HSPA2 Chaperone Contributes to the Maintenance of Epithelial Phenotype of Human Bronchial Epithelial Cells but Has Non-Essential Role in Supporting Malignant Features of Non-Small Cell Lung Carcinoma, MCF7, and HeLa Cancer Cells
Simple Summary
Abstract
1. Introduction
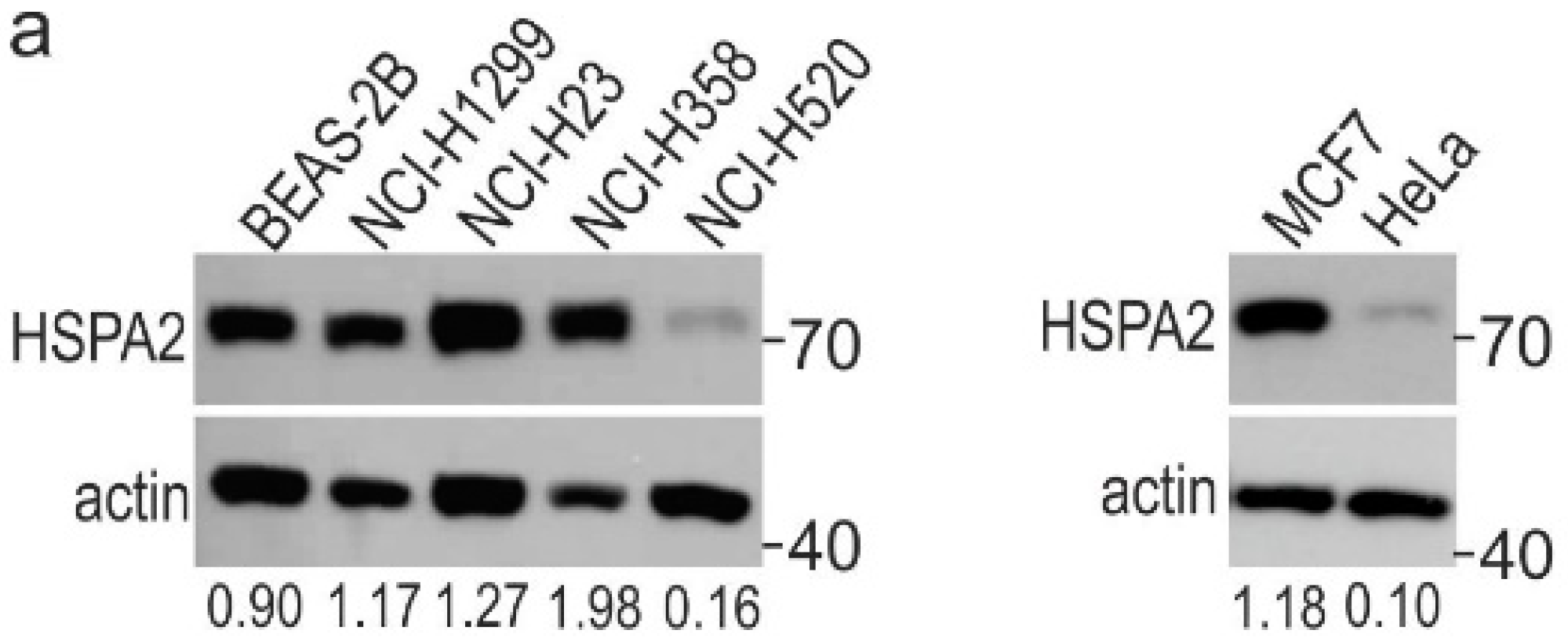
2. Results
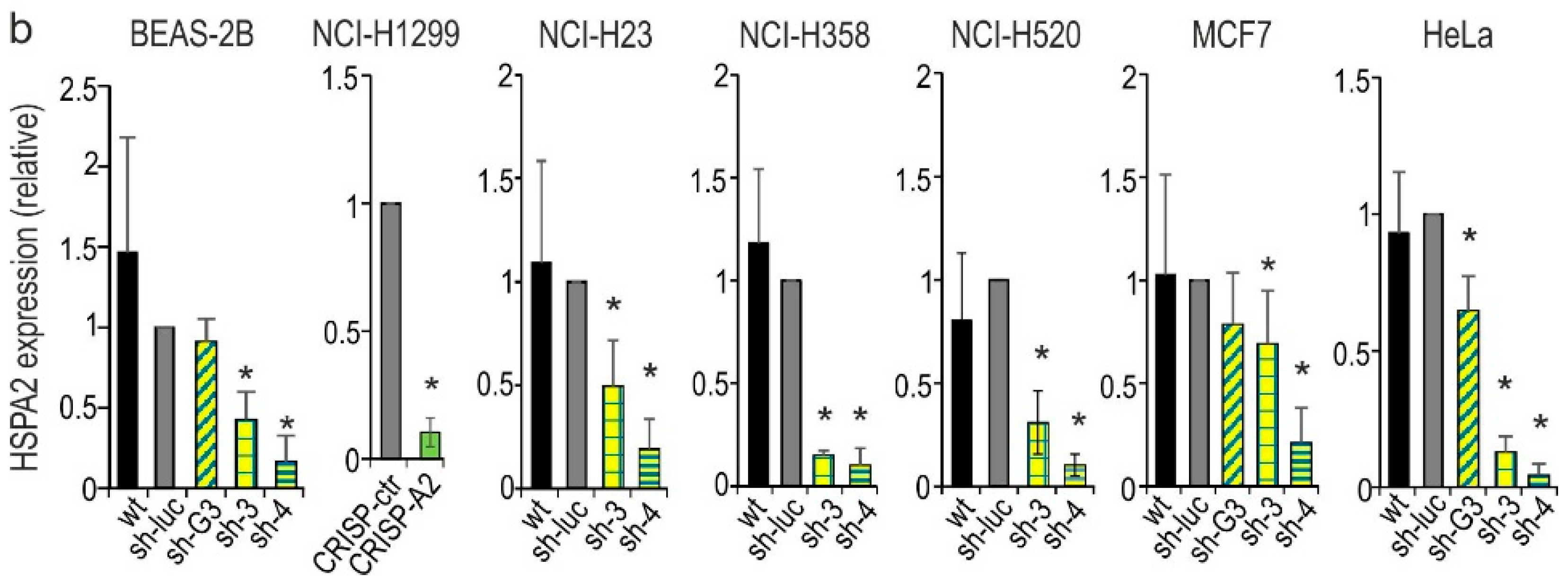
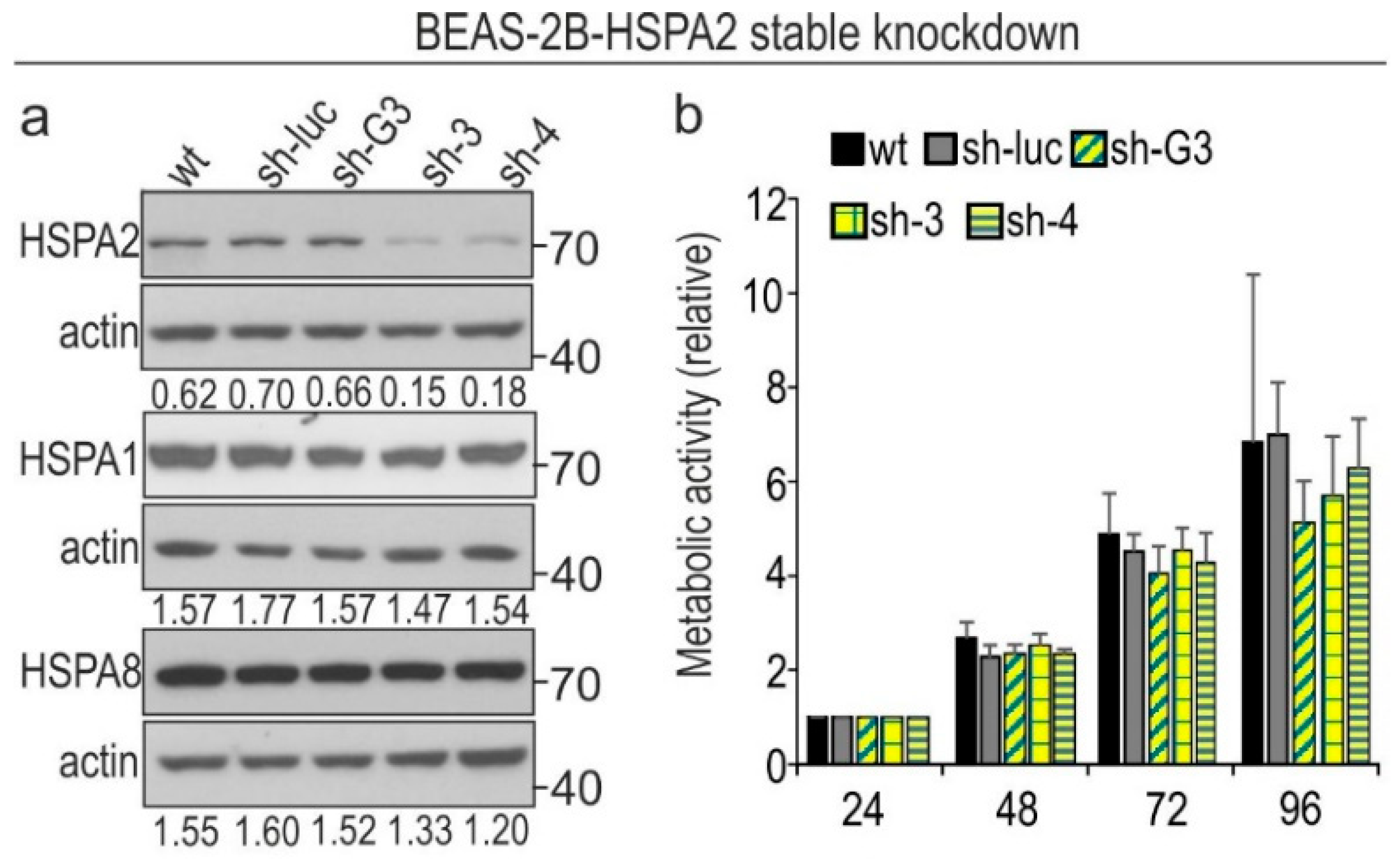
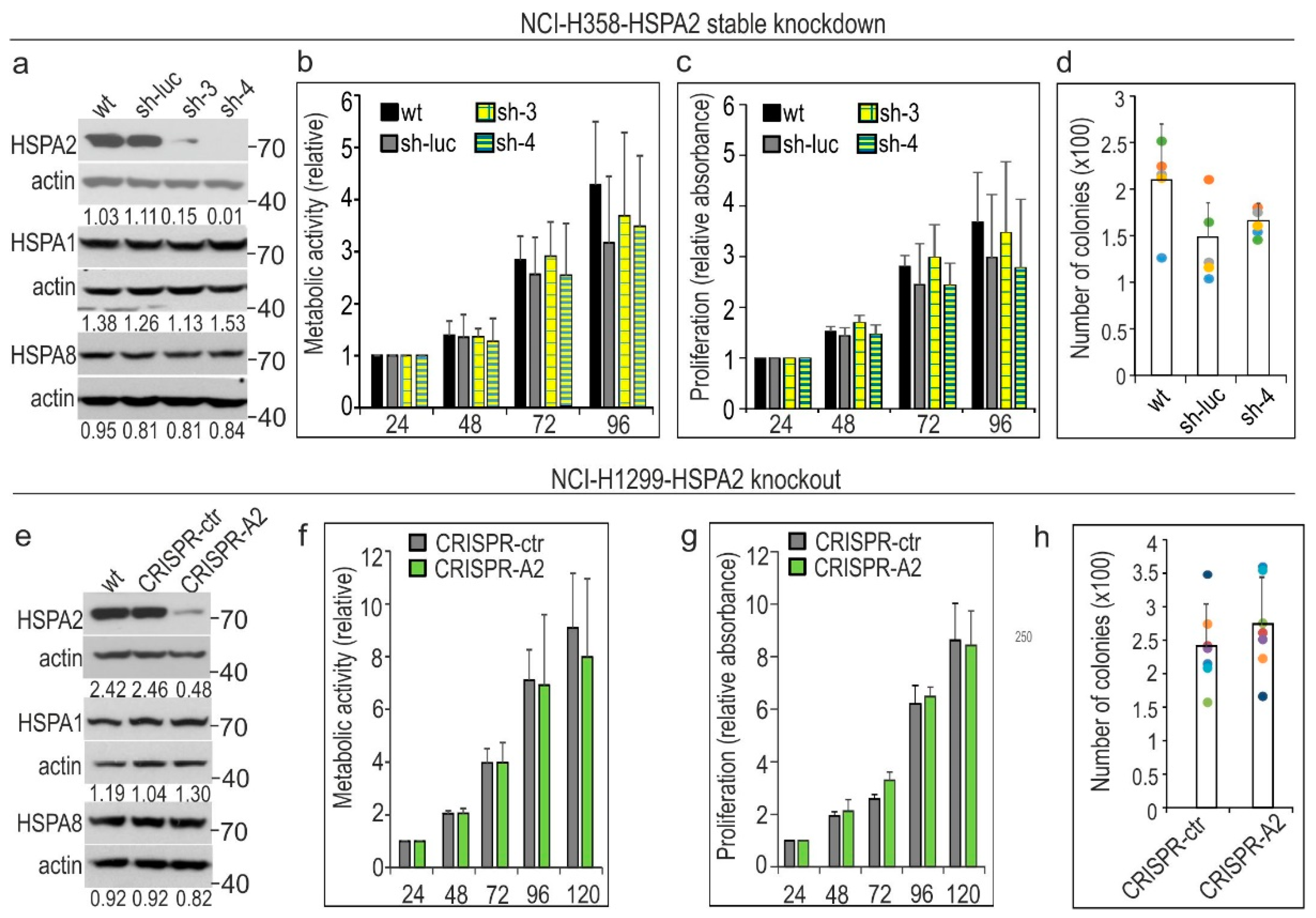
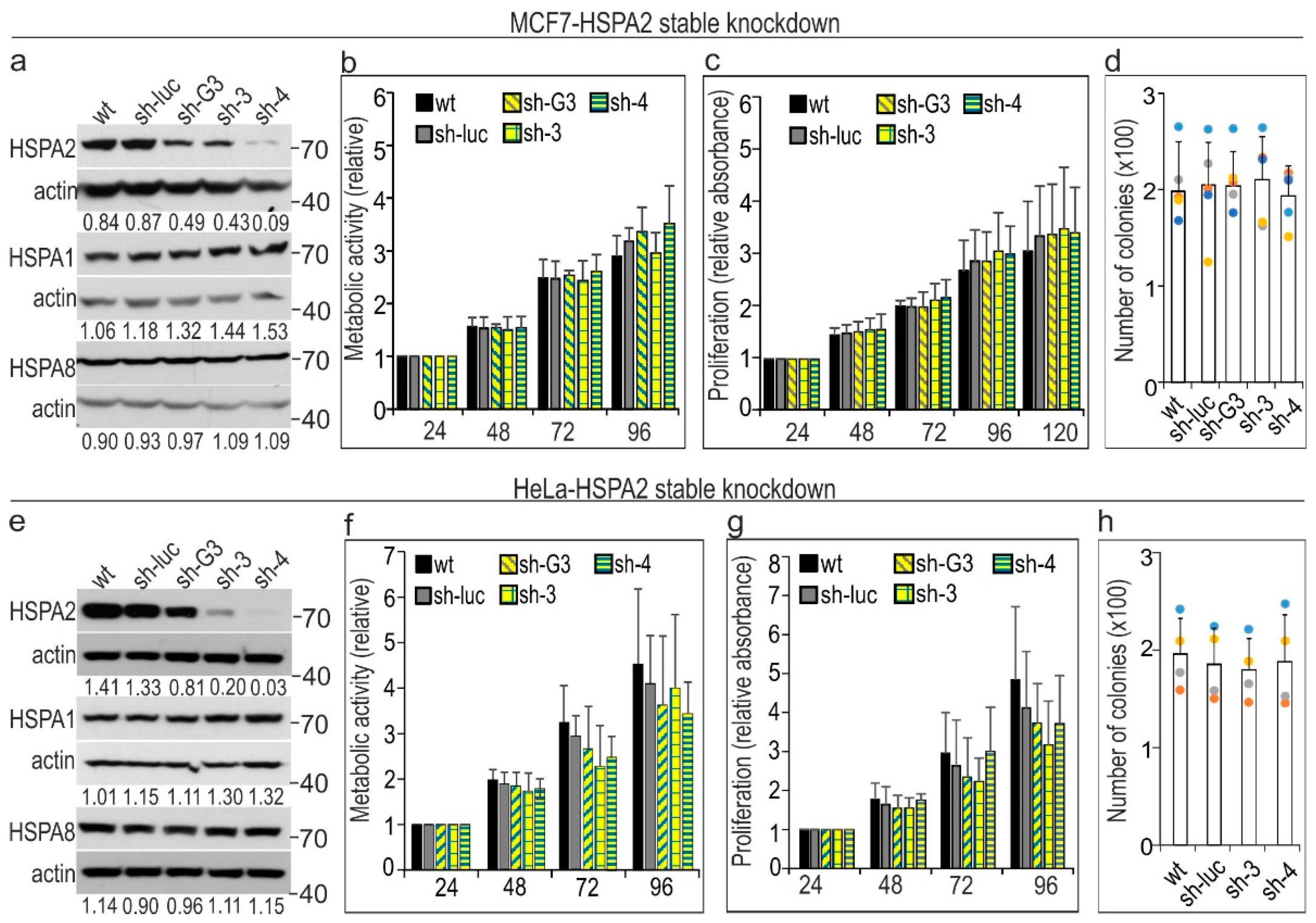
2.1. HSPA2 Knockdown Reduces Colony-Forming Capacity of HBEC but Not NSCLC Cells, although It Influences neither Proliferation nor Metabolic Activity of HBEC and NSCLC Cells
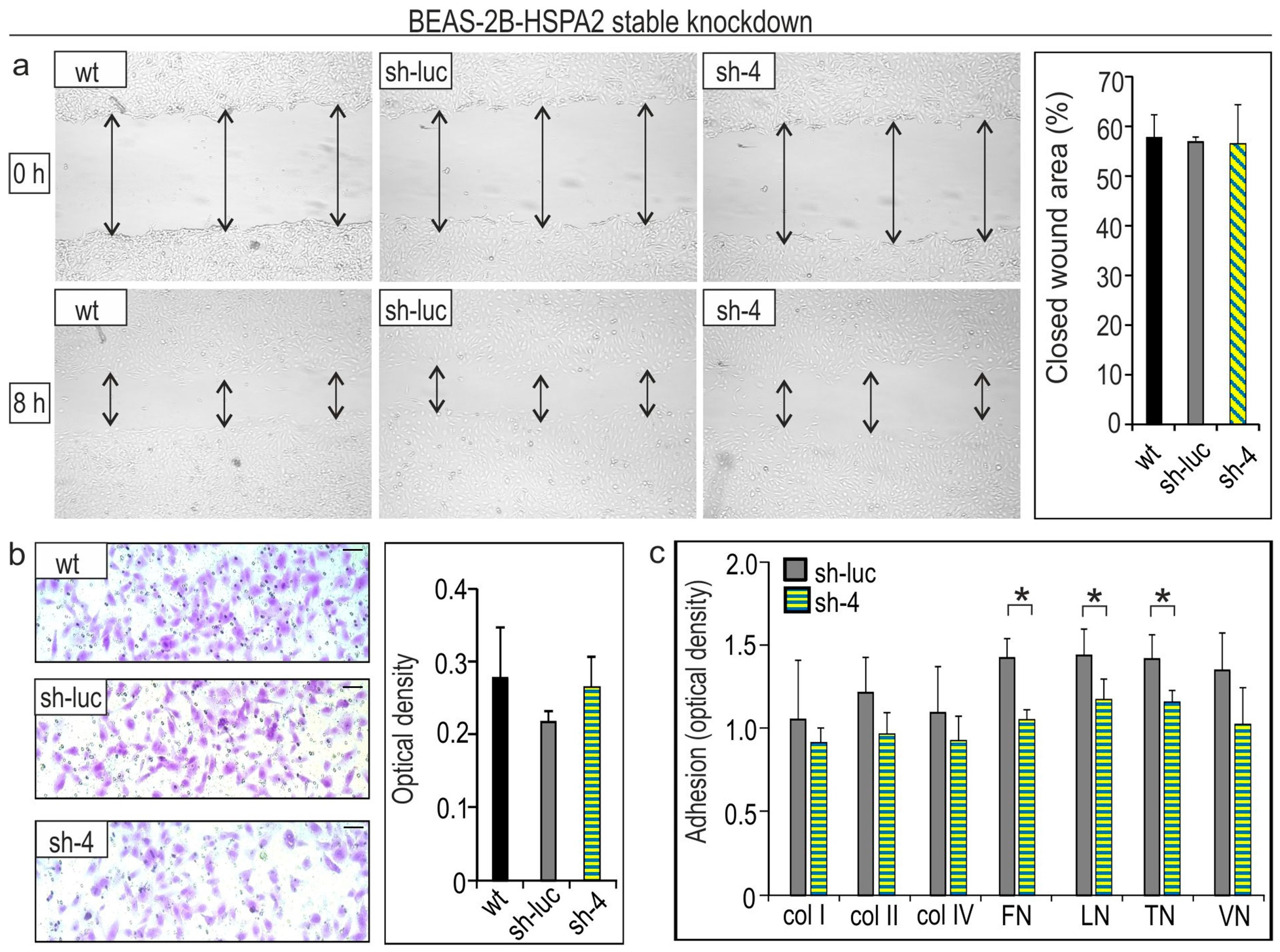
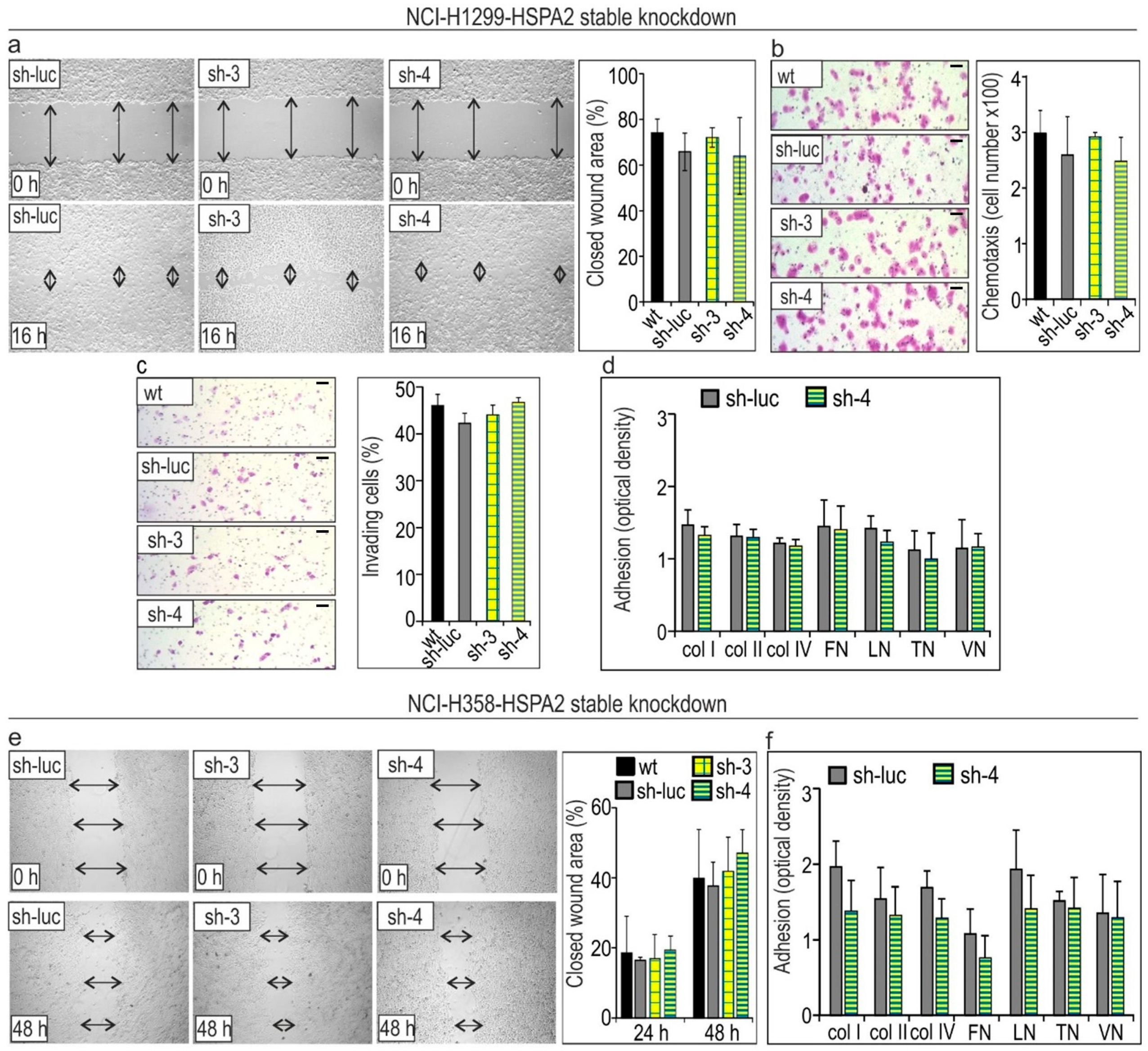
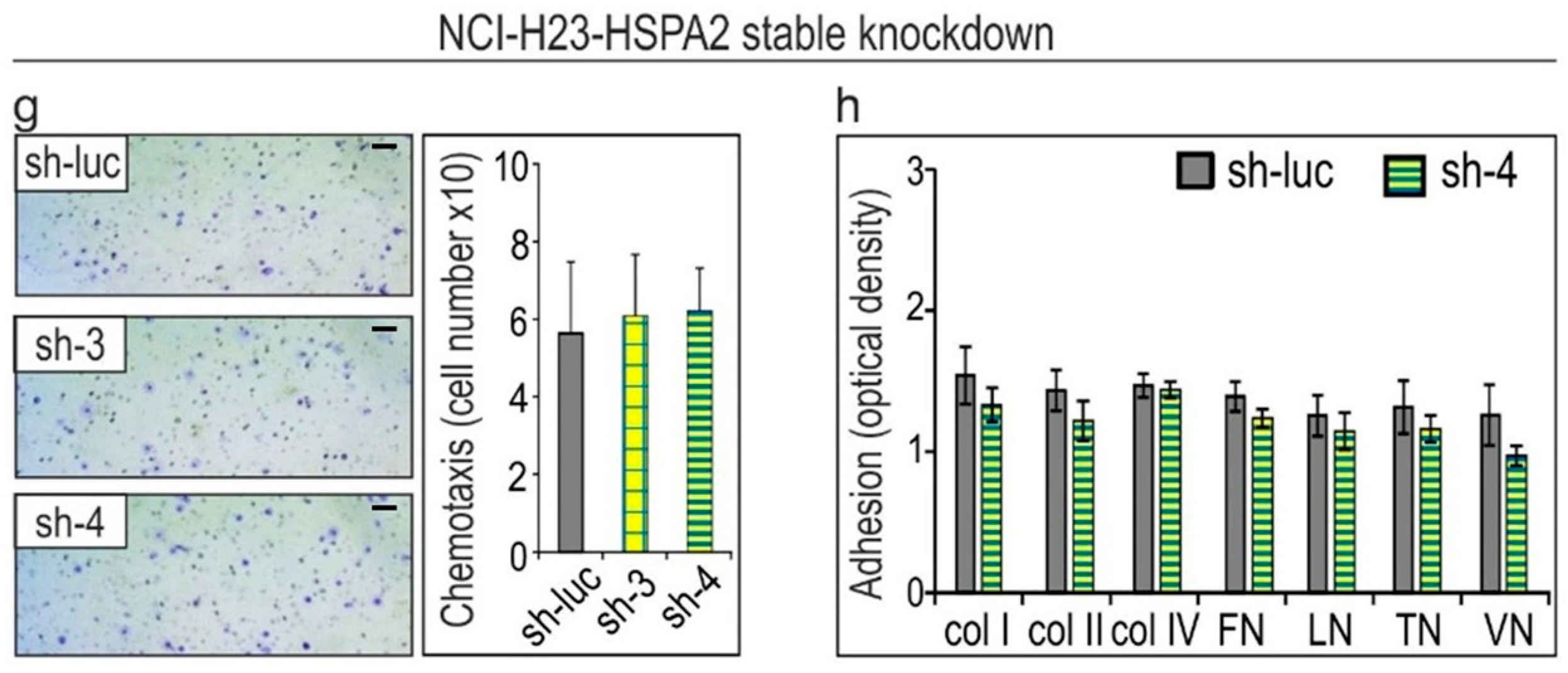
2.2. HSPA2 Knockdown Reduces Adhesiveness of HBEC but Not NSCLC Cells, While Having No Impact on Their Motility
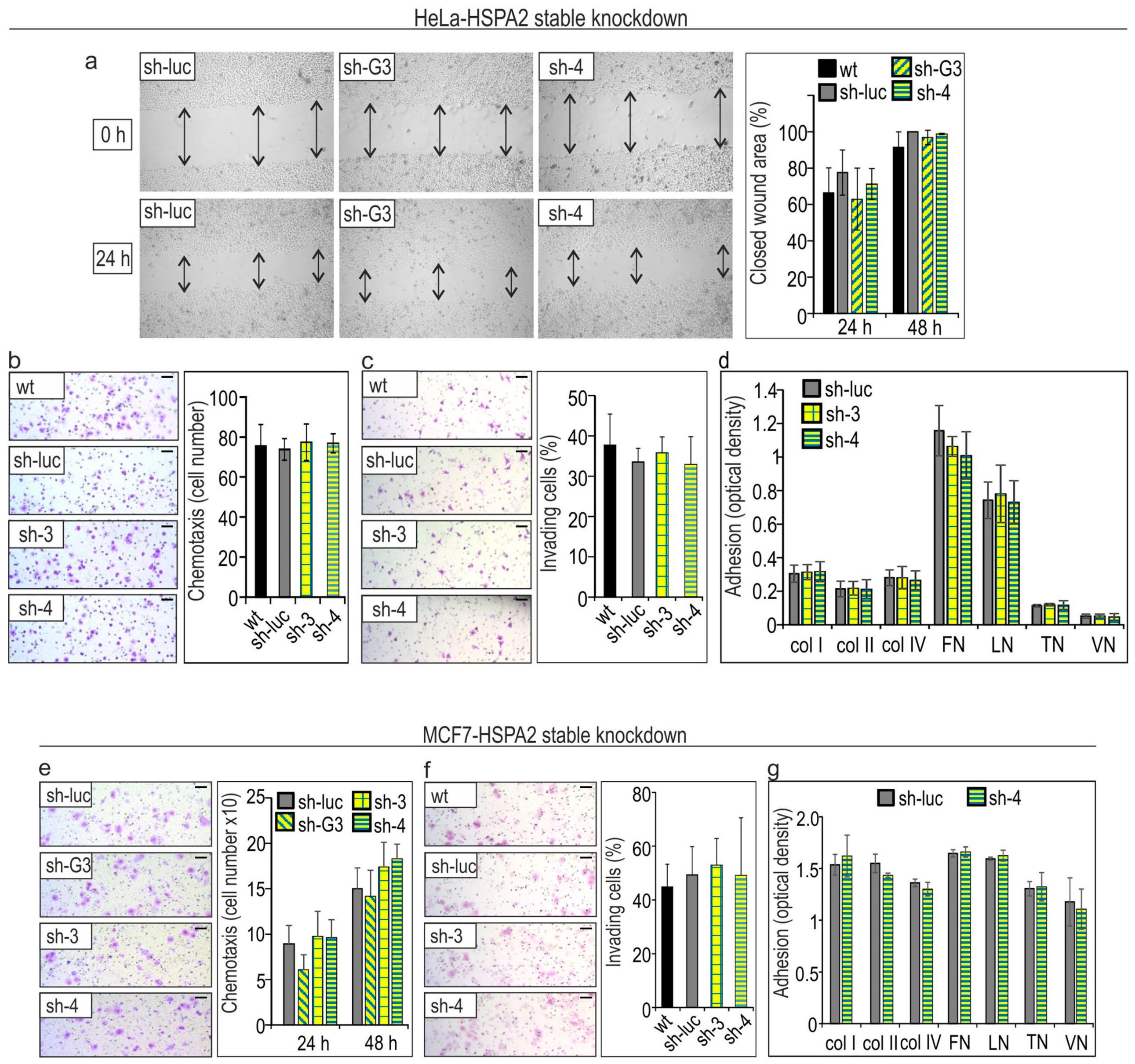
2.3. HSPA2 Knockdown Has No Impact on Growth, Migration, Invasion, and Adhesion to ECM Components of Breast (MCF7) and Cervical Cancer (HeLa) Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Experimental Conditions
4.2. Protein Extraction and Western Blot Analysis
4.3. Generation of HSPA2-Deficient Cell Lines
4.3.1. Generation of Lentivirally Modified Cell Lines
4.3.2. Generation of CRISPR/Cas9-Modified Cell Lines
4.4. Cell Proliferation, Metabolic Activity, and Clonogenic Assay
4.5. Adhesion Assay
4.6. In Vitro Wound-Healing Assay
4.7. Boyden Chamber Chemotaxis and Invasion Assays
4.7.1. Chemotactic Migration
4.7.2. Invasion Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gupta, R.S.; Singh, B. Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr. Biol. 1994, 4, 1104–1114. [Google Scholar] [CrossRef]
- Radons, J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2009, 14, 105–111. [Google Scholar] [CrossRef]
