Direct Transfer of Mesoporous Silica Nanoparticles between Macrophages and Cancer Cells
Simple Summary
Abstract
1. Introduction
2. Result and Discussion
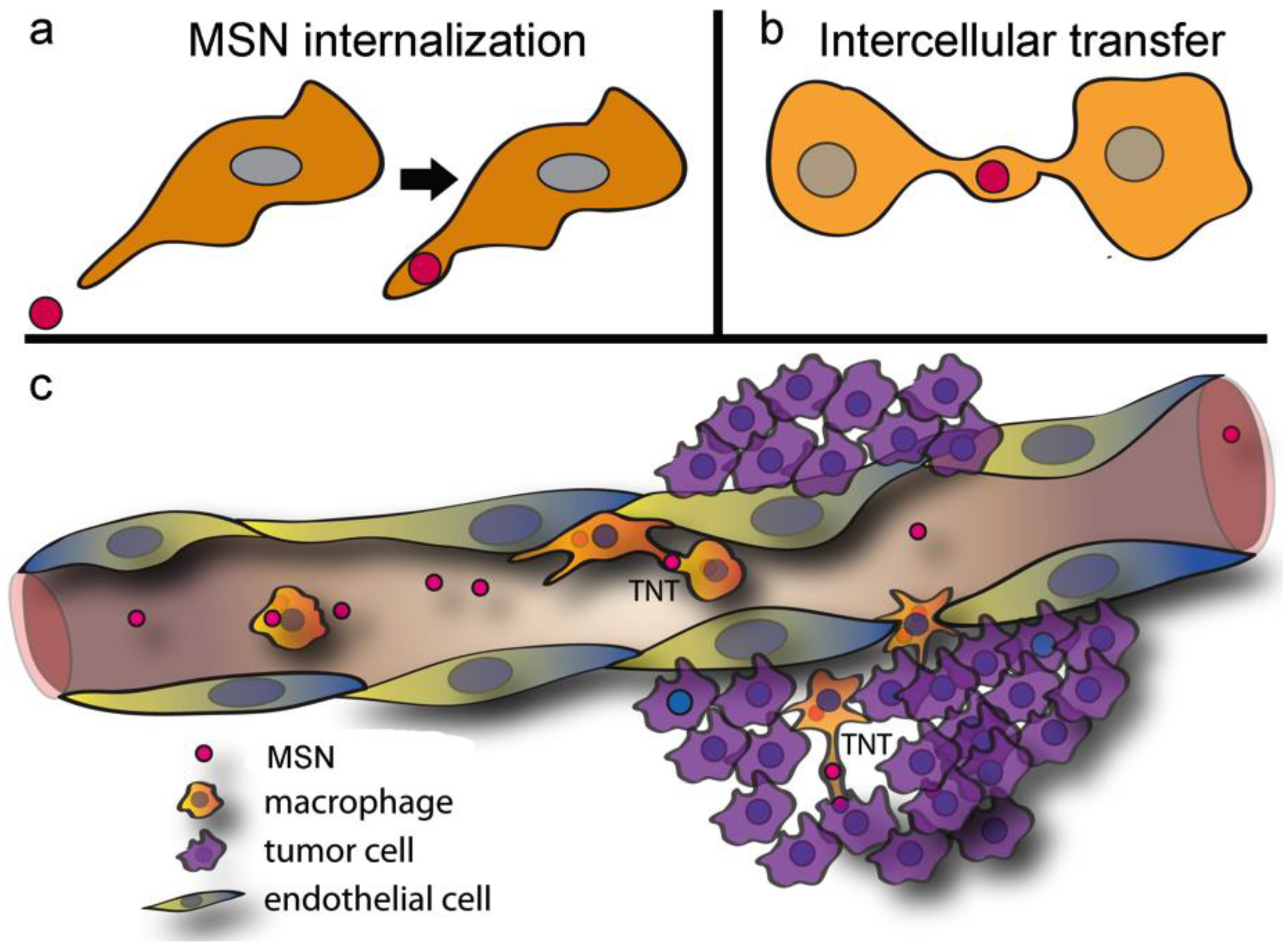
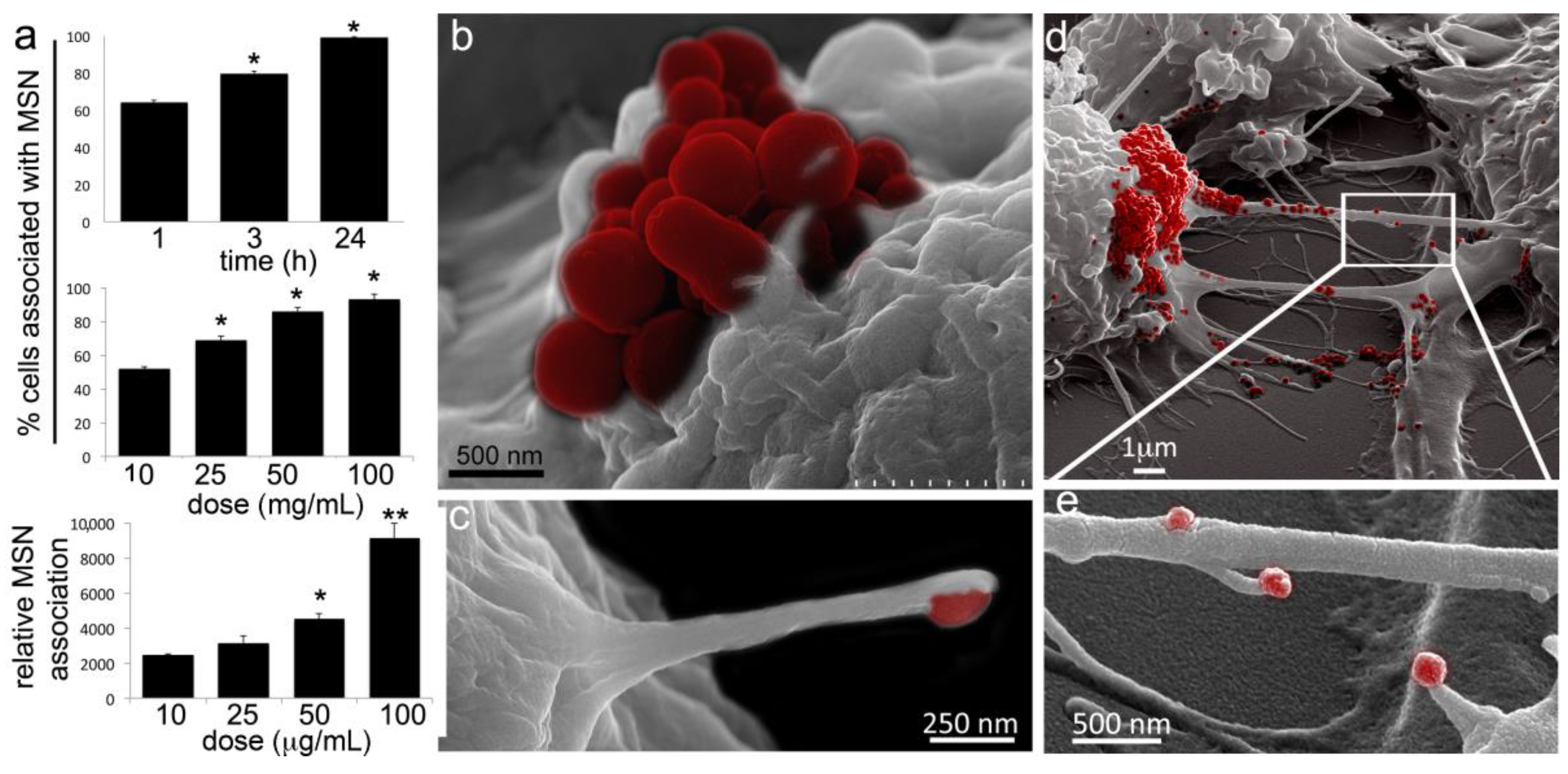
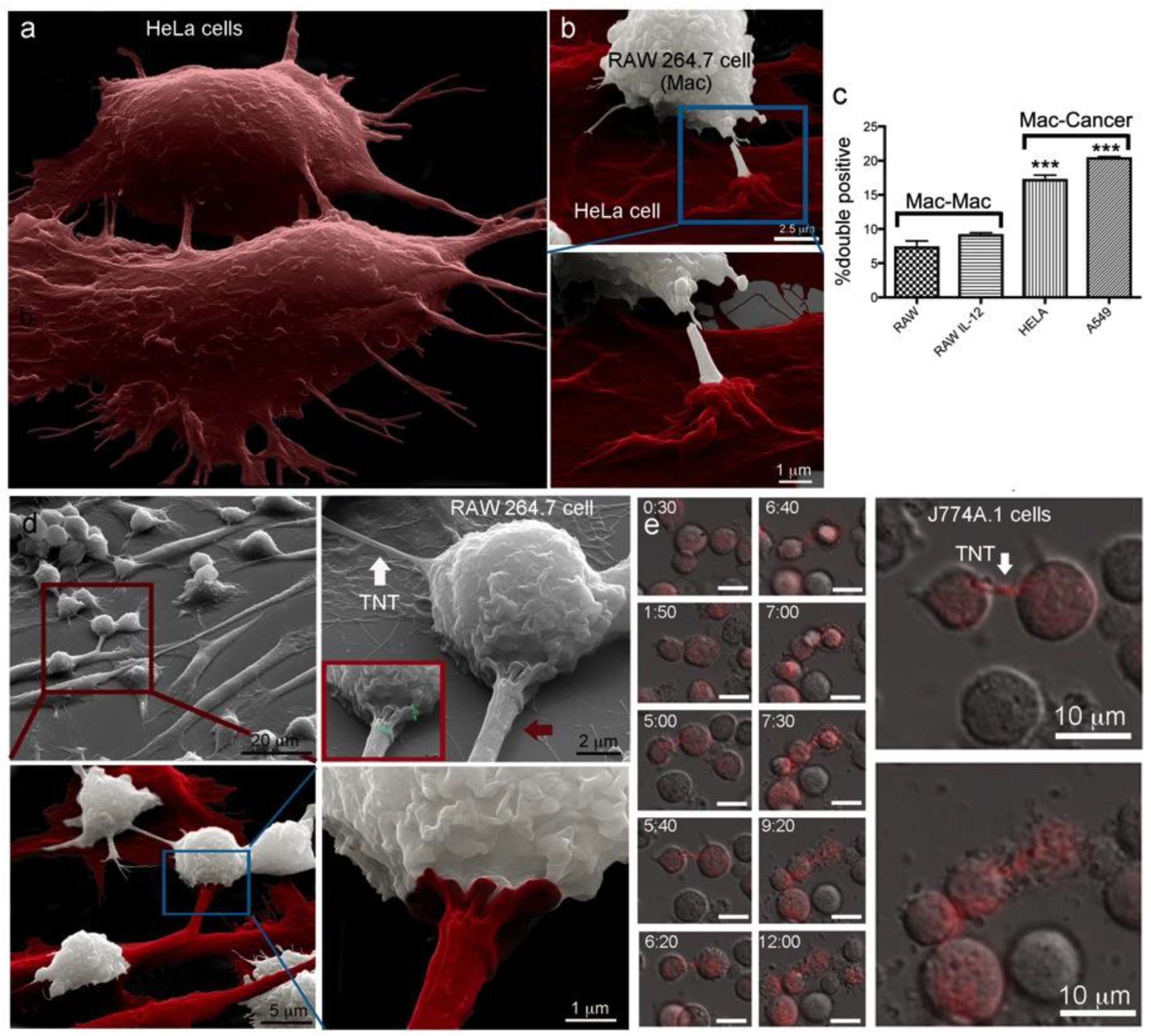
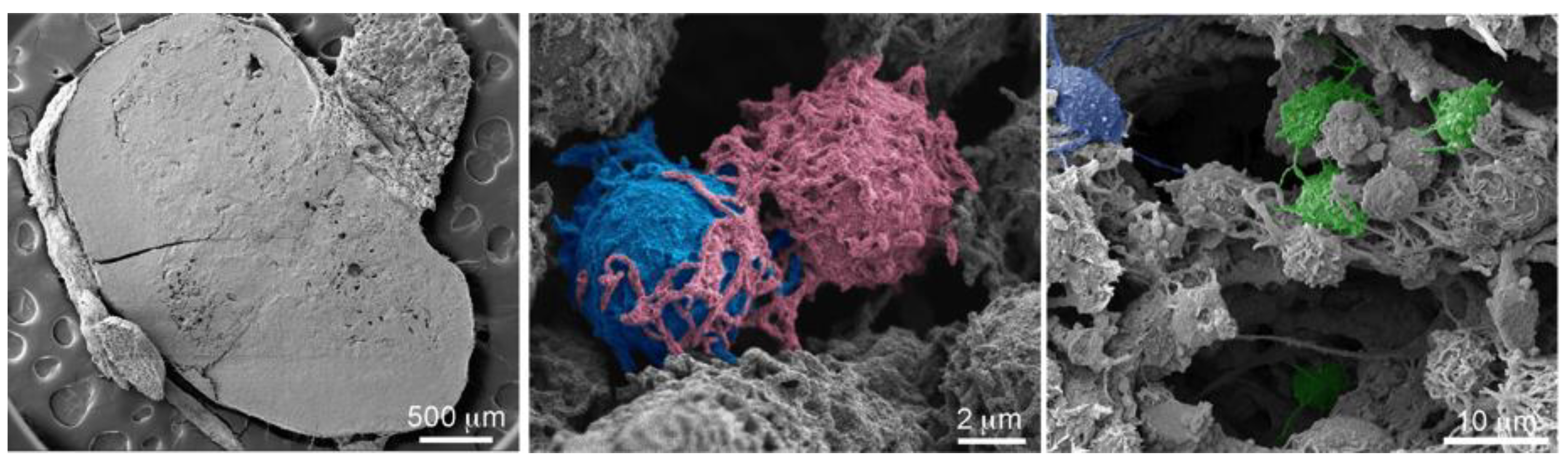
2.1. Macrophages Internalize and Transport MSNs through Extensive Crosstalk
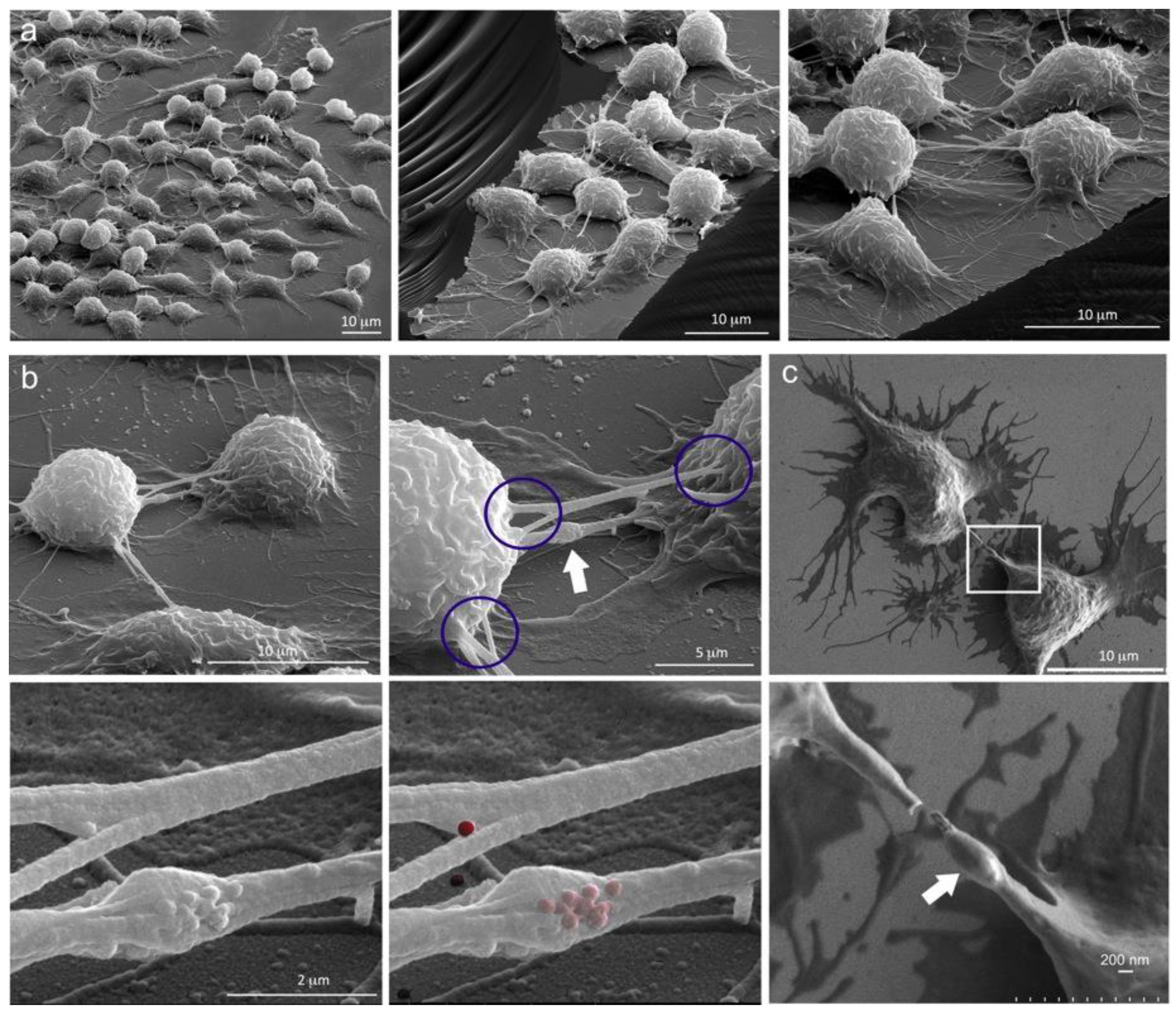
2.2. Diverse TNT Assemblies and Gondolas
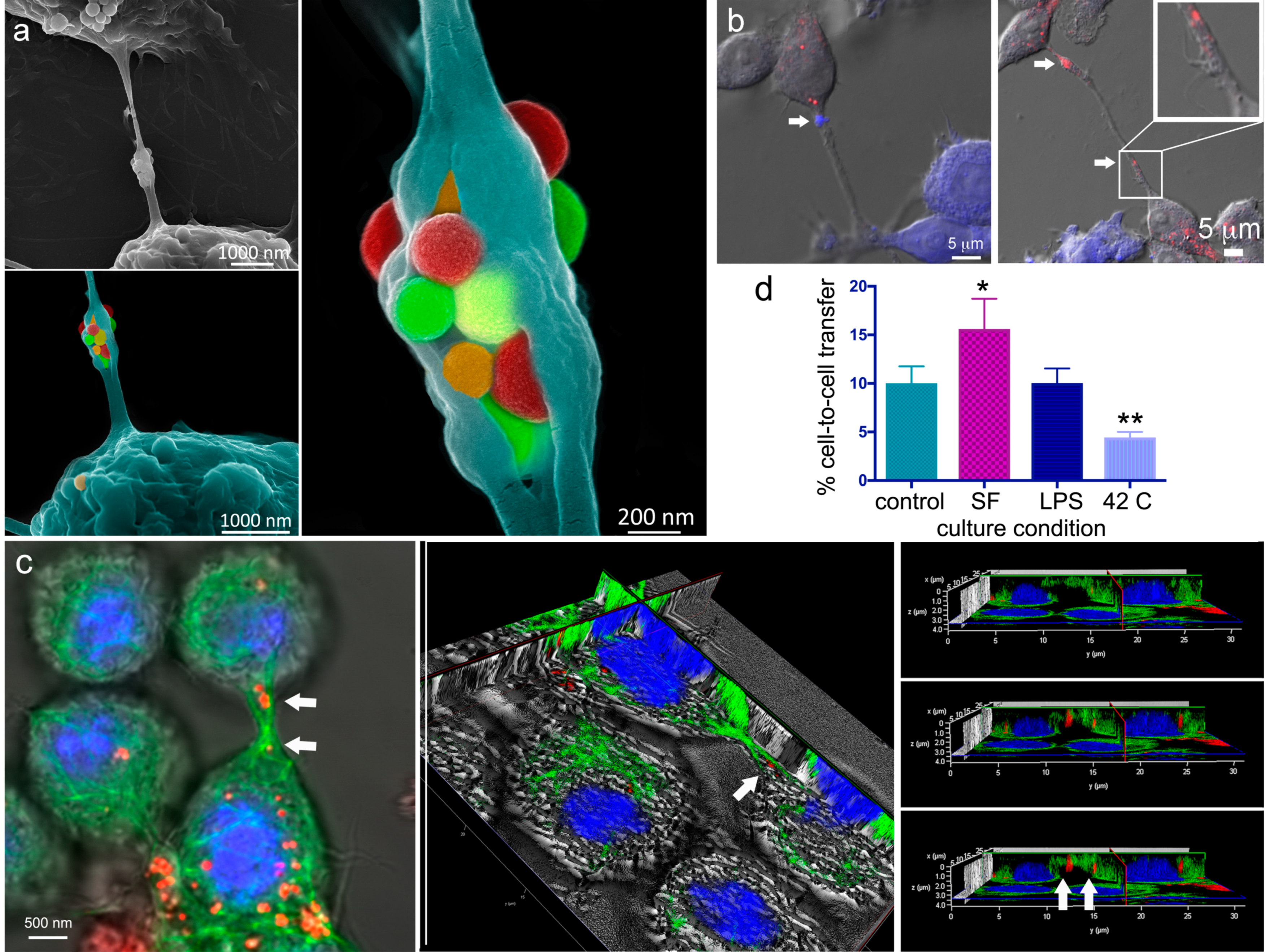
2.3. A Look Inside TNTs and Gondolas
2.4. The Impact of Environmental Triggers on Intercellular Transfer of MSN through TNT
2.5. Trans-Species Transfer of MSN from Murine Macrophages to Human Cancer Cells
2.6. Transfer of Carrier-Delivered Chemotherapeutics between Macrophages
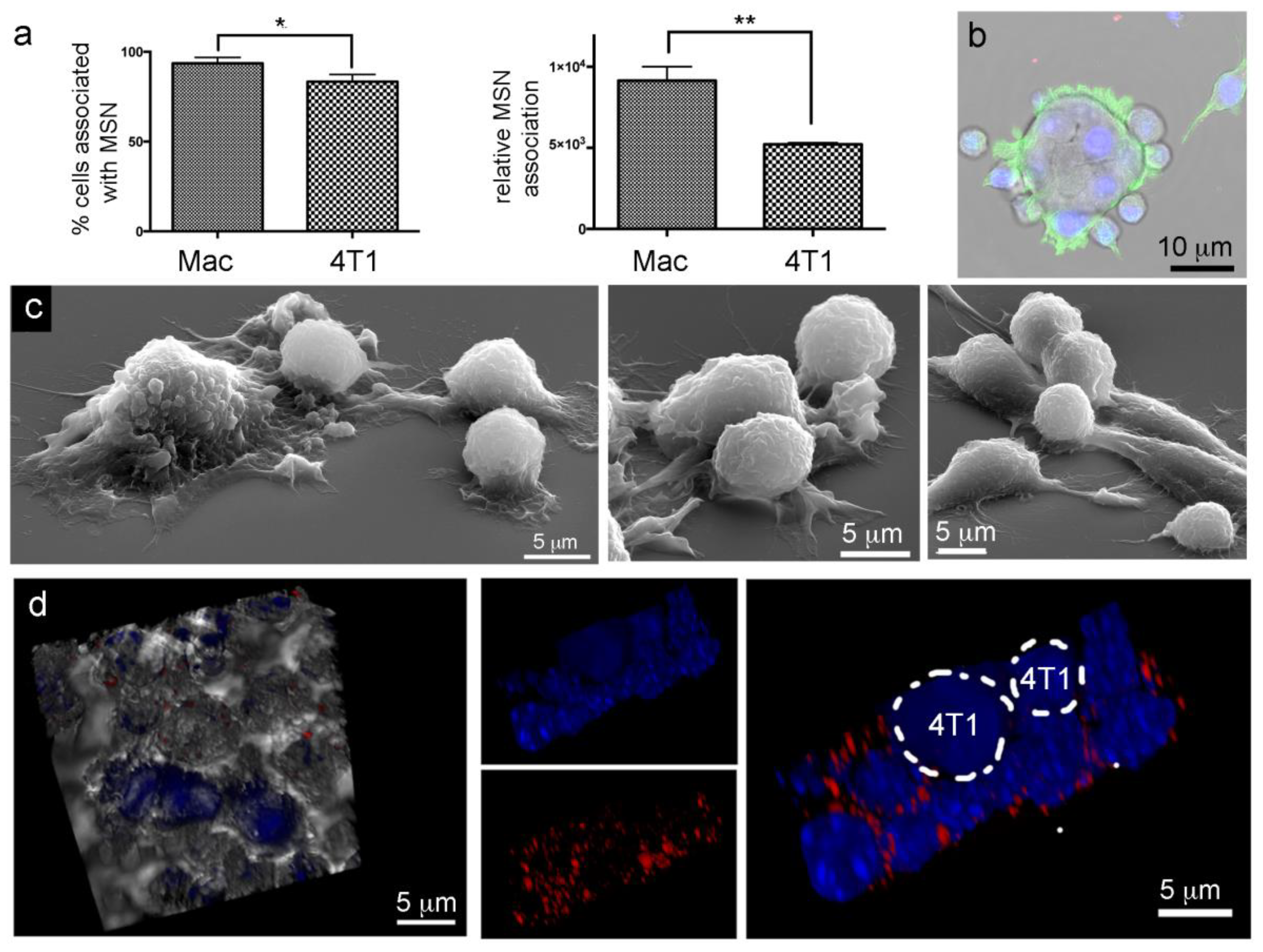
2.7. Lack of Murine Intraspecies MSN Heterotypic Transfer from Macrophages to Breast Cancer Cells
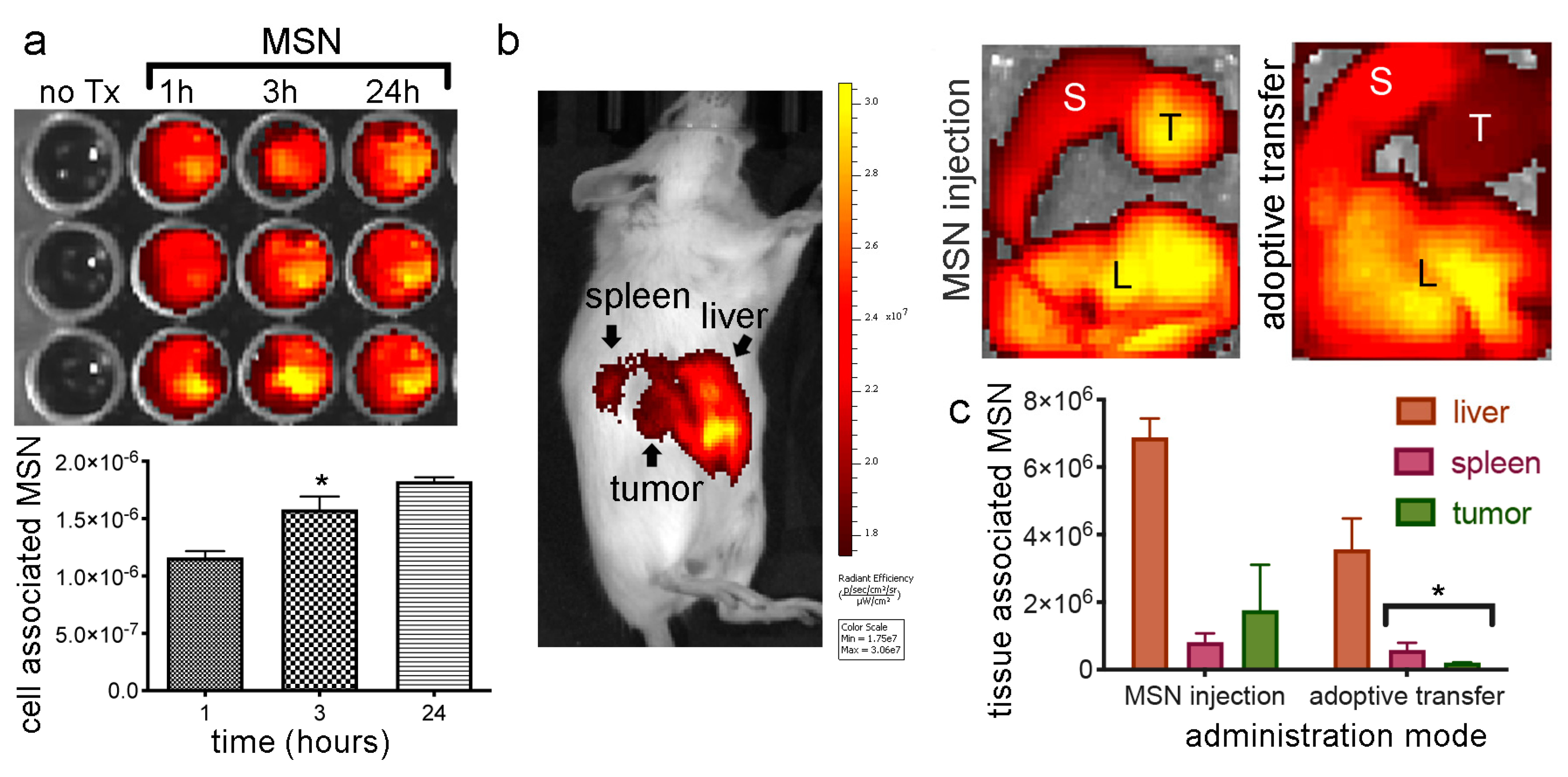
2.8. In Vivo MSN Biodistribution and Trafficking in TNTs
2.8.1. Biodistribution of Free MSN and Splenocytes-Loaded MSN
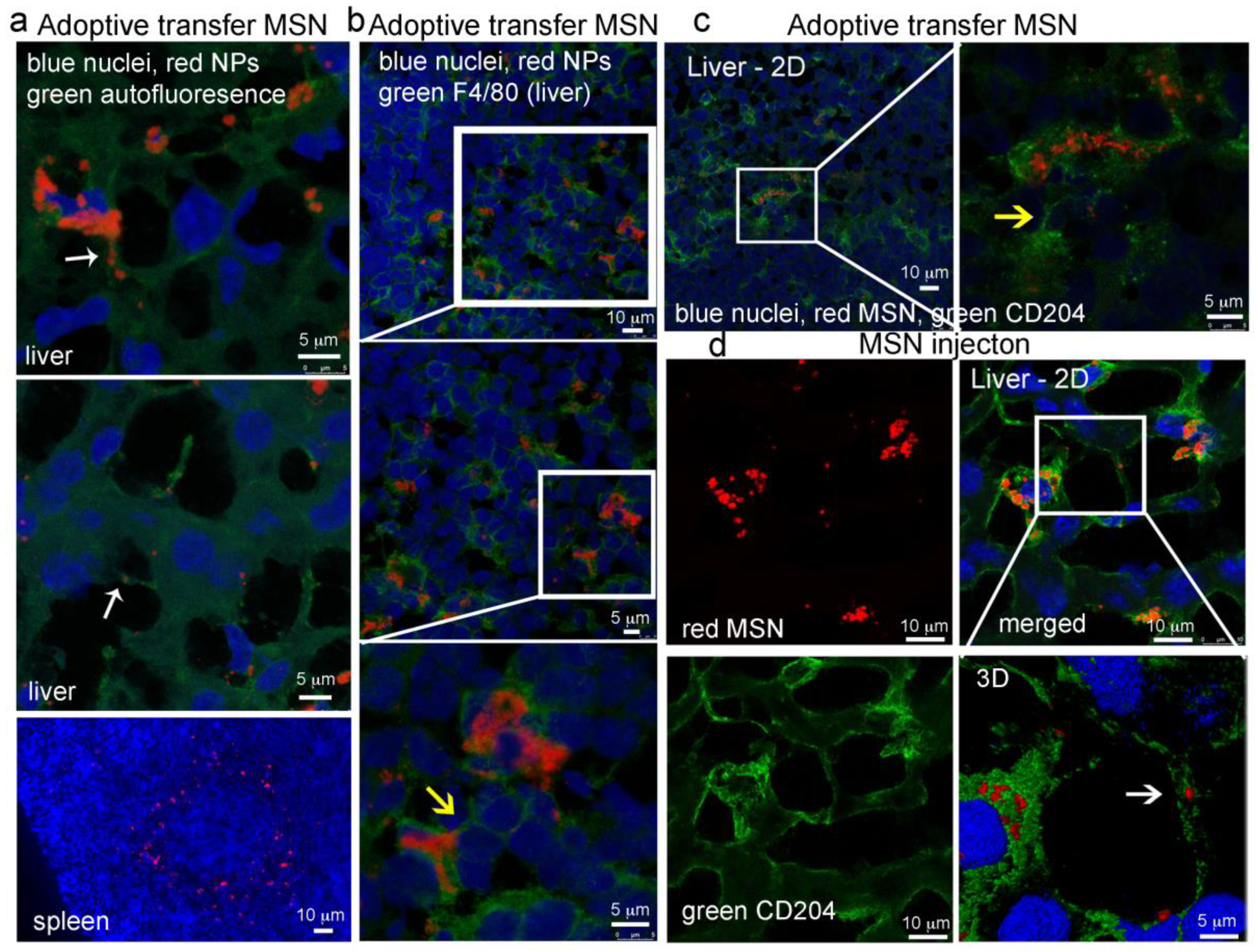
2.8.2. MSN Association with Hepatic Macrophages and In Vivo Homotypic MSN Transfer via TNTs
3. Materials and Methods
3.1. Cell Culture
3.2. MSN Fabrication
3.3. Confocal Microscopy Imaging of TNT Transport of Nanoparticles
3.4. Live-Cell Microscopy
3.5. Scanning Electron Microscopy (SEM) Imaging of Tissue and Cells
3.6. Flow Cytometry Analysis of Particle Association with Cells and Transfer between Cells
3.7. Biodistribution and Tissue Confocal Microscopy
3.8. Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grabowska, J.; Lopez-Venegas, M.; Affandi, A.J.; Haan, J.M.M.D. CD169+ Macrophages Capture and Dendritic Cells Instruct: The Interplay of the Gatekeeper and the General of the Immune System. Front. Immunol. 2018, 9, 2472. [Google Scholar] [CrossRef] [PubMed]
