Ubiquitin and Ubiquitin-Like Proteins Are Essential Regulators of DNA Damage Bypass
Simple Summary
Abstract
1. Introduction
2. Ubiquitin and Ubiquitin-Like Modifiers
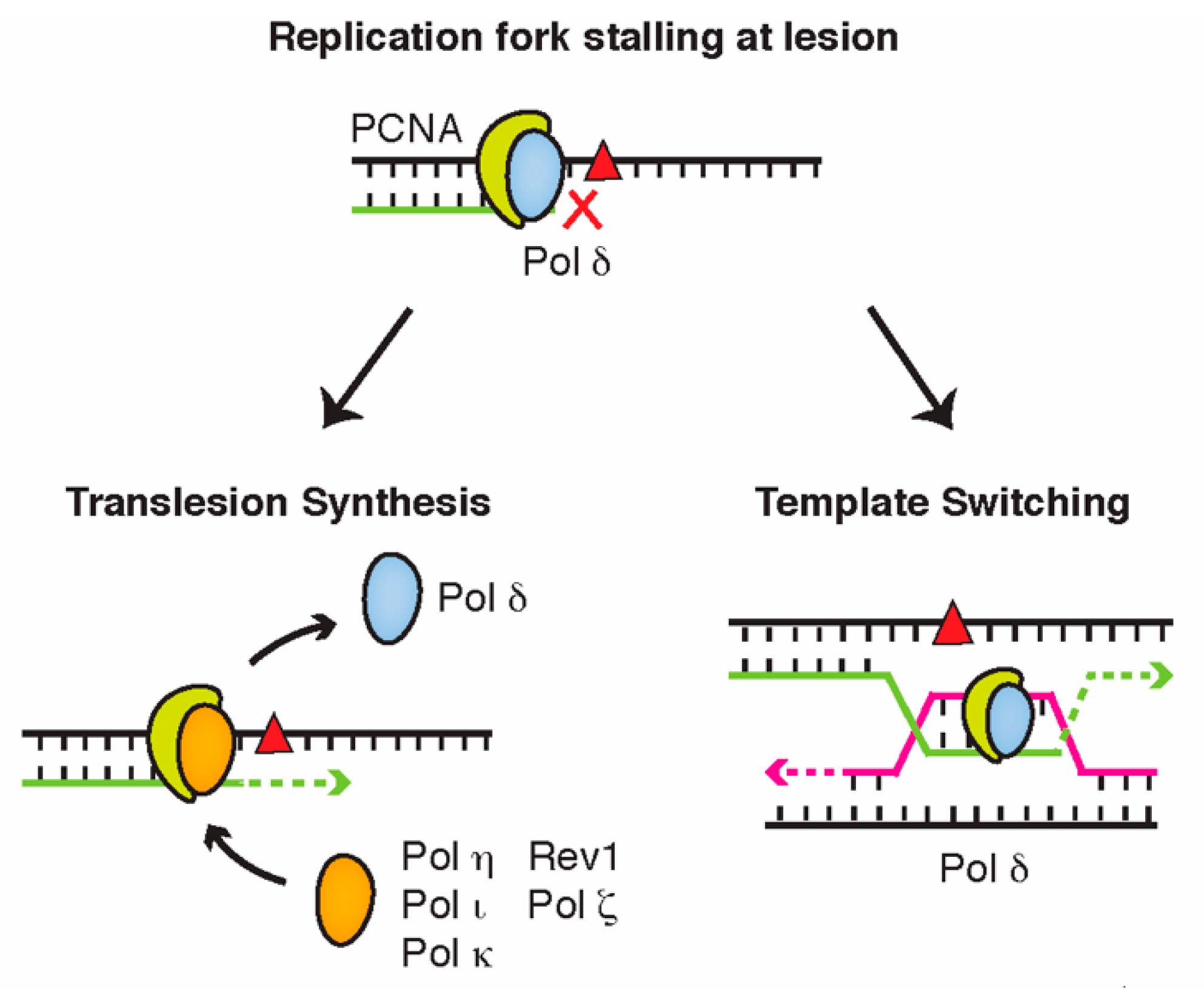
3. DNA Damage Bypass
4. Ubiquitin and Ubiquitin-Like Modifiers in DNA Damage Bypass
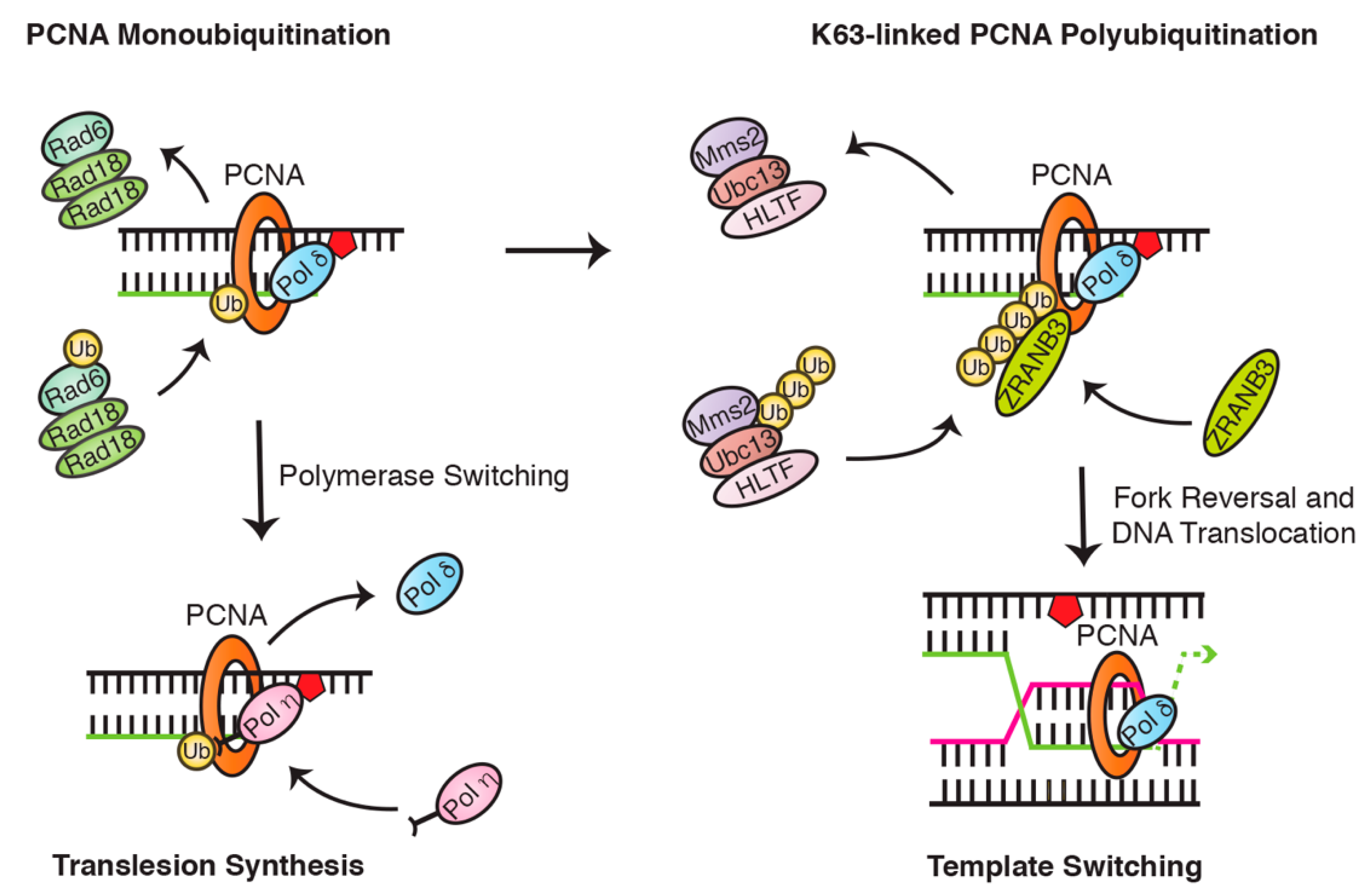
4.1. PCNA Ubiquitination Is a Central Regulator of Translesion Synthesis and Template Switching
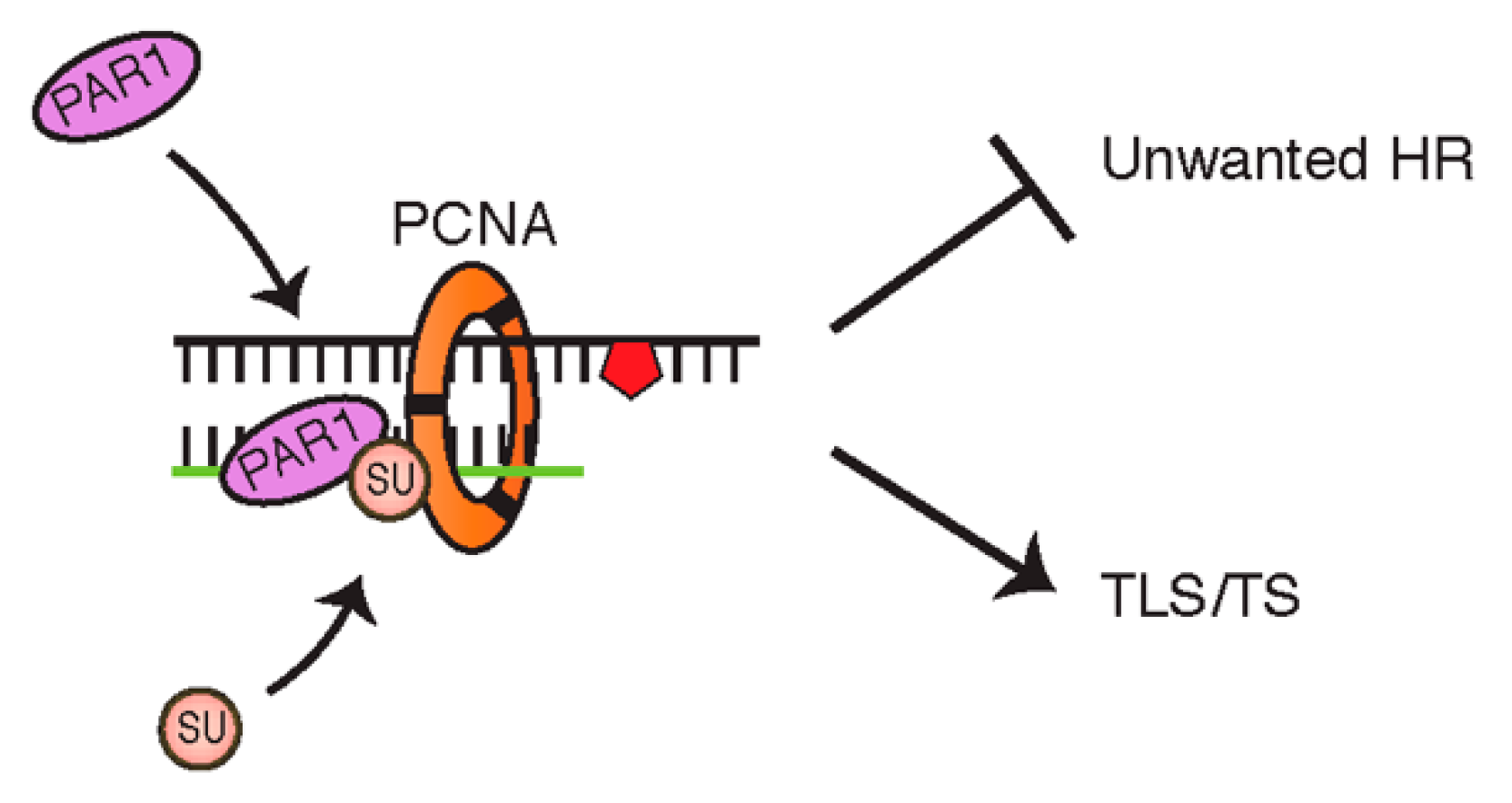
4.2. PCNA Can Also Be Modified by SUMOylation
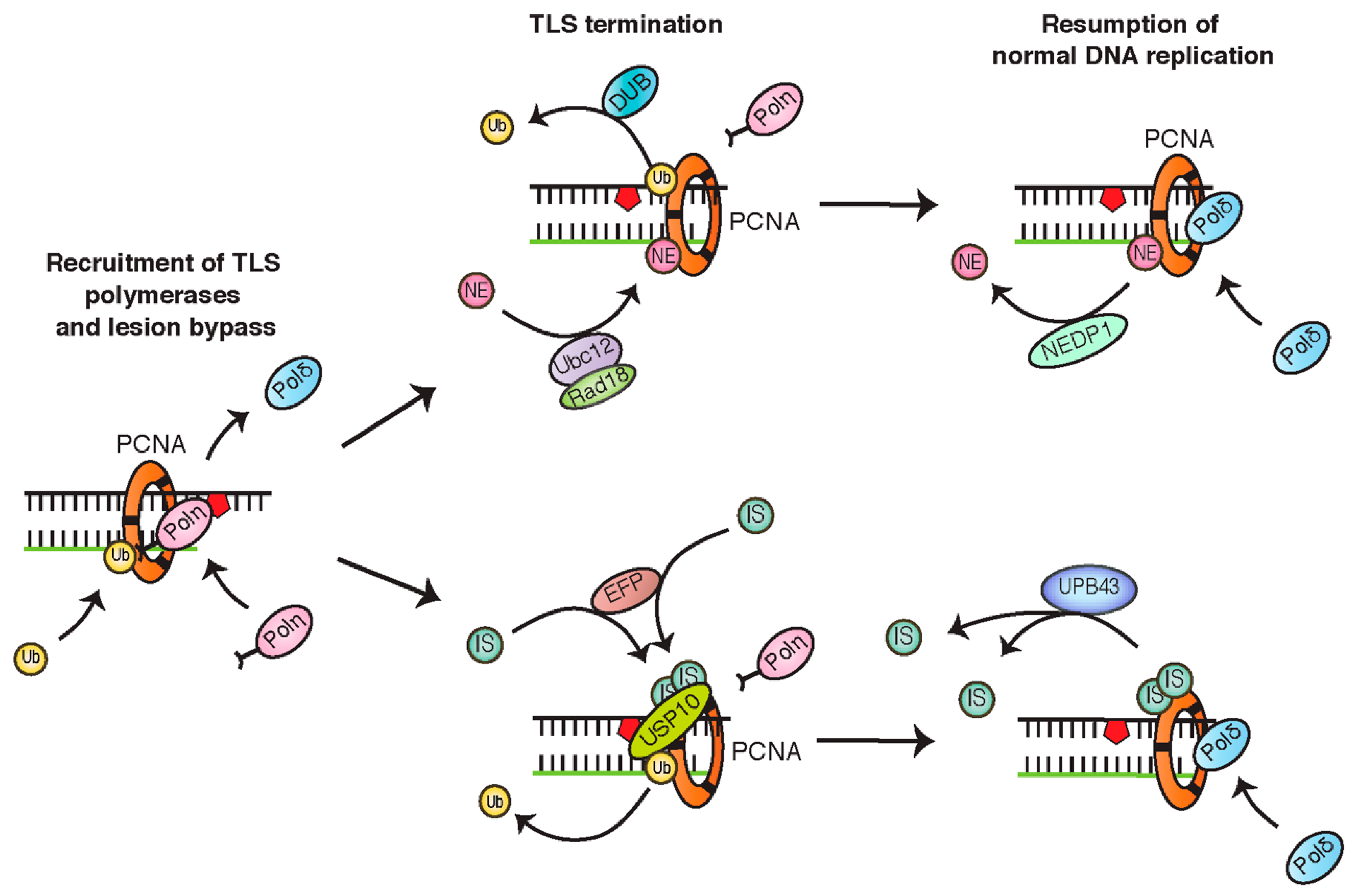
4.3. PCNA Monoubiquitination Is Negatively Regulated by NEDDylation and ISGylation
4.4. PCNA Monoubiquitination Is Opposed by Deubiquitinating Enzymes
4.5. TLS Polymerases Are also Regulated by Ubiquitin and Ubiquitin-Like Proteins
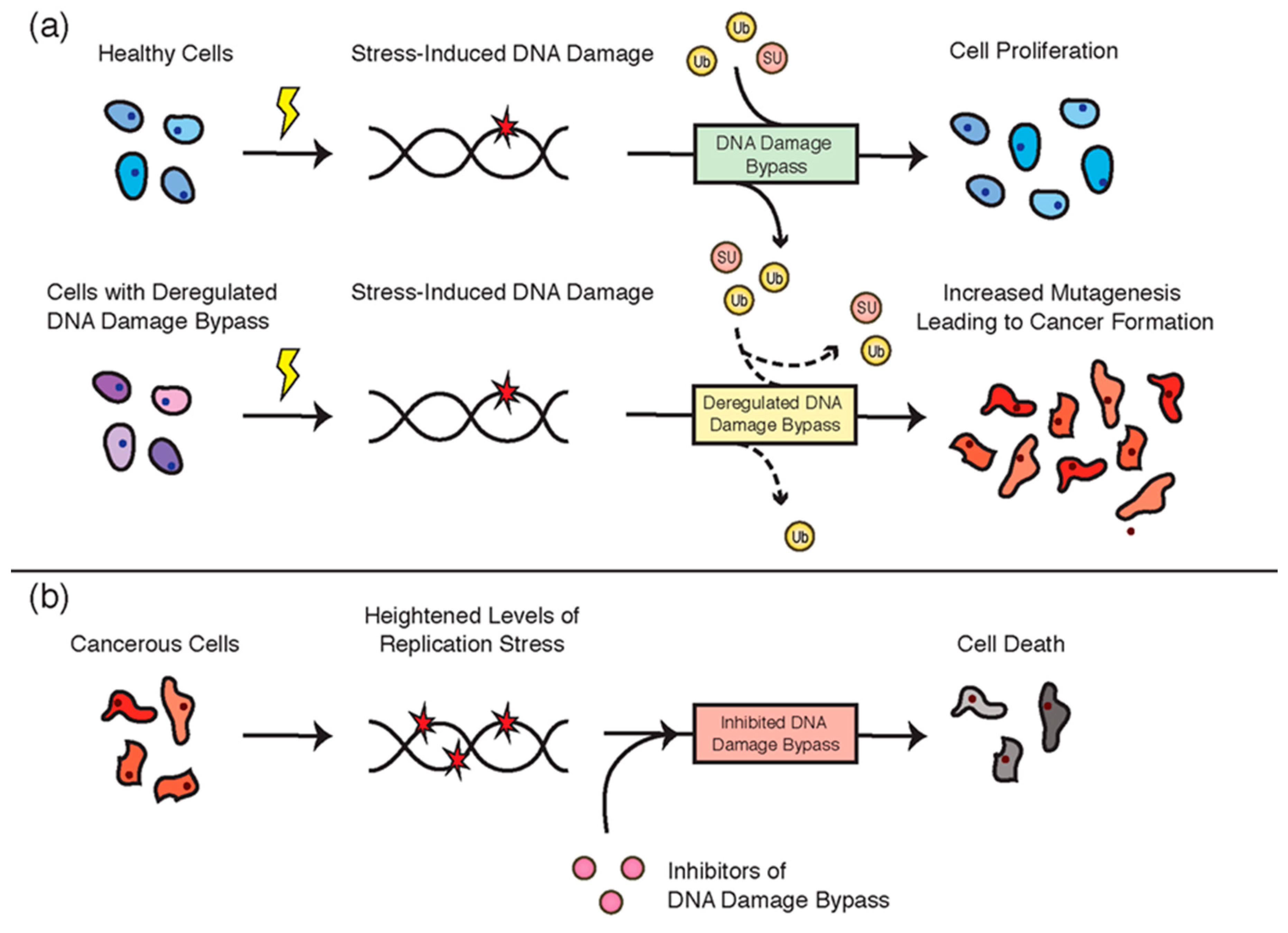
5. DNA Damage Bypass and Cancer
5.1. Deregulation of DNA Damage Bypass in Cancer
5.2. Targeting DNA Damage Bypass in Cancer Therapies
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Basu, A.K. DNA damage, mutagenesis and cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef] [PubMed]
- Marnett, L.J.; Plastaras, J.P. Endogenous DNA damage and mutation. Trends Genet. 2001, 17, 214–221. [Google Scholar] [CrossRef]
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 2015, 15, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015, 10, 425–448. [Google Scholar] [CrossRef]
- Shi, D.; Grossman, S.R. Ubiquitin becomes ubiquitous in cancer: Emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets. Cancer Biol. Ther. 2010, 10, 737–747. [Google Scholar] [CrossRef]
- Chang, H.-M.; Yeh, E.T.H. SUMO: From bench to bedside. Physiol. Rev. 2020, 100, 1599–1619. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.C.; Ding, J.L. Ubiquitination and SUMOylation in the chronic inflammatory tumor microenvironment. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2018, 1870, 165–175. [Google Scholar] [CrossRef]
- Vijay-Kumar, S.; Bugg, C.E.; Wilkinson, K.D.; Vierstra, R.D.; Hatfield, P.M.; Cook, W.J. Comparison of the three-dimensional structures of human, yeast, and oat ubiquitin. J. Biol. Chem. 1987, 262, 6396–6399. [Google Scholar]
- Zheng, N.; Shabek, N. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef]
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 enzymes: More than just middle men. Cell Res. 2016, 26, 423–440. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Hoeller, D.; Hecker, C.M.; Wagner, S.; Rogov, V.; Dötsch, V.; Dikic, I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol. Cell 2007, 26, 891–898. [Google Scholar] [CrossRef]
- Qiu, J.; Sheedlo, M.J.; Yu, K.; Tan, Y.; Nakayasu, E.S.; Das, C.; Liu, X.; Luo, Z.-Q. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 2016, 533, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Farshi, P.; Deshmukh, R.R.; Nwankwo, J.O.; Arkwright, R.T.; Cvek, B.; Liu, J.; Dou, Q.P. Deubiquitinases (DUBs) and DUB inhibitors: A patent review. Expert Opin. Ther. Pat. 2015, 25, 1191–1208. [Google Scholar] [CrossRef]
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Dwane, L.; Gallagher, W.M.; Chonghaile, T.N.; O’Connor, D.P. The emerging role of non-traditional ubiquitination in oncogenic pathways. J. Biol. Chem. 2017, 292, 3543–3551. [Google Scholar] [CrossRef]
- Li, W.; Ye, Y. Polyubiquitin chains: Functions, structures, and mechanisms. Cell. Mol. Life Sci. 2008, 65, 2397–2406. [Google Scholar] [CrossRef]
- Rao, H.; Sastry, A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 2002, 277, 11691–11695. [Google Scholar] [CrossRef]
- Cipolla, L.; Maffia, A.; Bertoletti, F.; Sabbioneda, S. The regulation of DNA damage tolerance by ubiquitin and ubiquitin-like modifiers. Front. Genet. 2016, 7, 105. [Google Scholar] [CrossRef] [PubMed]
- Nick McElhinny, S.A.; Gordenin, D.A.; Stith, C.M.; Burgers, P.M.; Kunkel, T.A. Division of labor at the eukaryotic replication fork. Mol. Cell 2008, 30, 137–144. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, S.D.; Kunkel, T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008, 18, 148–161. [Google Scholar] [CrossRef]
- Beard, W.A.; Wilson, S.H. Structural insights into the origins of DNA polymerase fidelity. Structure 2003, 11, 489–496. [Google Scholar] [CrossRef]