- Scieglinska, D.; Krawczyk, Z. Expression, function, and regulation of the testis-enriched heat shock HSPA2 gene in rodents and humans. Cell Stress Chaperones 2015, 20, 221–235. [Google Scholar] [CrossRef]
- Daugaard, M.; Rohde, M.; Jäättelä, M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef]
- Deane, C.A.S.; Brown, I.R. Differential Targeting of Hsp70 Heat Shock Proteins HSPA6 and HSPA1A with Components of a Protein Disaggregation/Refolding Machine in Differentiated Human Neuronal Cells following Thermal Stress. Front. Neurosci. 2017, 11, 227. [Google Scholar] [CrossRef]
- Hageman, J.; van Waarde, M.A.W.H.; Zylicz, A.; Walerych, D.; Kampinga, H.H. The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J. 2011, 435, 127–142. [Google Scholar] [CrossRef]
- Serlidaki, D.; van Waarde, M.; Rohland, L.; Wentink, A.S.; Dekker, S.L.; Kamphuis, M.J.; Boertien, J.M.; Brunsting, J.F.; Nillegoda, N.B.; Bukau, B.; et al. Functional diversity between HSP70 paralogs caused by variable interactions with specific co-chaperones. J. Biol. Chem. 2020, 295, 7301–7316. [Google Scholar] [CrossRef]
- Shevtsov, M.; Huile, G.; Multhoff, G. Membrane heat shock protein 70: A theranostic target for cancer therapy. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160526. [Google Scholar] [CrossRef]
- Boudesco, C.; Cause, S.; Jego, G.; Garrido, C. Hsp70: A Cancer Target Inside and Outside the Cell. Methods Mol. Biol. 2018, 1709, 371–396. [Google Scholar]
- Sherman, M.Y.; Gabai, V.L. Hsp70 in cancer: Back to the future. Oncogene 2015, 34, 4153–4161. [Google Scholar] [CrossRef]
- Gabai, V.L.; Yaglom, J.A.; Waldman, T.; Sherman, M.Y. Heat Shock Protein Hsp72 Controls Oncogene-Induced Senescence Pathways in Cancer Cells. Mol. Cell. Biol. 2009, 29, 559–569. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Zhou, Y.; Wang, Y.; Wang, S.; Zhang, W. Hsp70 promotes chemoresistance by blocking Bax mitochondrial translocation in ovarian cancer cells. Cancer Lett. 2012, 321, 137–143. [Google Scholar] [CrossRef]