- Weissleder, R.; Nahrendorf, M.; Pittet, M.J. Imaging macrophages with nanoparticles. Nat. Mater. 2014, 13, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Batrakova, E.V.; Gendelman, H.E.; Kabanov, A.V. Cell-mediated drug delivery. Expert. Opin. Drug. Deliv. 2011, 8, 415–433. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, S.; Warren, G.; Wei, X. Macrophages associated with tumors as potential targets and therapeutic intermediates. Nanomedicine 2014, 9, 695–707. [Google Scholar] [CrossRef] [PubMed]
- De Palma, M.; Biziato, D.; Petrova, T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 16014. [Google Scholar] [CrossRef]
- Nowacek, A.S.; Miller, R.L.; McMillan, J.; Kanmogne, G.; Kanmogne, M.; Mosley, R.L.; Ma, Z.; Graham, S.; Chaubal, M.; Werling, J.; et al. NanoART synthesis, characterization, uptake, release and toxicology for human monocyte–macrophage drug delivery. Nanomedicine 2009, 4, 903–917. [Google Scholar] [CrossRef]
- Liu, T.; Li, R.; Pan, T.; Liu, D.; Petersen, R.B.; Wong, B.-S.; Gambetti, P.; Sy, M.S. Intercellular Transfer of the Cellular Prion Protein. J. Biol. Chem. 2002, 277, 47671–47678. [Google Scholar] [CrossRef]
- Hanger, D.P.; Lau, D.H.; Phillips, E.C.; Bondulich, M.K.; Guo, T.; Woodward, B.W.; Pooler, A.M.; Noble, W. Intracellular and Extracellular Roles for Tau in Neurodegenerative Disease. J. Alzheimer’s Dis. 2014, 40, S37–S45. [Google Scholar] [CrossRef]
- Abounit, S.; Zurzolo, C. Wiring through tunneling nanotubes—From electrical signals to organelle transfer. J. Cell. Sci. 2012, 125, 1089–1098. [Google Scholar] [CrossRef]
- Watkins, S.C.; Salter, R.D. Functional Connectivity between Immune Cells Mediated by Tunneling Nanotubules. Immunity 2005, 23, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Hudrisier, D.; Riond, J.; Mazarguil, H.; Gairin, J.E.; Joly, E. Cutting edge: CTLs rapidly capture membrane fragments from target cells in a TCR signaling-dependent manner. J. Immunol. 2001, 166, 3645–3649. [Google Scholar] [CrossRef] [PubMed]
- Tatari-Calderone, Z.; Semnani, R.T.; Nutman, T.B.; Schlom, J.; Sabzevari, H. Acquisition of CD80 by Human T Cells at Early Stages of Activation: Functional Involvement of CD80 Acquisition in T Cell to T Cell Interaction. J. Immunol. 2002, 169, 6162–6169. [Google Scholar] [CrossRef] [PubMed]
