PIWI-Like 1 and PIWI-Like 2 Expression in Breast Cancer
Simple Summary
Abstract
1. Introduction
2. Results
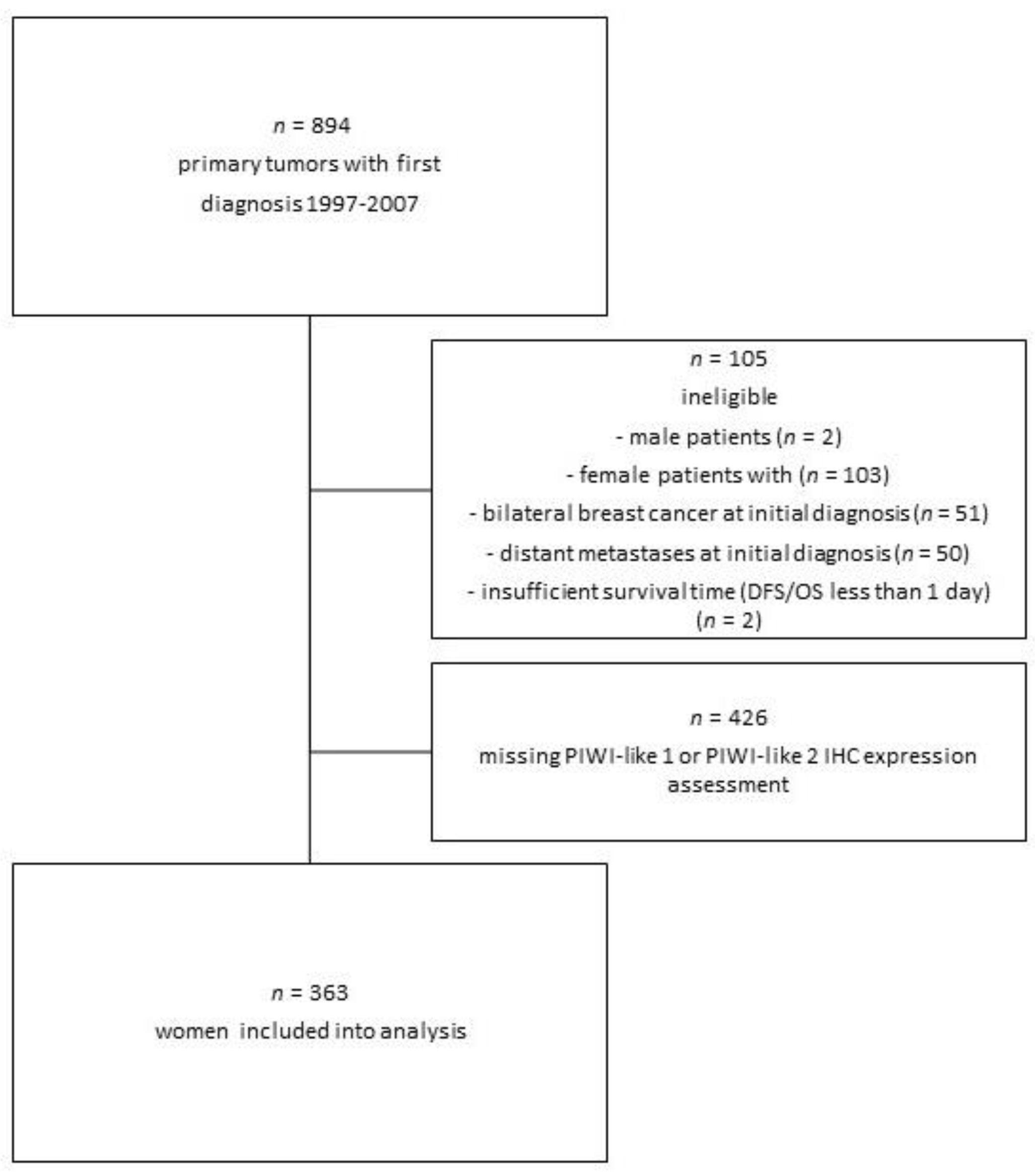
2.1. Patients
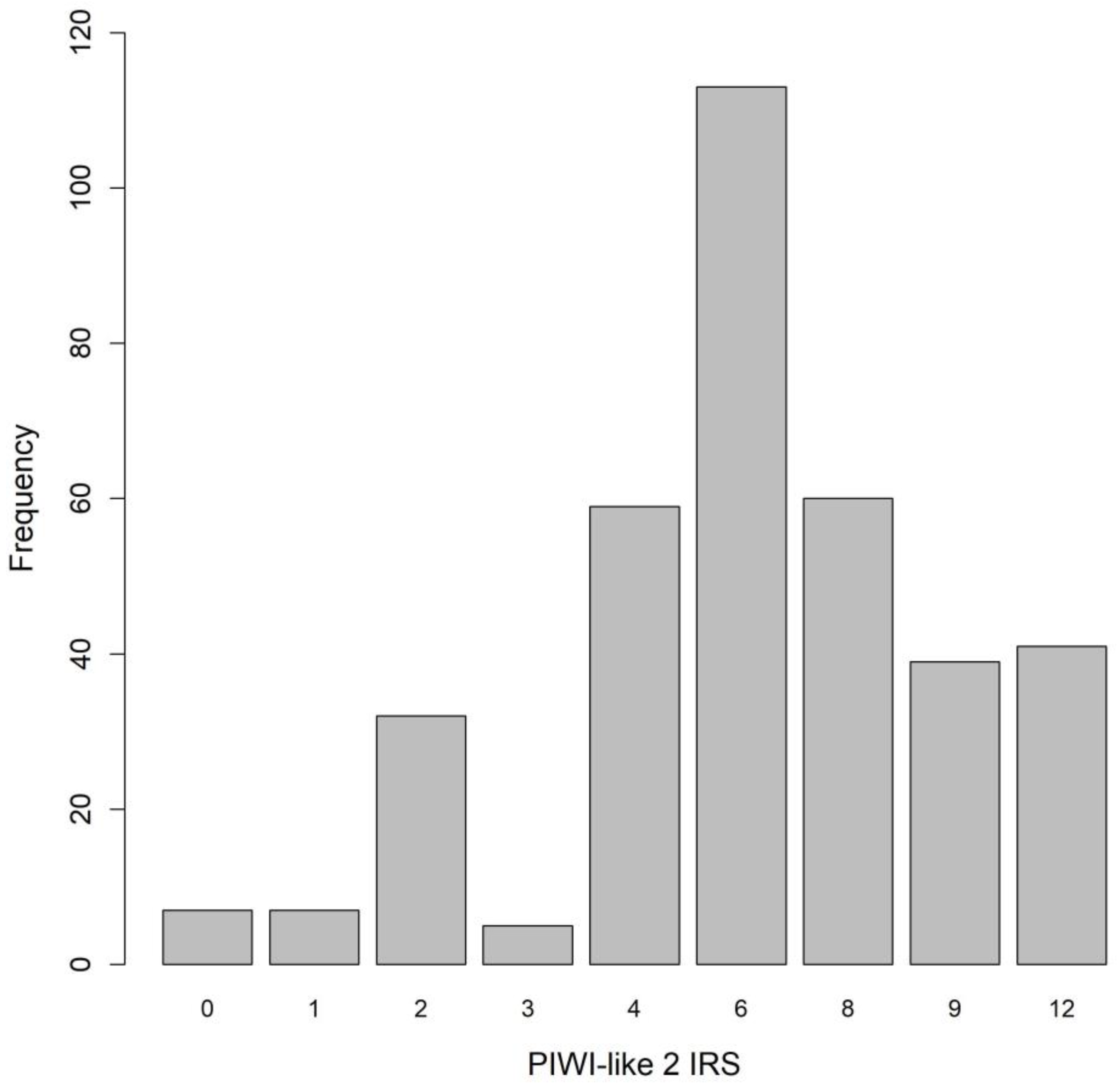
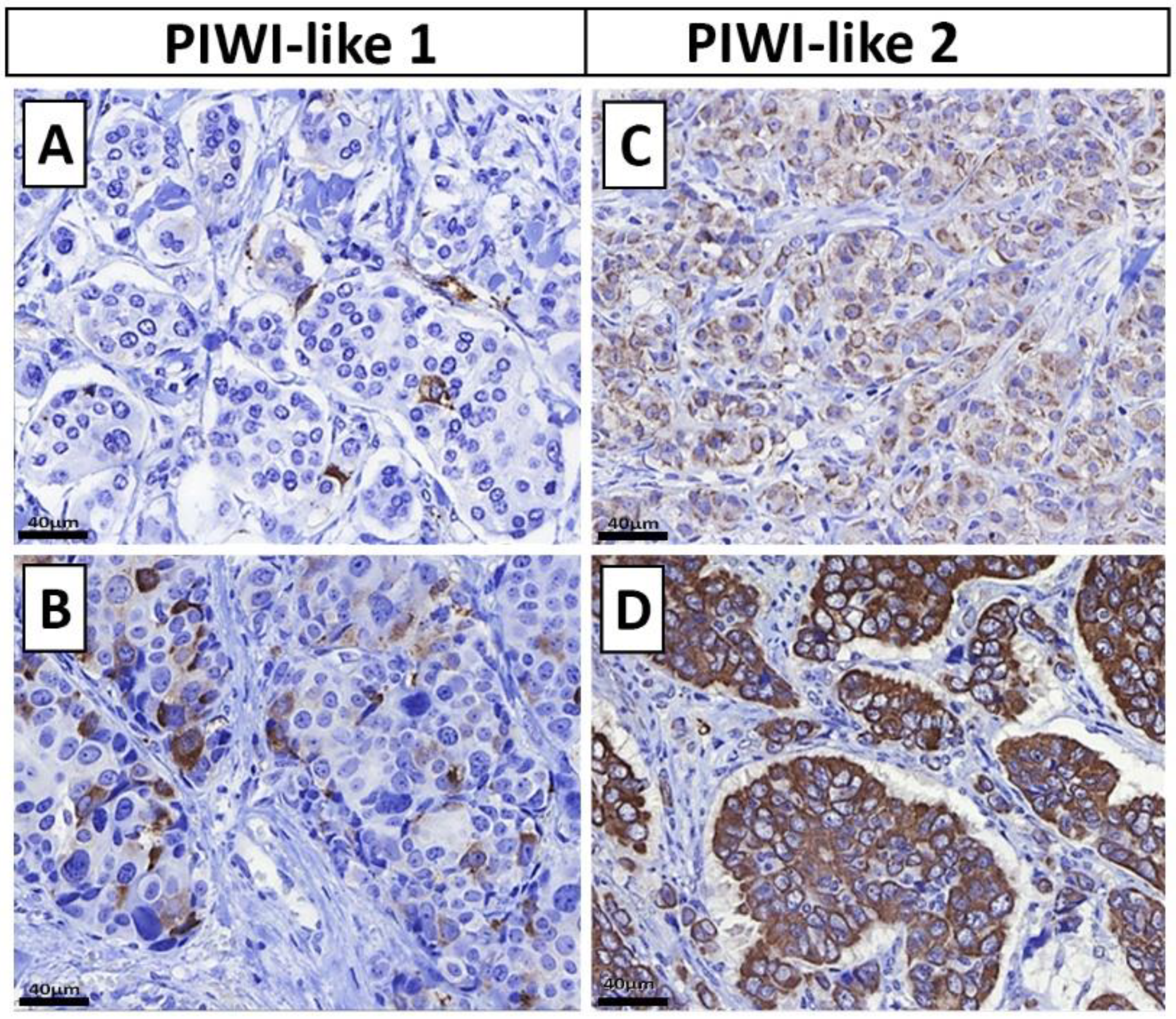
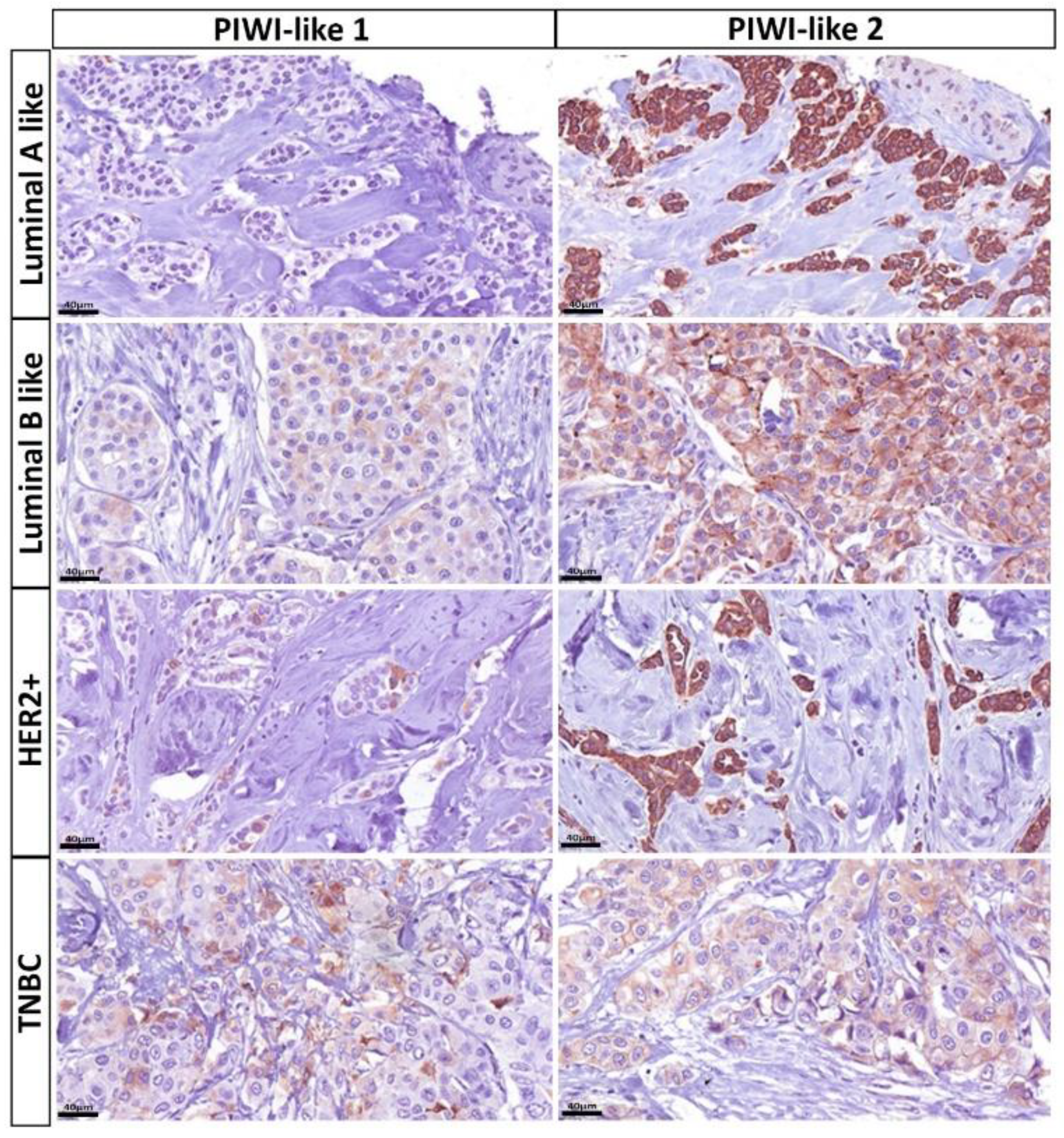
2.2. Distribution of PIWI-Like 1 and PIWI-Like 2
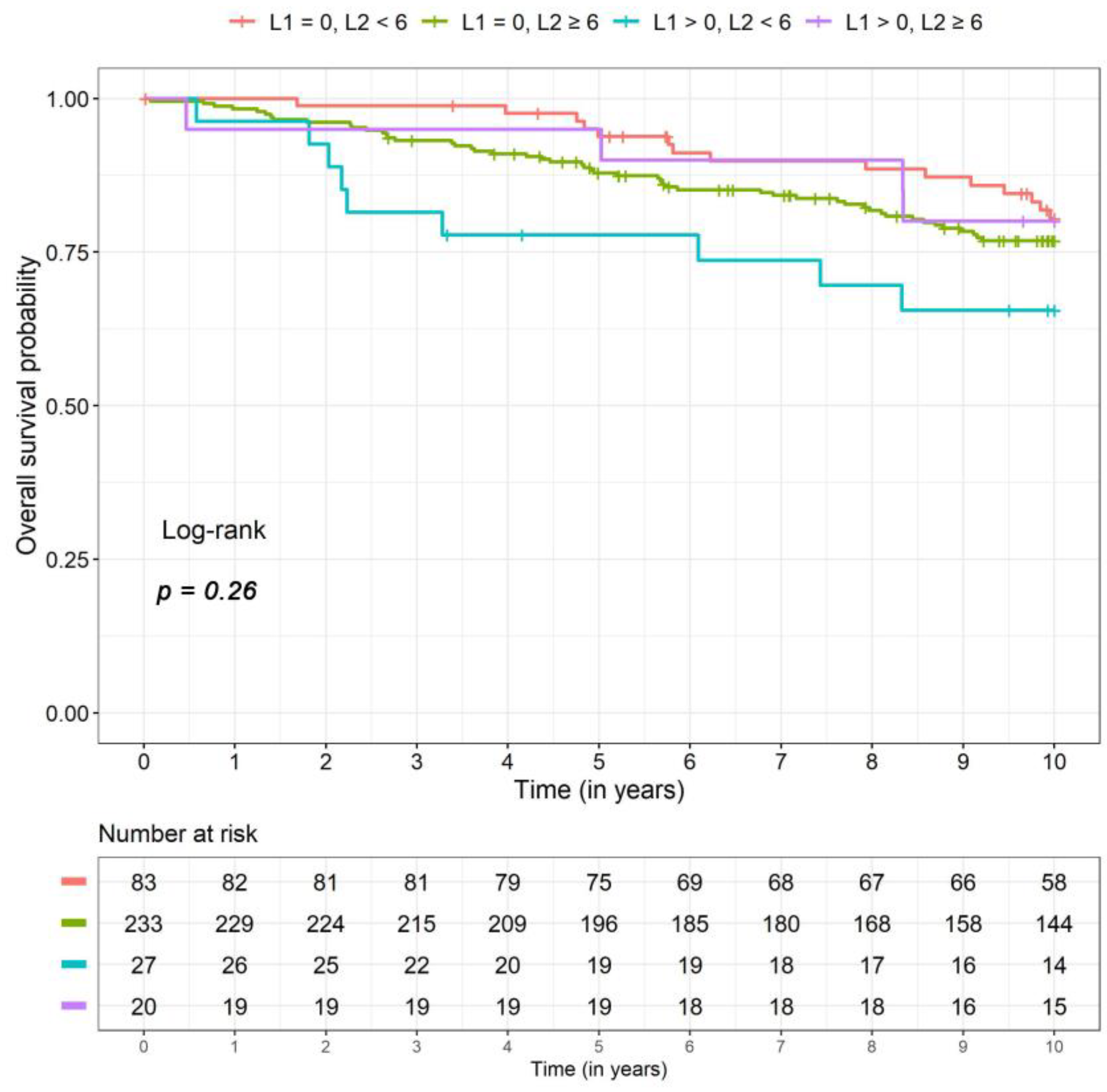
2.3. Overall Survival
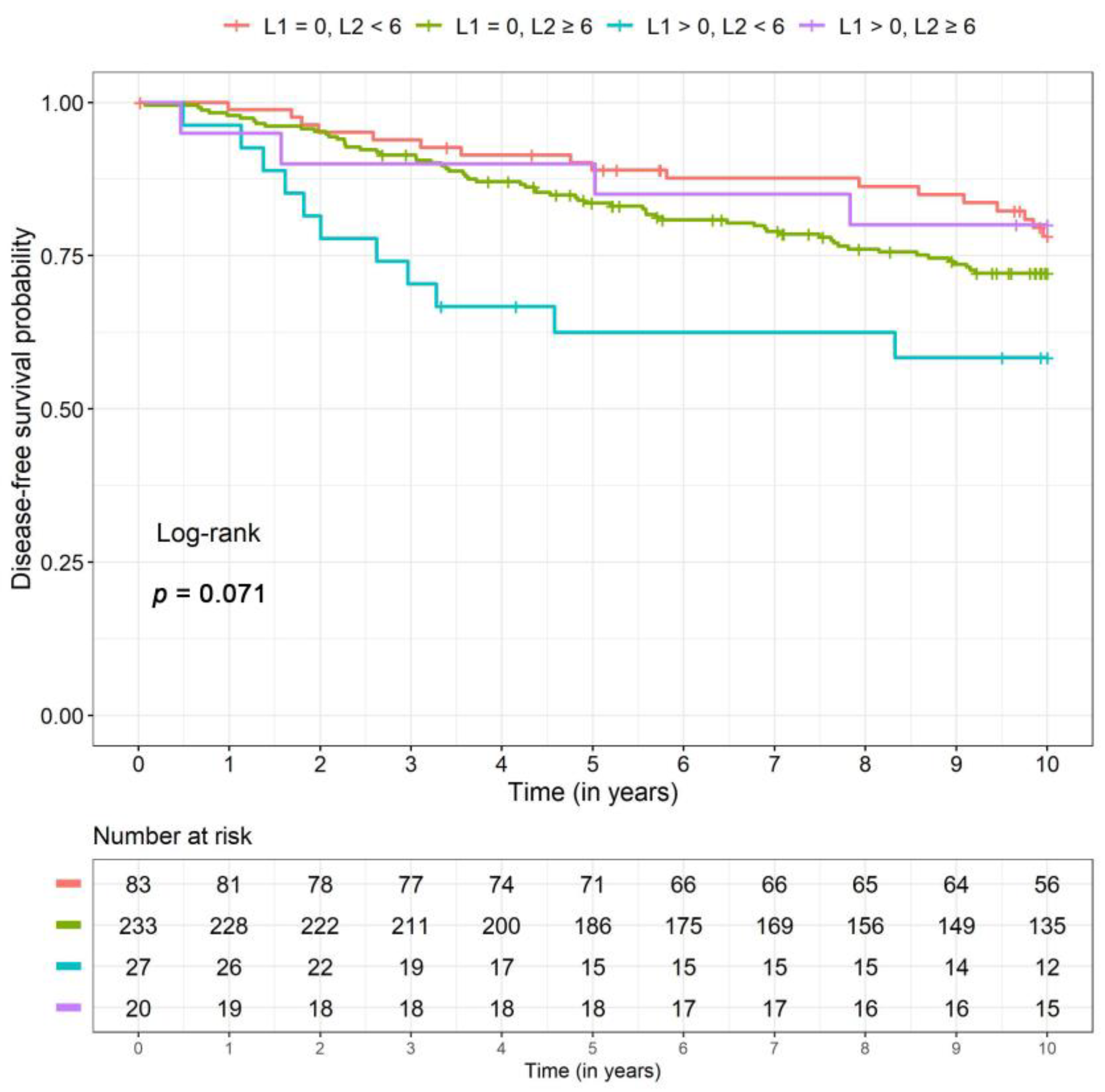
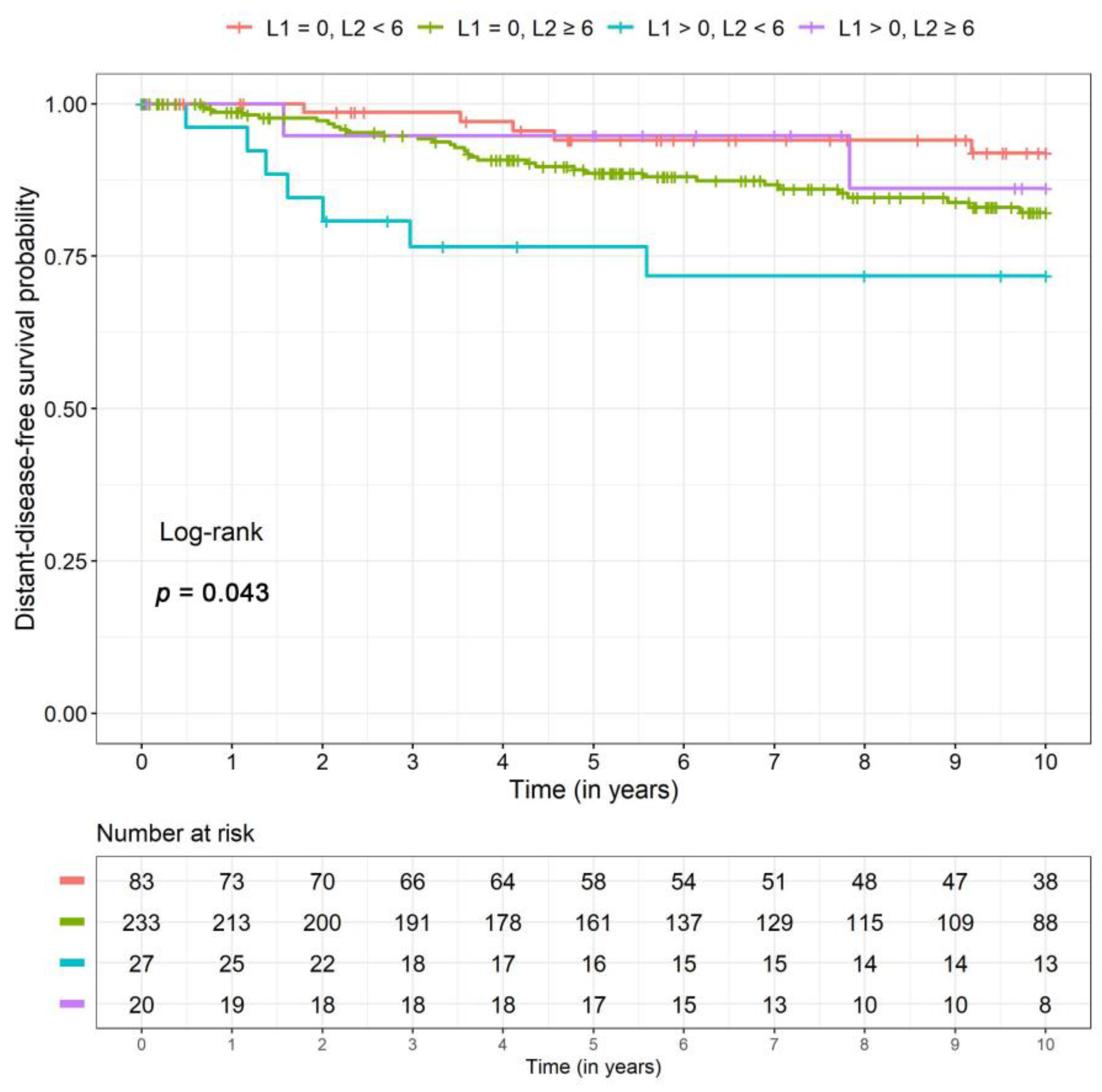
2.4. Disease-Free and Distant Disease-Free Survival
2.5. Associations between PIWI-Like 1-IRS and Breast Cancer Subclasses
2.6. Associations between PIWI-Like 2 IRS and Breast Cancer Subclasses
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. Clinical Data
4.3. Histopathological Assessment
4.4. Immunohistochemical Staining of ER, PR, HER2, and Ki-67 and Molecular (-Like) Subtyping
4.5. Construction of Tissue Microarrays (BBCC TMA)
4.6. PIWI-Like 1 and PIWI-Like 2 Immunohistochemistry
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sasaki, T.; Shiohama, A.; Minoshima, S.; Shimizu, N. Identification of eight members of the Argonaute family in the human genome. Genomics 2003, 82, 323–330. [Google Scholar] [CrossRef]
- Litwin, M.; Szczepańska-Buda, A.; Piotrowska, A.; Dzięgiel, P.; Witkiewicz, W. The meaning of PIWI proteins in cancer development. Oncol. Lett. 2017, 13, 3354–3362. [Google Scholar] [CrossRef]
- Livak, K.J. Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics 1990, 124, 303–316. [Google Scholar] [PubMed]
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998, 12, 3715–3727. [Google Scholar] [CrossRef]
- Ishizu, H.; Siomi, H.; Siomi, M.C. Biology of PIWI-interacting RNAs: New insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012, 26, 2361–2373. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef]
- Hashim, A.; Rizzo, F.; Marchese, G.; Ravo, M.; Tarallo, R.; Nassa, G.; Giurato, G.; Santamaria, G.; Cordella, A.; Cantarella, C.; et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 2014, 5, 9901–9910. [Google Scholar] [CrossRef]
- Ye, Y.; Yin, D.T.; Chen, L.; Zhou, Q.; Shen, R.; He, G.; Yan, Q.; Tong, Z.; Issekutz, A.C.; Shapiro, C.L.; et al. Identification of Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS ONE 2010, 5, e13406. [Google Scholar] [CrossRef]