- Endo, H.; Yano, M.; Okumura, Y.; Kido, H. Ibuprofen enhances the anticancer activity of cisplatin in lung cancer cells by inhibiting the heat shock protein 70. Cell Death Dis. 2014, 5, e1027. [Google Scholar] [CrossRef]
- Meng, L.; Hunt, C.; Yaglom, J.A.; Gabai, V.L.; Sherman, M.Y. Heat shock protein Hsp72 plays an essential role in Her2-induced mammary tumorigenesis. Oncogene 2011, 30, 2836–2845. [Google Scholar] [CrossRef]
- Powers, M.V.; Clarke, P.A.; Workman, P. Dual Targeting of HSC70 and HSP72 Inhibits HSP90 Function and Induces Tumor-Specific Apoptosis. Cancer Cell 2008, 14, 250–262. [Google Scholar] [CrossRef]
- Schlecht, R.; Scholz, S.R.; Dahmen, H.; Wegener, A.; Sirrenberg, C.; Musil, D.; Bomke, J.; Eggenweiler, H.M.; Mayer, M.P.; Bukau, B. Functional Analysis of Hsp70 Inhibitors. PLoS ONE 2013, 8, e78443. [Google Scholar] [CrossRef]
- Sojka, D.R.; Gogler-Pigłowska, A.; Vydra, N.; Cortez, A.J.; Filipczak, P.T.; Krawczyk, Z.; Scieglinska, D. Functional redundancy of HSPA1, HSPA2 and other HSPA proteins in non-small cell lung carcinoma (NSCLC); an implication for NSCLC treatment. Sci. Rep. 2019, 9, 14394. [Google Scholar] [CrossRef] [PubMed]
- Nixon, B.; Bromfield, E.G.; Cui, J.; De Iuliis, G.N. Heat Shock Protein A2 (HSPA2): Regulatory Roles in Germ Cell Development and Sperm Function. Adv. Anat. Embryol. Cell Biol. 2017, 222, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Scieglinska, D.; Sojka, D.R.; Gogler-Pigłowska, A.; Chumak, V.; Krawczyk, Z. Various Anti-HSPA2 Antibodies Yield Different Results in Studies on Cancer-Related Functions of Heat Shock Protein A2. Int. J. Mol. Sci. 2020, 21, 4296. [Google Scholar] [CrossRef] [PubMed]
- Scieglinska, D.; Piglowski, W.; Chekan, M.; Mazurek, A.; Krawczyk, Z. Differential expression of HSPA1 and HSPA2 proteins in human tissues; tissue microarray-based immunohistochemical study. Histochem. Cell Biol. 2011, 135, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Scieglinska, D.; Gogler-Piglowska, A.; Butkiewicz, D.; Chekan, M.; Malusecka, E.; Harasim, J.; Habryka, A.; Krawczyk, Z. HSPA2 is expressed in human tumors and correlates with clinical features in non-small cell lung carcinoma patients. Anticancer Res. 2014, 34, 2833–2840. [Google Scholar] [PubMed]