- Saintigny, Y.; Delacôte, F.; Varès, G.; Petitot, F.; Lambert, S.; Averbeck, D.; Lopez, B.S. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001, 20, 3861–3870. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Woodgate, R. What a difference a decade makes: Insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 15591–15598. [Google Scholar] [CrossRef] [PubMed]
- Waters, L.S.; Minesinger, B.K.; Wiltrout, M.E.; D’Souza, S.; Woodruff, R.V.; Walker, G.C. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009, 73, 134–154. [Google Scholar] [CrossRef]
- Masutani, C.; Kusumoto, R.; Iwai, S.; Hanaoka, F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000, 19, 3100–3109. [Google Scholar] [CrossRef]
- Choi, J.H.; Besaratinia, A.; Lee, D.H.; Lee, C.S.; Pfeifer, G.P. The role of DNA polymerase ι in UV mutational spectra. Mutat. Res. 2006, 599, 58–65. [Google Scholar] [CrossRef]
- Choi, J.Y.; Angel, K.C.; Guengerich, F.P. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J. Biol. Chem. 2006, 281, 21062–21072. [Google Scholar] [CrossRef]
- Ogi, T.; Shinkai, Y.; Tanaka, K.; Ohmori, H. Polκ protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc. Natl. Acad. Sci. USA 2002, 99, 15548–15553. [Google Scholar] [CrossRef] [PubMed]
- Sale, J.E. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012708. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio, M.; Zwicky, K.; Follonier, C.; Foiani, M.; Lopes, M.; Branzei, D. Visualization of recombination-mediated damage bypass by template switching. Nat. Struct. Mol. Biol. 2014, 21, 884–892. [Google Scholar] [CrossRef] [PubMed]
- Guilliam, T.A.; Doherty, A.J. PrimPol-prime time to reprime. Genes 2017, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Hedglin, M.; Benkovic, S.J. Eukaryotic translesion DNA synthesis on the leading and lagging strands: Unique detours around the same obstacle. Chem. Rev. 2017, 117, 7857–7877. [Google Scholar] [CrossRef]
- Ripley, B.M.; Gildenberg, M.S.; Washington, M.T. Control of DNA damage bypass by ubiquitylation of PCNA. Genes 2020, 11, 138. [Google Scholar] [CrossRef]
- Krishna, T.S.; Kong, X.P.; Gary, S.; Burgers, P.M.; Kuriyan, J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 1994, 79, 1233–1243. [Google Scholar] [CrossRef]
- Acharya, N.; Klassen, R.; Johnson, R.E.; Prakash, L.; Prakash, S. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc. Natl. Acad. Sci. USA 2011. [Google Scholar] [CrossRef]
- Hishiki, A.; Hashimoto, H.; Hanafusa, T.; Kamei, K.; Ohashi, E.; Shimizu, T.; Ohmori, H.; Sato, M. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J. Biol. Chem. 2009, 284, 10552–10560. [Google Scholar] [CrossRef]
- Davies, A.A.; Huttner, D.; Daigaku, Y.; Chen, S.; Ulrich, H.D. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell 2008, 29, 625–636. [Google Scholar] [CrossRef]
- Hedglin, M.; Aitha, M.; Pedley, A.; Benkovic, S.J. Replication protein A dynamically regulates monoubiquitination of proliferating cell nuclear antigen. J. Biol. Chem. 2019, 294, 5157–5168. [Google Scholar] [CrossRef] [PubMed]
- Hendel, A.; Krijger, P.H.L.; Diamant, N.; Goren, Z.; Langerak, P.; Kim, J.; Reißner, T.; Lee, K.-Y.; Geacintov, N.E.; Carell, T.; et al. PCNA ubiquitination Is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011, 7, e1002262. [Google Scholar] [CrossRef]
- Kannouche, P.L.; Wing, J.; Lehmann, A.R. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 2004, 14, 491–500. [Google Scholar] [CrossRef]
- Watanabe, K.; Tateishi, S.; Kawasuji, M.; Tsurimoto, T.; Inoue, H.; Yamaizumi, M. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004, 23, 3886–3896. [Google Scholar] [CrossRef]
- Bienko, M.; Green, C.M.; Crosetto, N.; Rudolf, F.; Zapart, G.; Coull, B.; Kannouche, P.; Wider, G.; Peter, M.; Lehmann, A.R.; et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 2005, 310, 1821–1824. [Google Scholar] [CrossRef]
- Plosky, B.S.; Vidal, A.E.; Fernández de Henestrosa, A.R.; McLenigan, M.P.; McDonald, J.P.; Mead, S.; Woodgate, R. Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. EMBO J. 2006, 25, 2847–2855. [Google Scholar] [CrossRef]
- Guo, C.; Tang, T.-S.; Bienko, M.; Parker, J.L.; Bielen, A.B.; Sonoda, E.; Takeda, S.; Ulrich, H.D.; Dikic, I.; Friedberg, E.C. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell. Biol. 2006, 26, 8892–8900. [Google Scholar] [CrossRef]
- Zhao, L.; Washington, M.T. Translesion synthesis: Insights into the selection and switching of DNA polymerases. Genes 2017, 8, 24. [Google Scholar] [CrossRef]