- Al-Janabi, O.; Wach, S.; Nolte, E.; Weigelt, K.; Rau, T.T.; Stohr, C.; Legal, W.; Schick, S.; Greither, T.; Hartmann, A.; et al. Piwi-like 1 and 4 gene transcript levels are associated with clinicopathological parameters in renal cell carcinomas. Biochim. Biophys. Acta 2014, 1842, 686–690. [Google Scholar] [CrossRef]
- Suzuki, R.; Honda, S.; Kirino, Y. PIWI Expression and Function in Cancer. Front. Genet. 2012, 3, 204. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Liu, L.; Liao, M.; Zhang, C.; Hu, S.; Zou, M.; Gu, M.; Li, X. Emerging roles for PIWI proteins in cancer. Acta Biochim. Biophys. Sin. 2015, 47, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Fathizadeh, H.; Asemi, Z. Epigenetic roles of PIWI proteins and piRNAs in lung cancer. Cell Biosci. 2019, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Qu, L.K.; Meng, L.; Liu, C.Y.; Dong, B.; Xing, X.F.; Wu, J.; Shou, C.C. HIWI expression profile in cancer cells and its prognostic value for patients with colorectal cancer. Chin. Med. J. 2011, 124, 2144–2149. [Google Scholar]
- Zhao, Y.M.; Zhou, J.M.; Wang, L.R.; He, H.W.; Wang, X.L.; Tao, Z.H.; Sun, H.C.; Wu, W.Z.; Fan, J.; Tang, Z.Y.; et al. HIWI is associated with prognosis in patients with hepatocellular carcinoma after curative resection. Cancer 2012, 118, 2708–2717. [Google Scholar] [CrossRef]
- Liu, W.; Gao, Q.; Chen, K.; Xue, X.; Li, M.; Chen, Q.; Zhu, G.; Gao, Y. Hiwi facilitates chemoresistance as a cancer stem cell marker in cervical cancer. Oncol. Rep. 2014, 32, 1853–1860. [Google Scholar] [CrossRef]
- Li, L.; Yu, C.; Gao, H.; Li, Y. Argonaute proteins: Potential biomarkers for human colon cancer. BMC Cancer 2010, 10, 38. [Google Scholar] [CrossRef]
- Li, D.; Sun, X.; Yan, D.; Huang, J.; Luo, Q.; Tang, H.; Peng, Z. Piwil2 modulates the proliferation and metastasis of colon cancer via regulation of matrix metallopeptidase 9 transcriptional activity. Exp. Biol. Med. (Maywood) 2012, 237, 1231–1240. [Google Scholar] [CrossRef]
- Lee, J.H.; Schutte, D.; Wulf, G.; Fuzesi, L.; Radzun, H.J.; Schweyer, S.; Engel, W.; Nayernia, K. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum. Mol. Genet. 2006, 15, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Ghosh, S.; Graham, K.; Mackey, J.R.; Kovalchuk, O.; Damaraju, S. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget 2016, 7, 37944–37956. [Google Scholar] [CrossRef]
- Wang, D.W.; Wang, Z.H.; Wang, L.L.; Song, Y.; Zhang, G.Z. Overexpression of hiwi promotes growth of human breast cancer cells. Asian Pac. J. Cancer Prev. APJCP 2014, 15, 7553–7558. [Google Scholar] [CrossRef] [PubMed]