- Filipczak, P.T.; Piglowski, W.; Glowala-Kosinska, M.; Krawczyk, Z.; Scieglinska, D. HSPA2 overexpression protects V79 fibroblasts against bortezomib-induced apoptosis. Biochem. Cell Biol. 2012, 90, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Petyuk, V.A.; Chang, R.; Ramirez-Restrepo, M.; Beckmann, N.D.; Henrion, M.Y.R.; Piehowski, P.D.; Zhu, K.; Wang, S.; Clarke, J.; Huentelman, M.J.; et al. The human brainome: Network analysis identifies HSPA2 as a novel Alzheimer’s disease target. Brain 2018. [Google Scholar] [CrossRef]
- Gogler-Pigłowska, A.; Klarzyńska, K.; Sojka, D.R.; Habryka, A.; Głowala-Kosińska, M.; Herok, M.; Kryj, M.; Halczok, M.; Krawczyk, Z.; Scieglinska, D. Novel role for the testis-enriched HSPA2 protein in regulating epidermal keratinocyte differentiation. J. Cell. Physiol. 2018, 233, 2629–2644. [Google Scholar] [CrossRef]
- Rohde, M.; Daugaard, M.; Jensen, M.H.; Helin, K.; Nylandsted, J.; Jäättelä, M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005, 19, 570–582. [Google Scholar] [CrossRef]
- Jagadish, N.; Agarwal, S.; Gupta, N.; Fatima, R.; Devi, S.; Kumar, V.; Suri, V.; Kumar, R.; Suri, V.; Sadasukhi, T.C.; et al. Heat shock protein 70-2 (HSP70-2) overexpression in breast cancer. J. Exp. Clin. Cancer Res. 2016, 35. [Google Scholar] [CrossRef]
- Garg, M.; Kanojia, D.; Saini, S.; Suri, S.; Gupta, A.; Surolia, A.; Suri, A. Germ cell-specific heat shock protein 70-2 is expressed in cervical carcinoma and is involved in the growth, migration, and invasion of cervical cells. Cancer 2010, 116, 3785–3796. [Google Scholar] [CrossRef]
- Barrandon, Y.; Green, H. Three clonal types of keratinocytes with different capacities for multiplication. Proc. Natl. Acad. Sci. USA 1987, 84, 2302–2306. [Google Scholar] [CrossRef]