- Branzei, D.; Seki, M.; Enomoto, T. Rad18/Rad5/Mms2-mediated polyubiquitination of PCNA is implicated in replication completion during replication stress. Genes Cells 2004, 9, 1031–1042. [Google Scholar] [CrossRef]
- Hedglin, M.; Benkovic, S.J. Regulation of Rad6/Rad18 activity during DNA damage tolerance. Annu. Rev. Biophys. 2015, 44, 207–228. [Google Scholar] [CrossRef]
- Hofmann, R.M.; Pickart, C.M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 1999, 96, 645–653. [Google Scholar] [CrossRef]
- Unk, I.; Hajdú, I.; Fátyol, K.; Szakál, B.; Blastyák, A.; Bermudez, V.; Hurwitz, J.; Prakash, L.; Prakash, S.; Haracska, L. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. USA 2006, 103, 18107–18112. [Google Scholar] [CrossRef] [PubMed]
- Motegi, A.; Liaw, H.J.; Lee, K.Y.; Roest, H.P.; Maas, A.; Wu, X.; Moinova, H.; Markowitz, S.D.; Ding, H.; Hoeijmakers, J.H.; et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. USA 2008, 105, 12411–12416. [Google Scholar] [CrossRef]
- Kang, H.J.; Park, H.; Yoo, E.J.; Lee, J.H.; Choi, S.Y.; Lee-Kwon, W.; Lee, K.Y.; Hur, J.H.; Seo, J.K.; Ra, J.S.; et al. TonEBP regulates PCNA polyubiquitination in response to DNA damage through interaction with SHPRH and USP1. iScience 2019, 19, 177–190. [Google Scholar] [CrossRef]
- Lin, J.-R.; Zeman, M.K.; Chen, J.-Y.; Yee, M.-C.; Cimprich, K.A. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol. Cell 2011, 42, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Nimonkar, A.V.; Hu, Y.; Hajdu, I.; Achar, Y.J.; Izhar, L.; Petit, S.A.; Adamson, B.; Yoon, J.C.; Kowalczykowski, S.C.; et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell 2012, 47, 396–409. [Google Scholar] [CrossRef]
- Gervai, J.Z.; Gálicza, J.; Szeltner, Z.; Zámborszky, J.; Szüts, D. A genetic study based on PCNA-ubiquitin fusions reveals no requirement for PCNA polyubiquitylation in DNA damage tolerance. DNA Repair 2017, 54, 46–54. [Google Scholar] [CrossRef]
- Mohiuddin, M.; Evans, T.J.; Rahman, M.M.; Keka, I.S.; Tsuda, M.; Sasanuma, H.; Takeda, S. SUMOylation of PCNA by PIAS1 and PIAS4 promotes template switch in the chicken and human B cell lines. Proc. Natl. Acad. Sci. USA 2018, 115, 12793–12798. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Yunus, A.A.; Lima, C.D. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 2009, 35, 669–682. [Google Scholar] [CrossRef]
- Papouli, E.; Chen, S.; Davies, A.A.; Huttner, D.; Krejci, L.; Sung, P.; Ulrich, H.D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 2005, 19, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Pfander, B.; Moldovan, G.L.; Sacher, M.; Hoege, C.; Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005, 436, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Arbel, M.; Bronstein, A.; Sau, S.; Liefshitz, B.; Kupiec, M. Access to PCNA by Srs2 and Elg1 controls the choice between alternative repair pathways in Saccharomyces cerevisiae. mBio 2020, 11, e00705-20. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Vanoli, F.; Foiani, M. SUMOylation regulates Rad18-mediated template switch. Nature 2008, 456, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Gali, H.; Juhasz, S.; Morocz, M.; Hajdu, I.; Fatyol, K.; Szukacsov, V.; Burkovics, P.; Haracska, L. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 2012, 40, 6049–6059. [Google Scholar] [CrossRef]
- Moldovan, G.-L.; Dejsuphong, D.; Petalcorin, M.I.R.; Hofmann, K.; Takeda, S.; Boulton, S.J.; D’Andrea, A.D. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 2012, 45, 75–86. [Google Scholar] [CrossRef]
- Barber, L.J.; Youds, J.L.; Ward, J.D.; McIlwraith, M.J.; O’Neil, N.J.; Petalcorin, M.I.; Martin, J.S.; Collis, S.J.; Cantor, S.B.; Auclair, M.; et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 2008, 135, 261–271. [Google Scholar] [CrossRef]
- Burkovics, P.; Dome, L.; Juhasz, S.; Altmannova, V.; Sebesta, M.; Pacesa, M.; Fugger, K.; Sorensen, C.S.; Lee, M.Y.; Haracska, L.; et al. The PCNA-associated protein PARI negatively regulates homologous recombination via the inhibition of DNA repair synthesis. Nucleic Acids Res. 2016, 44, 3176–3189. [Google Scholar] [CrossRef]
- Chiolo, I.; Saponaro, M.; Baryshnikova, A.; Kim, J.H.; Seo, Y.S.; Liberi, G. The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles. Mol. Cell. Biol. 2007, 27, 7439–7450. [Google Scholar] [CrossRef]