- Litwin, M.; Szczepanska-Buda, A.; Michalowska, D.; Grzegrzolka, J.; Piotrowska, A.; Gomulkiewicz, A.; Wojnar, A.; Dziegiel, P.; Witkiewicz, W. Aberrant Expression of PIWIL1 and PIWIL2 and Their Clinical Significance in Ductal Breast Carcinoma. Anticancer Res. 2018, 38, 2021–2030. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Taubert, H.; Wach, S.; Jung, R.; Pugia, M.; Keck, B.; Bertz, S.; Nolte, E.; Stoehr, R.; Lehmann, J.; Ohlmann, C.-H.; et al. Piwil 2 Expression Is Correlated with Disease-Specific and Progression-Free Survival of Chemotherapy-Treated Bladder Cancer Patients. Mol. Med. 2015, 21, 371–380. [Google Scholar] [CrossRef]
- Eckstein, M.; Jung, R.; Weigelt, K.; Sikic, D.; Stohr, R.; Geppert, C.; Agaimy, A.; Lieb, V.; Hartmann, A.; Wullich, B.; et al. Piwi-like 1 and -2 protein expression levels are prognostic factors for muscle invasive urothelial bladder cancer patients. Sci. Rep. 2018, 8, 17693. [Google Scholar] [CrossRef]
- Stöhr, C.G.; Steffens, S.; Polifka, I.; Jung, R.; Kahlmeyer, A.; Ivanyi, P.; Weber, F.; Hartmann, A.; Wullich, B.; Wach, S.; et al. Piwi-like 1 protein expression is a prognostic factor for renal cell carcinoma patients. Sci. Rep. 2019, 9, 1741. [Google Scholar] [CrossRef]
- Greither, T.; Koser, F.; Kappler, M.; Bache, M.; Lautenschläger, C.; Göbel, S.; Holzhausen, H.-J.; Wach, S.; Würl, P.; Taubert, H. Expression of human Piwi-like genes is associated with prognosis for soft tissue sarcoma patients. BMC Cancer 2012, 12, 272. [Google Scholar] [CrossRef]
- Liu, J.J.; Shen, R.; Chen, L.; Ye, Y.; He, G.; Hua, K.; Jarjoura, D.; Nakano, T.; Ramesh, G.K.; Shapiro, C.L.; et al. Piwil2 is expressed in various stages of breast cancers and has the potential to be used as a novel biomarker. Int. J. Clin. Exp. Pathol. 2010, 3, 328–337. [Google Scholar] [PubMed]
- Cao, J.; Xu, G.; Lan, J.; Huang, Q.; Tang, Z.; Tian, L. High expression of piwi-like RNA-mediated gene silencing 1 is associated with poor prognosis via regulating transforming growth factor-beta receptors and cyclin-dependent kinases in breast cancer. Mol. Med. Rep. 2016, 13, 2829–2835. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, N.; Shi, S.; Liu, S.; Lin, H. The Role of PIWIL4, an Argonaute Family Protein, in Breast Cancer. J. Biol. Chem. 2016, 291, 10646–10658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ren, Y.; Xu, H.; Pang, D.; Duan, C.; Liu, C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg. Oncol. 