- Banno, T.; Blumenberg, B. Keratinocyte detachment-differentiation connection revisited, or anoikis-pityriasi nexus redut. PLoS ONE 2014, 9, e100279. [Google Scholar] [CrossRef]
- Tata, P.R.; Rajagopal, J. Plasticity in the lung: Making and breaking cell identity. Development 2017, 144, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Crews, A.L.; Chen, W.; Park, J.; Yin, Q.; Ren, X.R.; Adler, K.B. MARCKS and HSP70 interactions regulate mucin secretion by human airway epithelial cells in vitro. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 304, L511–L518. [Google Scholar] [CrossRef] [PubMed]
- Raiford, K.L.; Park, J.; Lin, K.-W.; Fang, S.; Crews, A.L.; Adler, K.B. Mucin granule-associated proteins in human bronchial epithelial cells: The airway goblet cell “granulome”. Respir. Res. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Huang, S.; Yang, J.; Niu, P.; Gong, Z.; Liu, X.; Xin, L.; Currie, R.W.; Wu, T. HspA1A facilitates DNA repair in human bronchial epithelial cells exposed to Benzo[a]pyrene and interacts with casein kinase 2. Cell Stress Chaperones 2014, 19, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Guo, L.; Liao, Z.; Zhang, M.; Zhang, M.; Wang, T.; Chen, L.; Xu, D.; Feng, Y.; Wen, F. Increased expression of heat shock protein 70 in chronic obstructive pulmonary disease. Int. Immunopharmacol. 2013, 17, 885–893. [Google Scholar] [CrossRef]
- Hulina-Tomašković, A.; Heijink, I.H.; Jonker, M.R.; Somborac-Bačura, A.; Grdić Rajković, M.; Rumora, L. Pro-inflammatory effects of extracellular Hsp70 and cigarette smoke in primary airway epithelial cells from COPD patients. Biochimie 2019, 156, 47–58. [Google Scholar] [CrossRef]
- Wheeler, D.S.; Chase, M.A.; Senft, A.P.; Poynter, S.E.; Wong, H.R.; Page, K. Extracellular Hsp72, an endogenous DAMP, is released by virally infected airway epithelial cells and activates neutrophils via Toll-like receptor (TLR)-4. Respir. Res. 2009, 10, 31. [Google Scholar] [CrossRef]