- Ferrati, S.; McConnell, I.K.; Mack, A.C.; Sirisaengtaksin, N.; Díaz, R.; Bean, A.J.; Ferrari, M.; Serda, R.E. Cellular communication via nanoparticle-transporting biovesicles. Nanomedicine 2014, 9, 581–592. [Google Scholar] [CrossRef]
- Ferrati, S.; Mack, A.; Chiappini, C.; Liu, X.; Bean, A.J.; Ferrari, M.; Serda, R.E. Intracellular trafficking of silicon particles and logic-embedded vectors. Nanoscale 2010, 2, 1512–1520. [Google Scholar] [CrossRef]
- McCoy-Simandle, K.; Hanna, S.J.; Cox, D. Exosomes and nanotubes: Control of immune cell communication. Int. J. Biochem. Cell. Biol. 2016, 71, 44–54. [Google Scholar] [CrossRef]
- Ferrati, S.; Shamsudeen, S.; Summers, H.D.; Rees, P.; Abbey, J.V.A.; Schmulen, J.; Liu, X.; Wong, S.T.C.; Bean, A.J.; Ferrari, M.; et al. Inter-endothelial Transport of Microvectors using Cellular Shuttles and Tunneling Nanotubes. Small 2012, 8, 3151–3160. [Google Scholar] [CrossRef]
- Roehlecke, C.; Schmidt, M.H. Tunneling Nanotubes and Tumor Microtubes in Cancer. Cancers 2020, 12, 857. [Google Scholar] [CrossRef]
- Goodman, S.; Naphade, S.; Khan, M.; Sharma, J.; Cherqui, S. Macrophage polarization impacts tunneling nanotube formation and intercellular organelle trafficking. Sci. Rep. 2019, 9, 14529. [Google Scholar] [CrossRef]
- Carter, K.P.; Hanna, S.; Genna, A.; Lewis, D.; Segall, J.E.; Cox, D. Macrophages enhance 3D invasion in a breast cancer cell line by induction of tumor cell tunneling nanotubes. Cancer. Rep. 2019, 2, e1213. [Google Scholar] [CrossRef]
- Dubois, F.; Bénard, M.; Jean-Jacques, B.; Schapman, D.; Roberge, H.; Lebon, A.; Goux, D.; Monterroso, B.; Elie, N.; Komuro, H.; et al. Investigating Tunneling Nanotubes in Cancer Cells: Guidelines for Structural and Functional Studies through Cell Imaging. Biomed. Res. Int. 2020, 2020, 1–16. [Google Scholar] [CrossRef]
- Gousset, K.; Schiff, E.; Langevin, C.; Marijanovic, Z.; Caputo, A.; Browman, D.T.; Chenouard, N.; De Chaumont, F.; Martino, A.; Enninga, J.; et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat. Cell. Biol. 2009, 11, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Önfelt, B.; Nedvetzki, S.; Yanagi, K.; Davis, D.M. Cutting Edge: Membrane Nanotubes Connect Immune Cells. J. Immunol. 2004, 173, 1511–1513. [Google Scholar] [CrossRef]