- Fugger, K.; Mistrik, M.; Danielsen, J.R.; Dinant, C.; Falck, J.; Bartek, J.; Lukas, J.; Mailand, N. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 2009, 186, 655–663. [Google Scholar] [CrossRef]
- Guan, J.; Yu, S.; Zheng, X. NEDDylation antagonizes ubiquitination of proliferating cell nuclear antigen and regulates the recruitment of polymerase η in response to oxidative DNA damage. Protein Cell 2018, 9, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Yang, S.W.; Yu, K.R.; Ka, S.H.; Lee, S.W.; Seol, J.H.; Jeon, Y.J.; Chung, C.H. Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol. Cell 2014, 54, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.T.; Nijman, S.M.; Mirchandani, K.D.; Galardy, P.J.; Cohn, M.A.; Haas, W.; Gygi, S.P.; Ploegh, H.L.; Bernards, R.; D’Andrea, A.D. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006, 8, 339–347. [Google Scholar] [CrossRef]
- Kategaya, L.; Di Lello, P.; Rougé, L.; Pastor, R.; Clark, K.R.; Drummond, J.; Kleinheinz, T.; Lin, E.; Upton, J.P.; Prakash, S.; et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 2017, 550, 534–538. [Google Scholar] [CrossRef]
- Qing, P.; Han, L.; Bin, L.; Yan, L.; Ping, W.X. USP7 regulates the stability and function of HLTF through deubiquitination. J. Cell. Biochem. 2011, 112, 3856–3862. [Google Scholar] [CrossRef]
- Zlatanou, A.; Sabbioneda, S.; Miller, E.S.; Greenwalt, A.; Aggathanggelou, A.; Maurice, M.M.; Lehmann, A.R.; Stankovic, T.; Reverdy, C.; Colland, F.; et al. USP7 is essential for maintaining Rad18 stability and DNA damage tolerance. Oncogene 2016, 35, 965–976. [Google Scholar] [CrossRef]
- Bienko, M.; Green, C.M.; Sabbioneda, S.; Crosetto, N.; Matic, I.; Hibbert, R.G.; Begovic, T.; Niimi, A.; Mann, M.; Lehmann, A.R.; et al. Regulation of translesion synthesis DNA polymerase η by monoubiquitination. Mol. Cell 2010, 37, 396–407. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, J.; McLenigan, M.P.; Frank, E.G.; Dai, X.; Yang, W.; Wang, Y.; Woodgate, R. Posttranslational regulation of human DNA polymerase ι. J. Biol. Chem. 2015, 290, 27332–27344. [Google Scholar] [CrossRef]
- Jung, Y.-S.; Hakem, A.; Hakem, R.; Chen, X. Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase eta to suppress translesion DNA synthesis. Mol. Cell. Biol. 2011, 31, 3997–4006. [Google Scholar] [CrossRef]
- Jung, Y.-S.; Liu, G.; Chen, X. Pirh2 E3 ubiquitin ligase targets DNA polymerase eta for 20S proteasomal degradation. Mol. Cell. Biol. 2010, 30, 1041–1048. [Google Scholar] [CrossRef]
- McIntyre, J.; Vidal, A.E.; McLenigan, M.P.; Bomar, M.G.; Curti, E.; McDonald, J.P.; Plosky, B.S.; Ohashi, E.; Woodgate, R. Ubiquitin mediates the physical and functional interaction between human DNA polymerases η and ι. Nucleic Acids Res. 2013, 41, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Despras, E.; Sittewelle, M.; Pouvelle, C.; Delrieu, N.; Cordonnier, A.M.; Kannouche, P.L. Rad18-dependent SUMOylation of human specialized DNA polymerase eta is required to prevent under-replicated DNA. Nat. Commun. 2016, 7, 13326. [Google Scholar] [CrossRef] [PubMed]
- Guérillon, C.; Smedegaard, S.; Hendriks, I.A.; Nielsen, M.L.; Mailand, N. Multisite SUMOylation restrains DNA polymerase η interactions with DNA damage sites. J. Biol. Chem. 2020, 295, 8350–8362. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.K.; Eoff, R.L. Translesion DNA synthesis in cancer: Molecular mechanisms and therapeutic opportunities. Chem. Res. Toxicol. 2017, 30, 1942–1955. [Google Scholar] [CrossRef] [PubMed]
- Luedeke, M.; Linnert, C.M.; Hofer, M.D.; Surowy, H.M.; Rinckleb, A.E.; Hoegel, J.; Kuefer, R.; Rubin, M.A.; Vogel, W.; Maier, C. Predisposition for TMPRSS2-ERG fusion in prostate cancer by variants in DNA repair genes. Cancer Epidemiol. Biomark. Prev. 2009, 18, 3030. [Google Scholar] [CrossRef] [PubMed]
- Sakiyama, T.; Kohno, T.; Mimaki, S.; Ohta, T.; Yanagitani, N.; Sobue, T.; Kunitoh, H.; Saito, R.; Shimizu, K.; Hirama, C.; et al. Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int. J. Cancer 2005, 114, 730–737. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, Y.; Luo, Q.; Li, L.; Jia, L. Neddylation: A novel modulator of the tumor microenvironment. Mol. Cancer 2019, 18, 77. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, W.; Sun, Y.; Jia, L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell. Signal. 2018, 44, 92–102. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Guerra, S.; Sánchez-Madrid, F. ISGylation—A key to lock the cell gates for preventing the spread of threats. J. Cell Sci. 2017, 130, 2961–2969. [Google Scholar] [CrossRef]