- Hou, C.; Zhao, H.; Li, W.; Liang, Z.; Zhang, D.; Liu, L.; Tong, W.; Cai, S.X.; Zou, F. Increased heat shock protein 70 levels in induced sputum and plasma correlate with severity of asthma patients. Cell Stress Chaperones 2011, 16, 663–671. [Google Scholar] [CrossRef]
- Yaglom, J.A.; Wang, Y.; Li, A.; Li, Z.; Monti, S.; Alexandrov, I.; Lu, X.; Sherman, M.Y. Cancer cell responses to Hsp70 inhibitor JG-98: Comparison with Hsp90 inhibitors and finding synergistic drug combinations. Sci. Rep. 2018, 8, 3010. [Google Scholar] [CrossRef]
- Cao, L.; Yuan, X.; Bao, F.; Lv, W.; He, Z.; Tang, J.; Han, J.; Hu, J. Downregulation of HSPA2 inhibits proliferation via ERK1/2 pathway and endoplasmic reticular stress in lung adenocarcinoma. Ann. Transl. Med. 2019, 7, 540. [Google Scholar] [CrossRef]
- Clerico, E.M.; Meng, W.; Pozhidaeva, A.; Bhasne, K.; Petridis, C.; Gierasch, L.M. Hsp70 molecular chaperones: Multifunctional allosteric holding and unfolding machines. Biochem. J. 2019, 476, 1653–1677. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell. Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, W.; Duan, C.J.; Zhang, C.F. Overexpression of HSPA2 is correlated with poor prognosis in esophageal squamous cell carcinoma. World J. Surg. Oncol. 2013, 11, 141. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhao, H.; Li, X.S.; Kang, H.R.; Ma, J.X.; Yao, F.F.; Du, N. Expression of HSPA2 in human hepatocellular carcinoma and its clinical significance. Tumor Biol. 2014, 35, 11283–11287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, H.; Liu, C.; Kong, Y.; Wang, C.; Zhang, H. Expression and clinical significance of HSPA2 in pancreatic ductal adenocarcinoma. Diagn. Pathol. 2015, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.L.; Xie, Q.; Zhou, C.H.; Huang, D.W.; Tang, Z.G.; Ju, T.F. Overexpressed HSPA2 correlates with tumor angiogenesis and unfavorable prognosis in pancreatic carcinoma. Pancreatology 2017, 17, 457–463. [Google Scholar] [CrossRef]
