Clofarabine Improves Relapse-Free Survival of Acute Myeloid Leukemia in Younger Adults with Micro-Complex Karyotype
Abstract
:1. Introduction
2. Methods
2.1. Patients and Treatment
2.2. SNP-A Karyotyping
2.3. Mutational Analysis
2.4. Statistical Analysis
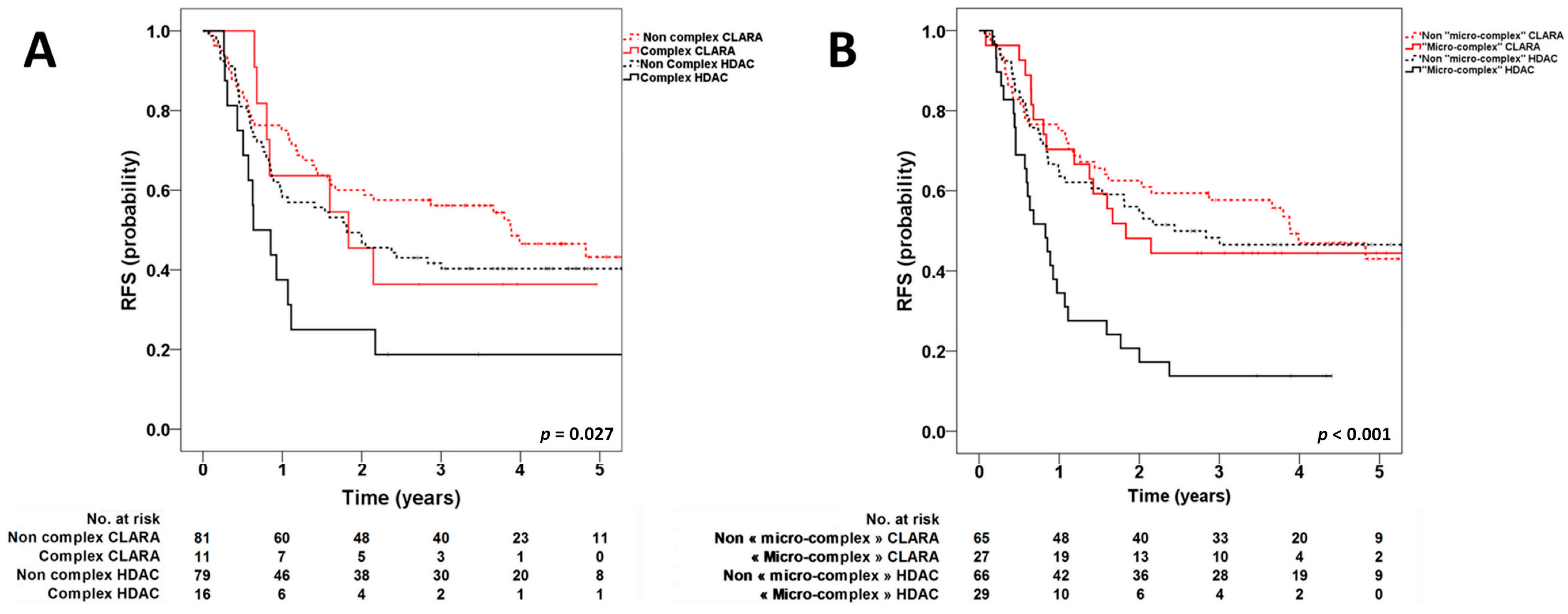
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimwade, D.; Hills, R.K.; Moorman, A.V.; Walker, H.; Chatters, S.; Goldstone, A.H.; Wheatley, K.; Harrison, C.J.; Burnett, A.K.; National Cancer Research Institute Adult Leukaemia Working Group. Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010, 116, 354–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renneville, A.; Abdelali, R.B.; Chevret, S.; Nibourel, O.; Cheok, M.; Pautas, C.; Duléry, R.; Boyer, T.; Cayuela, J.M.; Hayette, S.; et al. Clinical impact of gene mutations and lesions detected by SNP-array karyotyping in acute myeloid leukemia patients in the context of gemtuzumab ozogamicin treatment: Results of the ALFA-0701 trial. Oncotarget 2013, 5, 916–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bullinger, L.; Krönke, J.; Schön, C.; Radtke, I.; Urlbauer, K.; Botzenhardt, U.; Gaidzik, V.; Carió, A.; Senger, C.; Schlenk, R.F.; et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia 2010, 24, 438–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkin, B.; Erba, H.; Ouillette, P.; Roulston, D.; Purkayastha, A.; Karp, J.; Talpaz, M.; Kujawski, L.; Shakhan, S.; Li, C.; et al. Acquired genomic copy number aberrations and survival in adult acute myelogenous leukemia. Blood 2010, 116, 4958–4967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacoby, M.A.; Walter, M.J. Detection of copy number alterations in acute myeloid leukemia and myelodysplastic syndromes. Expert Rev. Mol. Diagn. 2012, 12, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Thomas, X.; de Botton, S.; Chevret, S.; Caillot, D.; Raffoux, E.; Lemasle, E.; Marolleau, J.P.; Berthon, C.; Pigneux, A.; Vey, N.; et al. Randomized Phase II Study of Clofarabine-Based Consolidation for Younger Adults With Acute Myeloid Leukemia in First Remission. J. Clin. Oncol. 