2013, 22, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Hu, H.; Xue, X.; Shen, S.; Gao, E.; Guo, G.; Shen, X.; Zhang, X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin. Transl. Oncol. 2013, 15, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Mai, D.; Zhang, B.; Jiang, X.; Zhang, J.; Bai, R.; Ye, Y.; Li, M.; Pan, L.; Su, J.; et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol. Cancer 2019, 18, 9. [Google Scholar] [CrossRef]
- Siddiqi, S.; Terry, M.; Matushansky, I. Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PLoS ONE 2012, 7, e33711. [Google Scholar] [CrossRef]
- Yin, D.-T.; Wang, Q.; Chen, L.; Liu, M.-Y.; Han, C.; Yan, Q.; Shen, R.; He, G.; Duan, W.; Li, J.-J.; et al. Germline stem cell gene PIWIL2 mediates DNA repair through relaxation of chromatin. PLoS ONE 2011, 6, e27154. [Google Scholar] [CrossRef]
- Wang, Q.-E.; Han, C.; Milum, K.; Wani, A.A. Stem cell protein Piwil2 modulates chromatin modifications upon cisplatin treatment. Mutat. Res. 2011, 708, 59–68. [Google Scholar] [CrossRef]
- Fasching, P.A.; Weihbrecht, S.; Haeberle, L.; Gasparyan, A.; Villalobos, I.E.; Ma, Y.; Ekici, A.B.; Wachter, D.L.; Hartmann, A.; Beckmann, M.W.; et al. HER2 and TOP2A amplification in a hospital-based cohort of breast cancer patients: Associations with patient and tumor characteristics. Breast Cancer Res. Treat. 2014, 145, 193–203. [Google Scholar] [CrossRef]
- Serce, N.B.; Boesl, A.; Klaman, I.; von Serenyi, S.; Noetzel, E.; Press, M.F.; Dimmler, A.; Hartmann, A.; Sehouli, J.; Knuechel, R.; et al. Overexpression of SERBP1 (Plasminogen activator inhibitor 1 RNA binding protein) in human breast cancer is correlated with favourable prognosis. BMC Cancer 2012, 12, 597. [Google Scholar] [CrossRef]
- Bektas, N.; Noetzel, E.; Veeck, J.; Press, M.F.; Kristiansen, G.; Naami, A.; Hartmann, A.; Dimmler, A.; Beckmann, M.W.; Knuchel, R.; et al. The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer. Breast Cancer Res. 2008, 10, R58. [Google Scholar] [CrossRef]
- Brennan, M.; Gass, P.; Häberle, L.; Wang, D.; Hartmann, A.; Lux, M.P.; Beckmann, M.W.; Untch, M.; Fasching, P.A. The effect of participation in neoadjuvant clinical trials on outcomes in patients with early breast cancer. Breast Cancer Res. Treat. 2018, 171, 747–758. [Google Scholar] [CrossRef]