- Yang, Y.L.; Zhang, Y.; Li, D.D.; Zhang, F.L.; Liu, H.Y.; Liao, X.H.; Xie, H.Y.; Lu, Q.; Zhang, L.; Hong, Q.; et al. RNF144A functions as a tumor suppressor in breast cancer through ubiquitin ligase activity-dependent regulation of stability and oncogenic functions of HSPA2. Cell Death Differ. 2020, 27, 1105–1118. [Google Scholar] [CrossRef]
- Klimczak, M.; Biecek, P.; Zylicz, A.; Zylicz, M. Heat shock proteins create a signature to predict the clinical outcome in breast cancer. Sci. Rep. 2019, 9, 7507. [Google Scholar] [CrossRef]
- Zoppino, F.C.M.; Guerrero-Gimenez, M.E.; Castro, G.N.; Ciocca, D.R. Comprehensive transcriptomic analysis of heat shock proteins in the molecular subtypes of human breast cancer. BMC Cancer 2018, 18. [Google Scholar] [CrossRef]
- Huang, Z.; Duan, H.; Li, H. Identification of Gene Expression Pattern Related to Breast Cancer Survival Using Integrated TCGA Datasets and Genomic Tools. BioMed. Res. Int. 2015, 2015, 878546. [Google Scholar] [CrossRef]
- Wen, W.; Liu, W.; Shao, Y.; Chen, L. VER-155008, a small molecule inhibitor of HSP70 with potent anti-cancer activity on lung cancer cell lines. Exp. Biol. Med. 2014, 239, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Gabai, V.L.; Budagova, K.R.; Sherman, M.Y. Increased expression of the major heat shock protein Hsp72 in human prostate carcinoma cells is dispensable for their viability but confers resistance to a variety of anticancer agents. Oncogene 2005, 24, 3328–3338. [Google Scholar] [CrossRef] [PubMed]
- Nylandsted, J.; Rohde, M.; Brand, K.; Bastholm, L.; Elling, F.; Jaattela, M. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2. Proc. Natl. Acad. Sci. USA 2000, 97, 7871–7876. [Google Scholar] [CrossRef] [PubMed]
- Multhoff, G.; Botzler, C. Heat-Shock Proteins and the Immune Response. Ann. N. Y. Acad. Sci. 1998, 851, 86–93. [Google Scholar] [CrossRef]