- Wu, B.; Wang, H.; Zhang, L.; Sun, C.; Li, H.; Jiang, C.; Liu, X. High expression of RAD18 in glioma induces radiotherapy resistance via down-regulating P53 expression. Biomed. Pharmacother. 2019, 112, 108555. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, Y.; Zlatanou, A.; Tateishi, S.; Yurchenko, V.; Rogozin, I.B.; Vaziri, C. Diverse roles of RAD18 and Y-family DNA polymerases in tumorigenesis. Cell Cycle 2018, 17, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Tateishi, S.; Vaziri, C. Pathological trans-lesion synthesis in cancer. Cell Cycle 2016, 15, 3005–3006. [Google Scholar] [CrossRef] [PubMed]
- Pilzecker, B.; Buoninfante, O.A.; Jacobs, H. DNA damage tolerance in stem cells, ageing, mutagenesis, disease and cancer therapy. Nucleic Acids Res. 2019, 47, 7163–7181. [Google Scholar] [CrossRef] [PubMed]
- Leung, W.; Baxley, R.M.; Moldovan, G.L.; Bielinsky, A.K. Mechanisms of DNA damage tolerance: Post-translational regulation of PCNA. Genes 2018, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Debauve, G.; Capouillez, A.; Belayew, A.; Saussez, S. The helicase-like transcription factor and its implication in cancer progression. Cell. Mol. Life Sci. 2008, 65, 591–604. [Google Scholar] [CrossRef]
- Inui, H.; Oh, K.S.; Nadem, C.; Ueda, T.; Khan, S.G.; Metin, A.; Gozukara, E.; Emmert, S.; Slor, H.; Busch, D.B.; et al. Xeroderma pigmentosum-variant patients from America, Europe, and Asia. J. Investig. Dermatol. 2008, 128, 2055–2068. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Han, C.; Zhao, R.; Cui, T.; Dai, Y.; Mao, C.; Zhao, W.; Zhang, X.; Yu, J.; Wang, Q.-E. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc. Natl. Acad. Sci. USA 2015, 112, 4411–4416. [Google Scholar] [CrossRef]
- Zhou, W.; Chen, Y.W.; Liu, X.; Chu, P.; Loria, S.; Wang, Y.; Yen, Y.; Chou, K.M. Expression of DNA translesion synthesis polymerase η in head and neck squamous cell cancer predicts resistance to gemcitabine and cisplatin-based chemotherapy. PLoS ONE 2013, 8, e83978. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, W.; Ren, C.; Kong, X.; Yan, W.; Chen, X. A PolH transcript with a short 3′UTR enhances PolH expression and mediates cisplatin resistance. Cancer Res. 2019, 79, 3714–3724. [Google Scholar] [CrossRef]
- Yuan, F.; Xu, Z.; Yang, M.; Wei, Q.; Zhang, Y.; Yu, J.; Zhi, Y.; Liu, Y.; Chen, Z.; Yang, J. Overexpressed DNA polymerase iota regulated by JNK/c-Jun contributes to hypermutagenesis in bladder cancer. PLoS ONE 2013, 8, e69317. [Google Scholar] [CrossRef]
- Yang, J.; Chen, Z.; Liu, Y.; Hickey, R.J.; Malkas, L.H. Altered DNA polymerase ι expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res. 2004, 64, 5597–5607. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.A.; Grishina, E.E.; Tarantul, V.Z.; Gening, L.V. Effect of human cell malignancy on activity of DNA polymerase ι. Biochemistry 2010, 75, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zou, S.; Zhang, S.; Liu, B.; Meng, X.; Li, X.; Yu, J.; Wu, J.; Zhou, J. Elevated DNA polymerase iota (Poli) is involved in the acquisition of aggressive phenotypes of human esophageal squamous cell cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 3591–3601. [Google Scholar] [PubMed]
- Wang, H.; Wu, W.; Wang, H.W.; Wang, S.; Chen, Y.; Zhang, X.; Yang, J.; Zhao, S.; Ding, H.F.; Lu, D. Analysis of specialized DNA polymerases expression in human gliomas: Association with prognostic significance. Neuro-Oncology 2010, 12, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Mukhopadhyay, S.; Anbalagan, M.; Makridakis, N. Somatic mutations in catalytic core of POLK reported in prostate cancer alter translesion DNA synthesis. Hum. Mutat. 2015, 36, 873–880. [Google Scholar] [CrossRef]
- Dai, Z.-J.; Liu, X.-H.; Ma, Y.-F.; Kang, H.-F.; Jin, T.-B.; Dai, Z.-M.; Guan, H.-T.; Wang, M.; Liu, K.; Dai, C.; et al. Association between single nucleotide polymorphisms in DNA polymerase kappa gene and breast cancer risk in Chinese han population: A STROBE-Compliant Observational Study. Medicine (Baltimore) 2016, 95, e2466. [Google Scholar] [CrossRef]
- Wang, J.; Kawamura, K.; Tada, Y.; Ohmori, H.; Kimura, H.; Sakiyama, S.; Tagawa, M. DNA polymerase κ, implicated in spontaneous and DNA damage-induced mutagenesis, is overexpressed in lung cancer. Cancer Res. 2001, 61, 5366–5369. [Google Scholar]