- Beckmann, M.W.; Brucker, C.; Hanf, V.; Rauh, C.; Bani, M.R.; Knob, S.; Petsch, S.; Schick, S.; Fasching, P.A.; Hartmann, A.; et al. Quality assured health care in certified breast centers and improvement of the prognosis of breast cancer patients. Onkologie 2011, 34, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Wockel, A.; Festl, J.; Stuber, T.; Brust, K.; Stangl, S.; Heuschmann, P.U.; Albert, U.S.; Budach, W.; Follmann, M.; Janni, W.; et al. Interdisciplinary Screening, Diagnosis, Therapy and Follow-up of Breast Cancer. Guideline of the DGGG and the DKG (S3-Level, AWMF Registry Number 032/045OL, December 2017)—Part 1 with Recommendations for the Screening, Diagnosis and Therapy of Breast Cancer. Geburtshilfe Frauenheilkd. 2018, 78, 927–948. [Google Scholar] [CrossRef] [PubMed]
- Wockel, A.; Festl, J.; Stuber, T.; Brust, K.; Krockenberger, M.; Heuschmann, P.U.; Jiru-Hillmann, S.; Albert, U.S.; Budach, W.; Follmann, M.; et al. Interdisciplinary Screening, Diagnosis, Therapy and Follow-up of Breast Cancer. Guideline of the DGGG and the DKG (S3-Level, AWMF Registry Number 032/045OL, December 2017)—Part 2 with Recommendations for the Therapy of Primary, Recurrent and Advanced Breast Cancer. Geburtshilfe Frauenheilkd. 2018, 78, 1056–1088. [Google Scholar] [CrossRef] [PubMed]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Wood, W.C.; Gelber, R.D.; Coates, A.S.; Thürlimann, B.; Senn, H.-J. Meeting Highlights: Updated International Expert Consensus on the Primary Therapy of Early Breast Cancer. J. Clin. Oncol. 2003, 21, 3357–3365. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Glick, J.H.; Gelber, R.D.; Coates, A.S.; Thurlimann, B.; Senn, H.J. Meeting highlights: International expert consensus on the primary therapy of early breast cancer 2005. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2005, 16, 1569–1583. [Google Scholar] [CrossRef]
- Hammond, M.E.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch. Pathol. Lab. Med. 2010, 134, e48–e72. [Google Scholar] [CrossRef]
- Hammond, M.E.; Hayes, D.F.; Wolff, A.C. Clinical Notice for American Society of Clinical Oncology-College of American Pathologists guideline recommendations on ER/PgR and HER2 testing in breast cancer. J. Clin. Oncol. 2011, 29, e458. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Glick, J.H.; Gelber, R.D.; Coates, A.S.; Senn, H.J. Meeting highlights: International Consensus Panel on the Treatment of Primary Breast Cancer. Seventh International Conference on Adjuvant Therapy of Primary Breast Cancer. J. Clin. Oncol. 2001, 19, 3817–3827. [Google Scholar] [CrossRef]