- De Maio, A.; Vazquez, D. Extracellular Heat Shock Proteins: A New Location, A New Function. Shock 2013, 40, 239–246. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sojka, D.R.; Gogler-Pigłowska, A.; Klarzyńska, K.; Klimczak, M.; Zylicz, A.; Głowala-Kosińska, M.; Krawczyk, Z.; Scieglinska, D. HSPA2 Chaperone Contributes to the Maintenance of Epithelial Phenotype of Human Bronchial Epithelial Cells but Has Non-Essential Role in Supporting Malignant Features of Non-Small Cell Lung Carcinoma, MCF7, and HeLa Cancer Cells. Cancers 2020, 12, 2749. https://doi.org/10.3390/cancers12102749
Sojka DR, Gogler-Pigłowska A, Klarzyńska K, Klimczak M, Zylicz A, Głowala-Kosińska M, Krawczyk Z, Scieglinska D. HSPA2 Chaperone Contributes to the Maintenance of Epithelial Phenotype of Human Bronchial Epithelial Cells but Has Non-Essential Role in Supporting Malignant Features of Non-Small Cell Lung Carcinoma, MCF7, and HeLa Cancer Cells. Cancers. 2020; 12(10):2749. https://doi.org/10.3390/cancers12102749
Chicago/Turabian StyleSojka, Damian Robert, Agnieszka Gogler-Pigłowska, Katarzyna Klarzyńska, Marta Klimczak, Alicja Zylicz, Magdalena Głowala-Kosińska, Zdzisław Krawczyk, and Dorota Scieglinska. 2020. "HSPA2 Chaperone Contributes to the Maintenance of Epithelial Phenotype of Human Bronchial Epithelial Cells but Has Non-Essential Role in Supporting Malignant Features of Non-Small Cell Lung Carcinoma, MCF7, and HeLa Cancer Cells" Cancers 12, no. 10: 2749. https://doi.org/10.3390/cancers12102749
APA StyleSojka, D. R., Gogler-Pigłowska, A., Klarzyńska, K., Klimczak, M., Zylicz, A., Głowala-Kosińska, M., Krawczyk, Z., & Scieglinska, D. (2020). HSPA2 Chaperone Contributes to the Maintenance of Epithelial Phenotype of Human Bronchial Epithelial Cells but Has Non-Essential Role in Supporting Malignant Features of Non-Small Cell Lung Carcinoma, MCF7, and HeLa Cancer Cells. Cancers, 12(10), 2749. https://doi.org/10.3390/cancers12102749