- Huang, Y.; Huang, H.; Liang, Y.; Qiu, S.; Li, W.; Zheng, Y.; Han, Z.-D.; Yuan, R. Elevated expression of REV 1 is a predictor of unfavorable prognosis in patients with prostate cancer. Int. J. Clin. Exp. Med. 2018, 11, 8412–8420. [Google Scholar]
- He, X.; Ye, F.; Zhang, J.; Cheng, Q.; Shen, J.; Chen, H. REV1 genetic variants associated with the risk of cervical carcinoma. Eur. J. Epidemiol. 2008, 23, 403–409. [Google Scholar] [CrossRef]
- Wang, X.; Hickey, R.J.; Malkas, L.H.; Koch, M.O.; Li, L.; Zhang, S.; Sandusky, G.E.; Grignon, D.J.; Eble, J.N.; Cheng, L. Elevated expression of cancer-associated proliferating cell nuclear antigen in high-grade prostatic intraepithelial neoplasia and prostate cancer. Prostate 2011, 71, 748–754. [Google Scholar] [CrossRef]
- Zheng, C.; Yang, R. RCD24, B7-H4 and PCNA expression and clinical significance in ovarian cancer. Off. J. Balk. Union Oncol. 2019, 24, 715–719. [Google Scholar]
- Wong, R.P.; Aguissa-Touré, A.H.; Wani, A.A.; Khosravi, S.; Martinka, M.; Martinka, M.; Li, G. Elevated expression of Rad18 regulates melanoma cell proliferation. Pigment Cell Melanoma Res. 2012, 25, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; He, C.; Gao, A.; Yan, X.; Xia, X.; Zhou, J.; Wu, J. RAD18 promotes colorectal cancer metastasis by activating the epithelial-mesenchymal transition pathway. Oncol. Rep. 2020, 44, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Moinova, H.R.; Chen, W.-D.; Shen, L.; Smiraglia, D.; Olechnowicz, J.; Ravi, L.; Kasturi, L.; Myeroff, L.; Plass, C.; Parsons, R.; et al. HLTF gene silencing in human colon cancer. Proc. Natl. Acad. Sci. USA 2002, 99, 4562–4567. [Google Scholar] [CrossRef]
- Hamai, Y.; Oue, N.; Mitani, Y.; Nakayama, H.; Ito, R.; Matsusaki, K.; Yoshida, K.; Toge, T.; Yasui, W. DNA hypermethylation and histone hypoacetylation of the HLTF gene are associated with reduced expression in gastric carcinoma. Cancer Sci. 2003, 94, 692–698. [Google Scholar] [CrossRef]
- Bai, A.H.; Tong, J.H.; To, K.F.; Chan, M.W.; Man, E.P.; Lo, K.W.; Lee, J.F.; Sung, J.J.; Leung, W.K. Promoter hypermethylation of tumor-related genes in the progression of colorectal neoplasia. Int. J. Cancer 2004, 112, 846–853. [Google Scholar] [CrossRef]
- Fukuoka, T.; Hibi, K.; Nakao, A. Aberrant methylation is frequently observed in advanced esophageal squamous cell carcinoma. Anticancer Res. 2006, 26, 3333–3335. [Google Scholar]
- Bell, D.W.; Sikdar, N.; Lee, K.-Y.; Price, J.C.; Chatterjee, R.; Park, H.-D.; Fox, J.; Ishiai, M.; Rudd, M.L.; Pollock, L.M.; et al. Predisposition to cancer caused by genetic and functional defects of mammalian Atad5. PLoS Genet. 2011, 7, e1002245. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; Ramus, S.J.; Tyrer, J.; Lee, A.; Shen, H.C.; Beesley, J.; Lawrenson, K.; McGuffog, L.; Healey, S.; Lee, J.M.; et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet. 2015, 47, 164–171. [Google Scholar] [CrossRef]
- Lessel, D.; Vaz, B.; Halder, S.; Lockhart, P.J.; Marinovic-Terzic, I.; Lopez-Mosqueda, J.; Philipp, M.; Sim, J.C.H.; Smith, K.R.; Oehler, J.; et al. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat. Genet. 2014, 46, 1239–1244. [Google Scholar] [CrossRef]
- Williams, S.A.; Maecker, H.L.; French, D.M.; Liu, J.; Gregg, A.; Silverstein, L.B.; Cao, T.C.; Carano, R.A.; Dixit, V.M. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 2011, 146, 918–930. [Google Scholar] [CrossRef] [PubMed]
- García-Santisteban, I.; Peters, G.J.; Giovannetti, E.; Rodríguez, J.A. USP1 deubiquitinase: Cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Mol. Cancer 2013, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Q.; Wang, Y.; Zhuang, H.; Chen, B. Clinical significance of ubiquitin specific protease 7 (USP7) in predicting prognosis of hepatocellular carcinoma and its functional mechanisms. Med Sci. Monit. 2018, 24, 1742–1750. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.-Y.; Lin, Z.-W.; Lu, C.-L.; Gu, J.; Yuan, Y.-F.; Xu, F.-K.; Liu, R.-H.; Ge, D.; Ding, J.-Y. USP7 overexpression predicts a poor prognosis in lung squamous cell carcinoma and large cell carcinoma. Tumor Biol. 2015, 36, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, H.; Tian, L.; Li, H. Expression of USP7 and MARCH7 is correlated with poor prognosis in epithelial ovarian cancer. Tohoku J. Exp. Med. 2016, 239, 165–175. [Google Scholar] [CrossRef]