- Sauter, G.; Lee, J.; Bartlett, J.M.; Slamon, D.J.; Press, M.F. Guidelines for human epidermal growth factor receptor 2 testing: Biologic and methodologic considerations. J. Clin. Oncol. 2009, 27, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Hammond, M.E.; Schwartz, J.N.; Hagerty, K.L.; Allred, D.C.; Cote, R.J.; Dowsett, M.; Fitzgibbons, P.L.; Hanna, W.M.; Langer, A.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch. Pathol. Lab. Med. 2007, 131, 18–43. [Google Scholar] [CrossRef] [PubMed]
- Wunderle, M.; Pretscher, J.; Brucker, S.Y.; Volz, B.; Hartmann, A.; Fiessler, C.; Hein, A.; Häberle, L.; Jud, S.M.; Lux, M.P.; et al. Association between breast cancer risk factors and molecular type in postmenopausal patients with hormone receptor-positive early breast cancer. Breast Cancer Res. Treat. 2019, 174, 453–461. [Google Scholar] [CrossRef]
- Bethune, G.C.; Veldhuijzen van Zanten, D.; MacIntosh, R.F.; Rayson, D.; Younis, T.; Thompson, K.; Barnes, P.J. Impact of the 2013 American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 (HER2) testing of invasive breast carcinoma: A focus on tumours assessed as ‘equivocal’ for HER2 gene amplification by fluorescence in-situ hybridization. Histopathology 2015, 67, 880–887. [Google Scholar] [CrossRef]
- Nielsen, T.; Wallden, B.; Schaper, C.; Ferree, S.; Liu, S.; Gao, D.; Barry, G.; Dowidar, N.; Maysuria, M.; Storhoff, J. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer 2014, 14, 177. [Google Scholar] [CrossRef]
- Remmele, W.; Stegner, H.E. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 1987, 8, 138–140. [Google Scholar]
- Grambsch, P.M.; Therneau, T.M. Proportional Hazards Tests and Diagnostics Based on Weighted Residuals. Biometrika 1994, 81, 515–526. [Google Scholar] [CrossRef]
- Salmen, J.; Neugebauer, J.; Fasching, P.A.; Haeberle, L.; Huober, J.; Wöckel, A.; Rauh, C.; Schuetz, F.; Weissenbacher, T.; Kost, B.; et al. Pooled analysis of the prognostic relevance of progesterone receptor status in five German cohort studies. Breast Cancer Res. Treat. 2014, 148, 143–151. [Google Scholar] [CrossRef] [PubMed]

| Parameter | n (%) or Median (IQR) | |
|---|---|---|
| n | 363 (100%) | |
| Age (years) | 58 (49, 67) | |
| BMI (kg/m2) | 25.7 (23.1, 28.6) | |
| Ki-67 (%) | 15 (10, 30) | |
