotes autophagy in human umbilical vein endothelial cells by sponging miR-216a-5p and regulating Beclin-1 expression. Eur. J. Pharmacol. 2019, 858, 172338. [Google Scholar] [CrossRef]
- Huang, W.; Huang, F.-Z.; Lei, Z.; Luo, H. LncRNA snhg11 promotes proliferation, migration, apoptosis, and autophagy by regulating hsa-miR-184/AGO2 in HCC. OncoTargets Ther. 2020, 13, 413–421. [Google Scholar] [CrossRef]
- Liu, K.; Hou, Y.; Liu, Y.; Zheng, J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J. Biomed. Sci. 2017, 24, 46. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Jia, H.-H.; Xu, Y.-Q.; Zhou, X.; Zhao, X.-H.; Wang, Y.-F.; Song, X.; Zhu, Z.-Y.; Sun, T.; Dou, Y.; et al. Paracrine and epigenetic control of CAF-induced metastasis: The role of HOTAIR stimulated by TGF-ß1 secretion. Mol. Cancer 2018, 17, 5. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, B.; Khalaj-Kondori, M.; Asadollahi, E.; Sadeghizadeh, M. Cancer-associated fibroblasts enhance cell proliferation and metastasis of colorectal cancer SW480 cells by provoking long noncoding RNA UCA1. J. Cell Commun. Signal. 2018, 13, 53–64. [Google Scholar] [CrossRef]
- Su, Y.; Zhou, Y.; Sun, Y.-J.; Wang, Y.; Yin, J.-Y.; Huang, Y.-J.; Zhang, J.; He, A.-N.; Han, K.; Zhang, H.-Z.; et al. Macrophage-derived CCL18 promotes osteosarcoma proliferation and migration by upregulating the expression of UCA1. J. Mol. Med. 2019, 97, 49–61. [Google Scholar] [CrossRef]
- Li, W.; Xie, P.; Ruan, W.-H. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J. Bone Oncol. 2016, 5, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhang, J.; Xu, X.; Qu, Y.; Dong, H.; Dang, J.; Huo, Z.; Xu, G. LncRNA expression profile during autophagy and Malat1 function in macrophages. PLoS ONE 2019, 14, e0221104. [Google Scholar] [CrossRef] [PubMed]
- Maia, J.; Caja, S.; Moraes, M.C.S.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Qin, P.; Zhang, D.; Cui, X.; Gao, J.; Yu, Z.; Chai, Y.; Wang, J.; Li, J. Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging 2019, 11, 9581–9596. [Google Scholar] [CrossRef]
- Hardin, H.; Helein, H.; Meyer, K.; Robertson, S.; Zhang, R.; Zhong, W.; Lloyd, R.V. Thyroid cancer stem-like cell exosomes: Regulation of EMT via transfer of lncRNAs. Lab. Investig. 2018, 98, 1133–1142. [Google Scholar] [CrossRef]
- Xue, M.; Chen, W.; Xiang, A.; Wang, R.; Chen, H.; Pan, J.; Pang, H.; An, H.; Wang, X.; Hou, H.; et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol. Cancer 2017, 16, 143. [Google Scholar] [CrossRef]
- Zhang, W.; Yuan, W.; Song, J.; Wang, S.; Gu, X. LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α. Biochim. 2018, 144, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Sceneay, J.; Parker, B.S.; Smyth, M.J.; Möller, A. Hypoxia-driven immunosuppression contributes to the pre-metastatic niche. OncoImmunology 2013, 2, e2235. [Google Scholar] [CrossRef]
- Erler, J.T.; Bennewith, K.L.; Cox, T.R.; Lang, G.; Bird, D.; Koong, A.; Le, Q.-T.; Giaccia, A.J. Hypoxia-Induced Lysyl Oxidase Is a Critical Mediator of Bone Marrow Cell Recruitment to Form the Premetastatic Niche. Cancer Cell 2009, 15, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Dong, B.; Dong, P.; He, Y.; Zheng, J.; Xu, P. Hypoxia induces the activation of hepatic stellate cells through the PVT1-miR-152-ATG14 signaling pathway. Mol. Cell. Biochem. 2020, 465, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Su, D.W.; Li, X.; Chen, J.; Dou, J.; Fang, G.E.; Luo, C.J. MiR-543 inhibits proliferation and metastasis of human colorectal cancer cells by targeting PLAS3. Eur Rev Med Pharmacol Sci 2020, 24, 8812–8821. [Google Scholar]
- Cao, P.; Jin, Q.; Feng, L.; Li, H.; Qin, G.; Zhou, G. Emerging roles and potential clinical applications of noncoding RNAs in hepatocellular carcinoma. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, W.; Chen, X.; Wang, Q.; Li, C.; Chen, Q.; Zhang, Y.; Lu, Y.; Ding, X.; Jiang, C. Bone marrow mesenchymal stem cells-derived exosomes for penetrating and targeted chemotherapy of pancreatic cancer. Acta Pharm. Sin. B. 2020, 10, 1563–1575. [Google Scholar] [CrossRef]