- Noureddine, A.; Gary-Bobo, M.; Lichon, L.; Garcia, M.; Zink, J.I.; Man, M.W.C.; Cattoën, X. Bis-clickable Mesoporous Silica Nanoparticles: Straightforward Preparation of Light-Actuated Nanomachines for Controlled Drug Delivery with Active Targeting. Chem. A Eur. J. 2016, 22, 9624–9630. [Google Scholar] [CrossRef] [PubMed]
- Villegas, M.R.; Baeza, A.; Noureddine, A.; Durfee, P.N.; Butler, K.S.; Agola, J.O.; Brinker, C.J.; Vallet-Regí, M. Multifunctional Protocells for Enhanced Penetration in 3D Extracellular Tumoral Matrices. Chem. Mater. 2017, 30, 112–120. [Google Scholar] [CrossRef]
- Noureddine, A.; Lichon, L.; Maynadier, M.; Garcia, M.; Gary-Bobo, M.; Zink, J.I.; Cattoën, X.; Man, M.W.C. Controlled multiple functionalization of mesoporous silica nanoparticles: Homogeneous implementation of pairs of functionalities communicating through energy or proton transfers. Nanoscale 2015, 7, 11444–11452. [Google Scholar] [CrossRef]
- Giret, S.; Man, M.W.C.; Carcel, C. Mesoporous-Silica-Functionalized Nanoparticles for Drug Delivery. Chem. A Eur. J. 2015, 21, 13850–13865. [Google Scholar] [CrossRef]
- Slowing, I.; Vivero-Escoto, J.L.; Zhao, Y.; Kandel, K.; Peeraphatdit, C.; Trewyn, B.G.; Lin, V.S.-Y. Exocytosis of Mesoporous Silica Nanoparticles from Mammalian Cells: From Asymmetric Cell-to-Cell Transfer to Protein Harvesting. Small 2011, 7, 1526–1532. [Google Scholar] [CrossRef]
- Rehberg, M.; Nekolla, K.; Sellner, S.; Praetner, M.; Mildner, K.; Zeuschner, D.; Krombach, F. Intercellular Transport of Nanomaterials is Mediated by Membrane Nanotubes In Vivo. Small 2016, 12, 1882–1890. [Google Scholar] [CrossRef]
- Sakagami, H.; Kishino, K.; Amano, O.; Kanda, Y.; Kunii, S.; Yokote, Y.; Oizumi, H.; Oizumi, T. Cell death induced by nutritional starvation in mouse macrophage-like RAW264.7 cells. Anticancer. Res. 2009, 29, 343–347. [Google Scholar]