| ER | Positive | 298 (82.1%) |
| PR | Positive | 276 (76.0%) |
| HER2 | Positive | 42 (11.6%) |
| Grading (G) | G1 | 30 (8.3%) |
| G2 | 248 (68.3%) | |
| G3 | 85 (23.4%) | |
| Nodal status (N) | N+ | 148 (40.8%) |
| Tumor stage (T) | T1 | 186 (51.2%) |
| T2 | 138 (38.0%) | |
| T3 | 19 (5.2%) | |
| T4 | 20 (5.5%) | |
| Molecular-like subtype | TNBC | 41 (11.3%) |
| Luminal A like | 143 (39.4%) | |
| Luminal B like | 137 (37.7%) | |
| HER2+ | 42 (11.6%) | |
| PAM50 subtype | Basal-like | 56 (15.4%) |
| Luminal A | 208 (57.3%) | |
| Luminal B | 69 (19.0%) | |
| HER2-enriched | 30 (8.3%) | |
| PIWI-like 1 IRS | Median | 0 (0, 0) |
| IRS > 0 | 47 (12.9%) | |
| PIWI-like 2 IRS | Median | 6 (4, 8) |
| IRS ≥ 6 | 253 (69.7%) | |
| Combination of PIWI-Like 1 IRS (L1) and PIWI-Like 2 IRS (L2) | Adjusted | Unadjusted | |||
|---|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||
| OS 79 events | L1 = 0, L2 < 6 | Reference | – | Reference | – |
| L1 = 0, L2 ≥ 6 | 1.36 (0.75, 2.46) | 0.31 | 1.30 (0.73, 2.31) | 0.37 | |
| L1 > 0, L2 < 6 | 2.92 (1.24, 6.90) | 0.01 | 2.26 (0.99, 5.17) | 0.05 | |
| L1 > 0, L2 ≥ 6 | 0.89 (0.29, 2.69) | 0.83 | 1.08 (0.36, 3.27) | 0.89 | |
| DFS 94 events | L1 = 0, L2 < 6 | Reference | – | Reference | – |
| L1 = 0, L2 ≥ 6 | 1.34 (0.77, 2.31) | 0.30 | 1.38 (0.81, 2.36) | 0.24 | |
| L1 > 0, L2 < 6 | 3.27 (1.48, 7.20) | 0.003 | 2.61 (1.22, 5.59) | 0.01 | |
| L1 > 0, L2 ≥ 6 | 0.83 (0.28, 2.48) | 0.74 | 0.97 (0.33, 2.88) | 0.96 | |
| DDFS 46 events | L1 = 0, L2 < 6 | Reference | – | Reference | – |
| L1 = 0, L2 ≥ 6 | 2.73 (1.04, 7.18) | 0.04 | 2.33 (0.91, 5.98) | 0.08 | |
| L1 > 0, L2 < 6 | 7.64 (2.35, 24.82) | <0.001 | 4.72 (1.50, 14.88) | 0.008 | |
| L1 > 0, L2 ≥ 6 | 1.19 (0.23, 6.24) | 0.83 | 1.52 (0.29, 7.83) | 0.62 | |
| Combination of PIWI-Like 1 IRS (L1) and PIWI-Like 2 IRS (L2) | L1 = 0, L2 < 6 | L1 = 0, L2 ≥ 6 | L1 > 0, L2 < 6 | L1 > 0, L2 ≥ 6 | |
|---|---|---|---|---|---|
| Total | n | 83 | 233 | 27 | 20 |
| OS | n events | 15 | 51 | 9 | 4 |
| 5-year SR (CI) | 0.91 (0.87, 0.96) | 0.89 (0.85, 0.92) | 0.81 (0.70, 0.94) | 0.90 (0.82, 1.00) | |
| 10-year SR (CI) | 0.81 (0.73, 0.90) | 0.77 (0.71, 0.82) | 0.63 (0.46, 0.85) | 0.80 (0.64, 1.00) | |
| DFS | n events | 17 | 62 | 11 | 4 |
| 5-year SR (CI) | 0.88 (0.82, 0.94) | 0.83 (0.79, 0.88) | 0.71 (0.57, 0.87) | 0.88 (0.77, 1.00) | |
| 10-year SR (CI) | 0.79 (0.71, 0.88) | 0.72 (0.66, 0.78) | 0.54 (0.37, 0.78) | 0.80 (0.64, 1.00) | |