- He, Y.; Wang, S.; Tong, J.; Jiang, S.; Yang, Y.; Zhang, Z.; Xu, Y.; Zeng, Y.; Cao, B.; Moran, M.F.; et al. The deubiquitinase USP7 stabilizes Maf proteins to promote myeloma cell survival. J. Biol. Chem. 2020, 295, 2084–2096. [Google Scholar] [CrossRef]
- Takayama, K.-I.; Suzuki, T.; Fujimura, T.; Takahashi, S.; Inoue, S. Association of USP10 with G3BP2 inhibits p53 signaling and contributes to poor outcome in prostate cancer. Mol. Cancer Res. 2018, 16, 846–856. [Google Scholar] [CrossRef]
- Zhu, H.; Yan, F.; Yuan, T.; Qian, M.; Zhou, T.; Dai, X.; Cao, J.; Ying, M.; Dong, X.; He, Q.; et al. USP10 promotes proliferation of hepatocellular carcinoma by deubiquitinating and stabilizing YAP/TAZ. Cancer Res. 2020, 80, 2204–2216. [Google Scholar] [CrossRef]
- Bailly, A.P.; Perrin, A.; Serrano-Macia, M.; Maghames, C.; Leidecker, O.; Trauchessec, H.; Martinez-Chantar, M.L.; Gartner, A.; Xirodimas, D.P. The balance between mono- and NEDD8-chains controlled by NEDP1 upon DNA damage is a regulatory module of the HSP70 ATPase activity. Cell Rep. 2019, 29, 212–224.e8. [Google Scholar] [CrossRef]
- Andersen, J.B.; Aaboe, M.; Borden, E.C.; Goloubeva, O.G.; Hassel, B.A.; Orntoft, T.F. Stage-associated overexpression of the ubiquitin-like protein, ISG15, in bladder cancer. Br. J. Cancer 2006, 94, 1465–1471. [Google Scholar] [CrossRef]
- Desai, S.D.; Haas, A.L.; Wood, L.M.; Tsai, Y.C.; Pestka, S.; Rubin, E.H.; Saleem, A.; Nur, E.K.A.; Liu, L.F. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 2006, 66, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, A.; Hogrefe, C.; Erb, S.; Bobach, C.; Fuessel, S.; Wessjohann, L.; Seliger, B. Expression, regulation and function of the ISGylation system in prostate cancer. Oncogene 2009, 28, 2606–2620. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, J.; Zhang, H.; Zhu, M.; Chen, F.; Hu, Y.; Liu, H.; Zhu, H. Interferon-stimulated gene 15 (ISG15) is a trigger for tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget 2014, 5, 8429–8441. [Google Scholar] [CrossRef]
- Barbier-Torres, L.; Delgado, T.C.; García-Rodríguez, J.L.; Zubiete-Franco, I.; Fernández-Ramos, D.; Buqué, X.; Cano, A.; Gutiérrez-de Juan, V.; Fernández-Domínguez, I.; Lopitz-Otsoa, F.; et al. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget 2015, 6, 2509–2523. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Yang, J.-P.; Cao, Y.; Peng, L.-X.; Zheng, L.-S.; Sun, R.; Meng, D.-F.; Wang, M.-Y.; Mei, Y.; Qiang, Y.-Y.; et al. Promoting tumorigenesis in nasopharyngeal carcinoma, NEDD8 serves as a potential theranostic target. Cell Death Dis. 2017, 8, e2834. [Google Scholar] [CrossRef]
- Albertella, M.R.; Green, C.M.; Lehmann, A.R.; O’Connor, M.J. A role for polymerase η in the cellular tolerance to cisplatin-induced damage. Cancer Res. 2005, 65, 9799–9806. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Kikuchi, S.; Hishiki, A.; Shao, Y.; Heath, R.; Evison, B.J.; Actis, M.; Canman, C.E.; Hashimoto, H.; Fujii, N. A small molecule inhibitor of monoubiquitinated Proliferating Cell Nuclear Antigen (PCNA) inhibits repair of interstrand DNA cross-link, enhances DNA double strand break, and sensitizes cancer cells to cisplatin. J. Biol. Chem. 2014, 289, 7109–7120. [Google Scholar] [CrossRef]
- Tan, Z.; Wortman, M.; Dillehay, K.L.; Seibel, W.L.; Evelyn, C.R.; Smith, S.J.; Malkas, L.H.; Zheng, Y.; Lu, S.; Dong, Z. Small-molecule targeting of proliferating cell nuclear antigen chromatin association inhibits tumor cell growth. Mol. Pharmacol. 2012, 81, 811–819. [Google Scholar] [CrossRef]
- Dillehay, K.L.; Lu, S.; Dong, Z. Antitumor effects of a novel small molecule targeting PCNA chromatin association in prostate cancer. Mol. Cancer Ther. 2014, 13, 2817–2826. [Google Scholar] [CrossRef]
- Cui, G.; Botuyan, M.V.; Mer, G. Structural basis for the interaction of mutasome assembly factor REV1 with ubiquitin. J. Mol. Biol. 2018, 430, 2042–2050. [Google Scholar] [CrossRef]
- Vanarotti, M.; Evison, B.J.; Actis, M.L.; Inoue, A.; McDonald, E.T.; Shao, Y.; Heath, R.J.; Fujii, N. Small-molecules that bind to the ubiquitin-binding motif of REV1 inhibit REV1 interaction with K164-monoubiquitinated PCNA and suppress DNA damage tolerance. Bioorg. Med. Chem. 2018, 26, 2345–2353. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Fischhaber, P.L.; Luk-Paszyc, M.J.; Masuda, Y.; Zhou, J.; Kamiya, K.; Kisker, C.; Friedberg, E.C. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003, 22, 6621–6630. [Google Scholar] [CrossRef] [PubMed]