- Mattila, P.K.; Lappalainen, P. Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell. Biol. 2008, 9, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, H.-H.; Bukoreshtliev, N.V.; Barroso, J.F. Tunneling nanotubes: A new route for the exchange of components between animal cells. FEBS Lett. 2007, 581, 2194–2201. [Google Scholar] [CrossRef] [PubMed]
- Hilpelä, P.; Vartiainen, M.K.; Lappalainen, P. Regulation of the Actin Cytoskeleton by PI(4,5)P2 and PI(3,4,5)P3. In Phosphoinositides in Subcellular Targeting and Enzyme Activation. Current Topics in Microbiology and Immunology; Stenmark, H., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 282, pp. 117–163. [Google Scholar]
- Galbraith, C.G.; Yamada, K.M.; Galbraith, J.A. Polymerizing Actin Fibers Position Integrins Primed to Probe for Adhesion Sites. Science 2007, 315, 992–995. [Google Scholar] [CrossRef]
- Gerdes, H.-H.; Rustom, A.; Wang, X. Tunneling nanotubes, an emerging intercellular communication route in development. Mech. Dev. 2013, 130, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Delage, E.; Cervantes, D.C.; Pénard, E.; Schmitt, C.; Syan, S.; Disanza, A.; Scita, G.; Zurzolo, C. Differential identity of Filopodia and Tunneling Nanotubes revealed by the opposite functions of actin regulatory complexes. Sci. Rep. 2016, 6, 39632. [Google Scholar] [CrossRef]
- Hurtig, J.; Chiu, D.T.; Önfelt, B. Intercellular nanotubes: Insights from imaging studies and beyond. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 260–276. [Google Scholar] [CrossRef]
- Drab, M.; Stopar, D.; Kralj-Iglič, V.; Iglič, A. Inception Mechanisms of Tunneling Nanotubes. Cells 2019, 8, 626. [Google Scholar] [CrossRef]
- Onfelt, B.; Nedvetzki, S.; Benninger, R.K.P.; Purbhoo, M.A.; Sowinski, S.; Hume, A.N.; Seabra, M.C.; Neil, M.; French, P.M.W.; Davis, D.M. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J. Immunol. 2006, 177, 8476–8483. [Google Scholar] [CrossRef]
- Pritz, C.; Bitsche, M.; Salvenmoser, W.; Edudas, J.; Schrott-Fischer, A.; Glueckert, R. Endocytic trafficking of silica nanoparticles in a cell line derived from the organ of Corti. Nanomedicine 2013, 8, 239–252. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Neefjes, J. Moving and positioning the endolysosomal system. Curr. Opin. Cell. Biol. 2017, 47, 1–8. [Google Scholar] [CrossRef]