| DDFS | n events | 5 | 32 | 7 | 2 |
| 5-year SR (CI) | 0.95 (0.90, 0.99) | 0.88 (0.84, 0.93) | 0.78 (0.64, 0.94) | 0.92 (0.82, 1.00) | |
| 10-year SR (CI) | 0.92 (0.86, 0.99) | 0.83 (0.77, 0.88) | 0.68 (0.51, 0.90) | 0.88 (0.74, 1.00) | |
| Parameter | PIWI-Like 1 IRS = 0 | PIWI-Like 1 IRS > 0 | p-Value | |
| Molecular-like subtype | TNBC | 29 (9.2%) | 12 (25.5%) | <0.001 |
| Luminal A like | 137 (43.3%) | 6 (12.8%) | ||
| Luminal B like | 114 (36.1%) | 23 (48.9%) | ||
| HER2+ | 36(11.4%) | 6 (12.8%) | ||
| PAM50 subtype | Basal-like | 38 (12.0%) | 18 (38.3%) | <0.001 |
| Luminal A | 201 (63.6%) | 7 (14.9%) | ||
| Luminal B | 57 (18.0%) | 12 (25.5%) | ||
| HER2-enriched | 20 (6.3%) | 10 (21.3%) | ||
| Parameter | PIWI-Like 2 IRS < 6 | PIWI-Like 2 IRS ≥ 6 | p-Value | |
| Molecular-like subtype | TNBC | 30 (27.3%) | 11 (4.3%) | <0.001 |
| Luminal A like | 27 (24.5%) | 116 (45.8%) | ||
| Luminal B like | 42 (38.2%) | 95 (37.5%) | ||
| HER2+ | 11 (10.0%) | 31 (12.3%) | ||
| PAM50 subtype | Basal-like | 38 (34.5%) | 18 (7.1%) | <0.001 |
| Luminal A | 34 (30.9%) | 174 (68.8%) | ||
| Luminal B | 26 (23.6%) | 43 (17.0%) | ||
| HER2-enriched | 12 (10.9%) | 18 (7.1%) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erber, R.; Meyer, J.; Taubert, H.; Fasching, P.A.; Wach, S.; Häberle, L.; Gaß, P.; Schulz-Wendtland, R.; Landgraf, L.; Olbricht, S.; et al. PIWI-Like 1 and PIWI-Like 2 Expression in Breast Cancer. Cancers 2020, 12, 2742. https://doi.org/10.3390/cancers12102742
Erber R, Meyer J, Taubert H, Fasching PA, Wach S, Häberle L, Gaß P, Schulz-Wendtland R, Landgraf L, Olbricht S, et al. PIWI-Like 1 and PIWI-Like 2 Expression in Breast Cancer. Cancers. 2020; 12(10):2742. https://doi.org/10.3390/cancers12102742
Chicago/Turabian StyleErber, Ramona, Julia Meyer, Helge Taubert, Peter A. Fasching, Sven Wach, Lothar Häberle, Paul Gaß, Rüdiger Schulz-Wendtland, Laura Landgraf, Sabrina Olbricht, and et al. 2020. "PIWI-Like 1 and PIWI-Like 2 Expression in Breast Cancer" Cancers 12, no. 10: 2742. https://doi.org/10.3390/cancers12102742
APA StyleErber, R., Meyer, J., Taubert, H., Fasching, P. A., Wach, S., Häberle, L., Gaß, P., Schulz-Wendtland, R., Landgraf, L., Olbricht, S., Jung, R., Beckmann, M. W., Hartmann, A., & Ruebner, M. (2020). PIWI-Like 1 and PIWI-Like 2 Expression in Breast Cancer. Cancers, 12(10), 2742. https://doi.org/10.3390/cancers12102742