- Sail, V.; Rizzo, A.A.; Chatterjee, N.; Dash, R.C.; Ozen, Z.; Walker, G.C.; Korzhnev, D.M.; Hadden, M.K. Identification of small molecule translesion synthesis inhibitors that target the Rev1-CT/RIR protein-protein interaction. ACS Chem. Biol. 2017, 12, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Wojtaszek, J.L.; Chatterjee, N.; Najeeb, J.; Ramos, A.; Lee, M.; Bian, K.; Xue, J.Y.; Fenton, B.A.; Park, H.; Li, D.; et al. A small molecule targeting mutagenic translesion synthesis improves chemotherapy. Cell 2019, 178, 152–159.e11. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.L.; Dong, Y.; Deng, Y.Z.; Wang, W.J.; Li, W.D. Tumor suppressor miR-145 reverses drug resistance by directly targeting DNA damage-related gene RAD18 in colorectal cancer. Tumor Biol. 2015, 36, 5011–5019. [Google Scholar] [CrossRef]



| Protein | Role in DNA Damage Response | Associated Cancers |
|---|---|---|
| Pol η | Y family TLS polymerase | Mutations/defects implicated in XP-V syndrome (high cancer susceptibility) [96] Upregulated in: bladder, non-small cell lung cancer, head and neck squamous cell carcinoma, ovarian cancer stem cells [97,98,99] |
| Pol ι | Y family TLS polymerase | Upregulated in: bladder cancer, breast cancer, basal cell carcinoma, esophageal squamous cell carcinoma, glioma (correlated with lymph node metastasis) [100,101,102,103,104] SNPs associated with prostate cancer, adenocarcinoma, squamous cell carcinoma [85,86] |
| Pol κ | Y family TLS polymerase | Mutations/defects implicated in prostate cancer, breast cancer [105,106] Upregulated in: glioma, non-small cell lung cancer [104,107] |
| Rev1 | Y family TLS polymerase | Upregulated in: prostate cancer [108] SNPs associated with cervical squamous cell carcinoma [109] |
| PCNA | DNA sliding clamp | Upregulated in: prostate cancer, ovarian cancer (especially with lymph node metastasis) [110,111] |
| Rad18 | E3 ubiquitin ligase | Upregulated in: colorectal cancer, primary and metastatic melanoma, glioma [90,112,113] |
| HLTF | E3 ubiquitin ligase | Downregulated in: colorectal, colon, stomach cancer, esophageal squamous cell carcinoma [114,115,116,117] |
| ATAD5 | Unloading of PCNA from DNA strand | Mutations/defects associated with endometrial carcinoma [118] Upregulated in: epithelial ovarian carcinoma [119] |
| SPRTN | Stabilizing PCNA, resolving fork stalling DNA-protein crosslinks | Mutations/defects implicated in hepatocellular carcinoma [120] |
| USP1 | Deubiquitinating enzyme | Upregulated in: cervical, stomach cancer, melanoma, sarcoma, osteosarcoma [121,122] |
| USP7 | Deubiquitinating enzyme | Upregulated in: hepatocellular carcinoma, non-small cell lung cancer, epithelial ovarian cancer, myeloma [123,124,125,126] |
| USP10 | Deubiquitinating enzyme | Upregulated in: prostate cancer, hepatocellular carcinoma [127,128] |
| NEDP1 | De-NEDDylating enzyme | Downregulated in: hepatocellular carcinoma [129] |
| ISG15 | Ubiquitin-like protein | Upregulated in: bladder, breast, endometrium, prostate cancer, hepatocellular carcinoma [130,131,132,133] |
| NEDD8 | Ubiquitin-like protein | Upregulated in: hepatocellular carcinoma, nasopharyngeal carcinoma (correlated with lymph node metastasis) [134,135] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilkinson, N.A.; Mnuskin, K.S.; Ashton, N.W.; Woodgate, R. Ubiquitin and Ubiquitin-Like Proteins Are Essential Regulators of DNA Damage Bypass. Cancers 2020, 12, 2848. https://doi.org/10.3390/cancers12102848
Wilkinson NA, Mnuskin KS, Ashton NW, Woodgate R. Ubiquitin and Ubiquitin-Like Proteins Are Essential Regulators of DNA Damage Bypass. Cancers. 2020; 12(10):2848. https://doi.org/10.3390/cancers12102848
Chicago/Turabian StyleWilkinson, Nicole A., Katherine S. Mnuskin, Nicholas W. Ashton, and Roger Woodgate. 2020. "Ubiquitin and Ubiquitin-Like Proteins Are Essential Regulators of DNA Damage Bypass" Cancers 12, no. 10: 2848. https://doi.org/10.3390/cancers12102848
APA StyleWilkinson, N. A., Mnuskin, K. S., Ashton, N. W., & Woodgate, R. (2020). Ubiquitin and Ubiquitin-Like Proteins Are Essential Regulators of DNA Damage Bypass. Cancers, 12(10), 2848. https://doi.org/10.3390/cancers12102848