- Zhu, D.; Tan, K.S.; Zhang, X.; Sun, A.Y.; Lee, J.C.-M. Hydrogen peroxide alters membrane and cytoskeleton properties and increases intercellular connections in astrocytes. J. Cell Sci. 2005, 118, 3695–3703. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gerdes, H.-H. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell. Death. Differ. 2015, 22, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cui, J.; Sun, X.; Zhang, Y. Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ. 2010, 18, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xue, C.; Xu, X.; Guo, Y.; Li, X.; Lu, J.; Ju, S.; Wang, Y.; Cao, Z.; Gu, X. Rab8a/Rab11a regulate intercellular communications between neural cells via tunneling nanotubes. Cell. Death. Dis. 2016, 7, e2523. [Google Scholar] [CrossRef]
- Kumar, A.; Kim, J.H.; Ranjan, P.; Metcalfe, M.G.; Cao, W.; Mishina, M.; Gangappa, S.; Guo, Z.; Boyden, E.S.; Zaki, S.; et al. Influenza virus exploits tunneling nanotubes for cell-to-cell spread. Sci. Rep. 2017, 7, 40360. [Google Scholar] [CrossRef]
- Osswald, M.; Jung, E.; Sahm, F.; Solecki, G.; Venkataramani, V.; Blaes, J.; Weil, S.; Horstmann, H.; Wiestler, B.; Syed, M.; et al. Brain tumour cells interconnect to a functional and resistant network. Nat. Cell. Biol. 2015, 528, 93–98. [Google Scholar] [CrossRef]
- Hanna, S.J.; McCoy-Simandle, K.; Miskolci, V.; Guo, P.; Cammer, M.; Hodgson, L.; Cox, D. The Role of Rho-GTPases and actin polymerization during Macrophage Tunneling Nanotube Biogenesis. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Nishimura, Y.; Itoh, K.; Yoshioka, K.; Ikeda, K.; Himeno, M. A role for small GTPase RhoA in regulating intracellular membrane traffic of lysosomes in invasive rat hepatoma cells. J. Mol. Histol. 2002, 34, 189–213. [Google Scholar]
- De Santa-María, I.S.; Bernardo-Castiñeira, C.; Enciso, E.; Garcia-Moreno, I.; Chiara, J.-L.; Suárez, C.; Chiara, M.-D. Control of long-distance cell-to-cell communication and autophagosome transfer in squamous cell carcinoma via tunneling nanotubes. Oncotarget 2017, 8, 20939–20960. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, Y.; Yang, Z.; Peng, H.; Wei, R.; Wang, C.; Feng, M. Tunneling Nanotubular Expressways for Ultrafast and Accurate M1 Macrophage Delivery of Anticancer Drugs to Metastatic Ovarian Carcinoma. ACS Nano 2019, 13, 1078–1096. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, M.; Tang, W.; Wen, R.; Zhou, S.; Lee, C.; Wang, H.; Jiang, W.; Delahunty, I.M.; Zhen, Z.; et al. Nanoparticle-Laden Macrophages for Tumor-Tropic Drug Delivery. Adv. Mater. 2018, 30, e1805557. [Google Scholar] [CrossRef] [PubMed]
- Tonkin, J.A.; Shamsudeen, S.; Brown, M.R.; Serda, R.E.; Rees, P.; Summers, H.D. Optical tracking of drug release from porous silicon delivery vectors. IET Optoelectron. 2014, 8, 113–116. [Google Scholar] [CrossRef]
- Desir, S.; O’Hare, P.; Vogel, R.I.; Sperduto, W.; Sarkari, A.; Dickson, E.L.; Wong, P.; Nelson, A.C.; Fong, Y.; Steer, C.J.; et al. Chemotherapy-Induced Tunneling Nanotubes Mediate Intercellular Drug Efflux in Pancreatic Cancer. Sci. Rep. 2018, 8, 9484. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wang, D.; Mei, D.; Zhang, H.; Wang, Z.; He, B.; Dai, W.; Zhang, H.; Wang, X.; Zhang, Q. Macrophage mediated biomimetic delivery system for the treatment of lung metastasis of breast cancer. J. Control. Release 2015, 204, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, H.H.; Holt-Casper, D.; Grainger, D.W.; Ghandehari, H. Nanoparticle uptake: The phagocyte problem. Nano. Today 2015, 10, 487–510. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Wilhelm, S.; Ding, D.; Syed, A.M.; Sindhwani, S.; Zhang, Y.; Chen, Y.Y.; Macmillan, P.; Chan, W.C.W. Quantifying the Ligand-Coated Nanoparticle Delivery to Cancer Cells in Solid Tumors. ACS Nano 2018, 12, 8423–8435. [Google Scholar] [CrossRef]
- Stouch, A.N.; Zaynagetdinov, R.; Barham, W.J.; Stinnett, A.M.; Slaughter, J.C.; Yull, F.E.; Hoffman, H.M.; Blackwell, T.S.; Prince, L.S. IkappaB kinase activity drives fetal lung macrophage maturation along a non-M1/M2 paradigm. J. Immunol. 2014, 193, 1184–1193. [Google Scholar] [CrossRef]
- Cassado, A.D.A. F4/80 as a Major Macrophage Marker: The Case of the Peritoneum and Spleen. In Macrophages; Springer: Cham, Switzerland, 2017; pp. 161–179. [Google Scholar]