- Cui, X.; Sun, Y.; Shen, M.; Song, K.; Yin, X.; Di, W.; Duan, Y. Enhanced chemotherapeutic efficacy of paclitaxel nanoparticles co-delivered with microRNA-7 by inhibiting paclitaxel-induced EGFR/ERK pathway activation for ovarian cancer therapy. ACS Appl. Mater. Interfaces 2018, 10, 7821–7831. [Google Scholar] [CrossRef]
- Tian, X.; Gu, T.; Patel, S.; Bode, A.M.; Lee, M.-H.; Surh, Y.-J. CRISPR/Cas9 – An evolving biological tool kit for cancer biology and oncology. NPJ Precis. Oncol. 2019, 3, 8. [Google Scholar] [CrossRef]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Rangwala, R.; Chang, Y.C.; Hu, J.; Algazy, K.M.; Evans, T.L.; A Fecher, L.; Schuchter, L.M.; A Torigian, D.; Panosian, J.T.; Troxel, A.B.; et al. Combined MTOR and autophagy inhibition. Autophagy 2014, 10, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, M.R.; Ye, X.; Supko, J.G.; Desideri, S.; A Grossman, S.; Brem, S.; Mikkelson, T.; Wang, D.; Chang, Y.C.; Hu, J.; et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 2014, 10, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Ojha, R.; Bhattacharyya, S.; Singh, S.K. Autophagy in cancer stem cells: A potential link between chemoresistance, recurrence, and metastasis. BioRes. Open Access 2015, 4, 97–108. [Google Scholar] [CrossRef]
- E Pollok, K.; Saadatzadeh, R.; Zd, N.; Gj, K.; Sk, G.; Ba, J.; Ia, C.; P, M.; Jn, S.; Ld, S. Faculty Opinions recommendation of DNA repair capacity in multiple pathways predicts chemoresistance in glioblastoma multiforme. Cancer Res. 2017, 77, 198–206. [Google Scholar]
- Abdullah, L.N.; Chow, E.K. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.-J.; Gao, J.-B. Molecular mechanisms of chemoresistance in gastric cancer. World J. Gastrointest. Oncol. 2016, 8, 673–681. [Google Scholar] [CrossRef]
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the Integrated Stress Response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Ogata, M.; Hino, S.-I.; Saito, A.; Morikawa, K.; Kondo, S.; Kanemoto, S.; Murakami, T.; Taniguchi, M.; Tanii, I.; Yoshinaga, K.; et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006, 26, 9220–9231. [Google Scholar] [CrossRef]
- Garbar, C.; Mascaux, C.; Giustiniani, J.; Merrouche, Y.; Bensussan, A. Chemotherapy treatment induces an increase of autophagy in the luminal breast cancer cell MCF7, but not in the triple-negative MDA-MB231. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Peng, Y.-F.; Shi, Y.; Ding, Z.-B.; Ke, A.-W.; Gu, C.-Y.; Hui, B.; Zhou, J.; Qiu, S.-J.; Dai, Z.; Fan, J. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy 2013, 9, 2056–2068. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell 2016, 30, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Celià-Terrassa, T.; Kang, Y. Metastatic niche functions and therapeutic opportunities. Nat. Cell Biol. 2018, 20, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, K.S.; Raza, A.; Karedath, T.; Raza, S.S.; Fathima, H.; Ahmed, E.I.; Kuttikrishnan, S.; Therachiyil, L.; Kulinski, M.; Dermime, S.; et al. Non-coding RNAs as regulators and markers for targeting of breast cancer and cancer stem cells. Cancers 2020, 351. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cheng, Z.; Pang, Y.; Cui, L.; Qian, T.; Quan, L.; Zhao, H.; Shi, J.; Ke, X.; Fu, L. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 2019, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ma, X.; Li, H. Intriguing circles: Conflicts and controversies in circular RNA research. Wiley Interdiscip. Rev. RNA 2019, 10, e1538. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Jara, C.A.C.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, P.; Wang, L.; Li, M.; Ge, Z.; Noordam, L.; Lieshout, R.; Verstegen, M.M.; Ma, B.; Su, J.; et al. Cancer-associated fibroblasts provide a stromal niche for liver cancer organoids that confers trophic effects and therapy resistance. Cell. Mol. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).