- Gordon, S.; Hamann, J.; Lin, H.H.; Stacey, M. F4/80 and the related adhesion-GPCRs. Eur. J. Immunol. 2011, 41, 2472–2476. [Google Scholar] [CrossRef]
- Kraal, G.; Janse, M. Marginal metallophilic cells of the mouse spleen identified by a monoclonal antibody. Immunology 1986, 58, 665–669. [Google Scholar]
- Noureddine, A.; Hjelvik, E.A.; Croissant, J.G.; Durfee, P.N.; Agola, J.O.; Brinker, C.J. Engineering of large-pore lipid-coated mesoporous silica nanoparticles for dual cargo delivery to cancer cells. J. Sol. Gel. Sci. Technol. 2018, 89, 78–90. [Google Scholar] [CrossRef]
- Serda, R.E.; Gu, J.; Bhavane, R.C.; Liu, X.; Chiappini, C.; Decuzzi, P.; Ferrari, M. The association of silicon microparticles with endothelial cells in drug delivery to the vasculature. Biomaterials 2009, 30, 2440–2448. [Google Scholar] [CrossRef] [PubMed]
- Lakkis, F.G.; Lechler, R.I. Origin and Biology of the Allogeneic Response. Cold Springer Harb. Perspect. Med. 2013, 3, a014993. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco, S.; Noureddine, A.; Guo, J.; Keth, J.; Paffett, M.L.; Brinker, C.J.; Serda, R.E. Direct Transfer of Mesoporous Silica Nanoparticles between Macrophages and Cancer Cells. Cancers 2020, 12, 2892. https://doi.org/10.3390/cancers12102892
Franco S, Noureddine A, Guo J, Keth J, Paffett ML, Brinker CJ, Serda RE. Direct Transfer of Mesoporous Silica Nanoparticles between Macrophages and Cancer Cells. Cancers. 2020; 12(10):2892. https://doi.org/10.3390/cancers12102892
Chicago/Turabian StyleFranco, Stefan, Achraf Noureddine, Jimin Guo, Jane Keth, Michael L. Paffett, C. Jeffrey Brinker, and Rita E. Serda. 2020. "Direct Transfer of Mesoporous Silica Nanoparticles between Macrophages and Cancer Cells" Cancers 12, no. 10: 2892. https://doi.org/10.3390/cancers12102892
APA StyleFranco, S., Noureddine, A., Guo, J., Keth, J., Paffett, M. L., Brinker, C. J., & Serda, R. E. (2020). Direct Transfer of Mesoporous Silica Nanoparticles between Macrophages and Cancer Cells. Cancers, 12(10), 2892. https://doi.org/10.3390/cancers12102892