2017, 35, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Duployez, N.; Boudry-Labis, E.; Roumier, C.; Boissel, N.; Petit, A.; Geffroy, S.; Helevaut, N.; Celli-Lebras, K.; Terré, C.; Fenneteau, O.; et al. SNP-array lesions in core binding factor acute myeloid leukemia. Oncotarget 2018, 9, 6478–6489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balsat, M.; Renneville, A.; Thomas, X.; de Botton, S.; Caillot, D.; Marceau, A.; Lemasle, E.; Marolleau, J.P.; Nibourel, O.; Berthon, C.; et al. Postinduction Minimal Residual Disease Predicts Outcome and Benefit From Allogeneic Stem Cell Transplantation in Acute Myeloid Leukemia With NPM1 Mutation: A Study by the Acute Leukemia French Association Group. J. Clin. Oncol. 2017, 35, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Kuykendall, A.; Duployez, N.; Boissel, N.; Lancet, J.E.; Welch, J.S. Acute Myeloid Leukemia: The Good, the Bad, and the Ugly. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 555–573. [Google Scholar] [CrossRef] [PubMed]
| Variables | Hazard Ratio | 95%CI | p-Value |
|---|---|---|---|
| HDAC | |||
| Micro-complex karyotype | 2.32 | 1.34–4.04 | <0.01 |
| Complex karyotype | 1.55 | 0.73–3.28 | 0.25 |
| TP53 mutation | 0.77 | 0.3–1.99 | 0.59 |
| CLARA | |||
| Micro-complex karyotype | 1.03 | 0.49–2.17 | 0.94 |
| Complex karyotype | 0.57 | 0.13–2.5 | 0.45 |
| TP53 mutation | 4.83 | 0.92–25.25 | 0.06 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fenwarth, L.; Duployez, N.; Thomas, X.; Boissel, N.; Geffroy, S.; Marceau-Renaut, A.; Caillot, D.; Raffoux, E.; Lemasle, E.; Marolleau, J.-P.; et al. Clofarabine Improves Relapse-Free Survival of Acute Myeloid Leukemia in Younger Adults with Micro-Complex Karyotype. Cancers 2020, 12, 88. https://doi.org/10.3390/cancers12010088
Fenwarth L, Duployez N, Thomas X, Boissel N, Geffroy S, Marceau-Renaut A, Caillot D, Raffoux E, Lemasle E, Marolleau J-P, et al. Clofarabine Improves Relapse-Free Survival of Acute Myeloid Leukemia in Younger Adults with Micro-Complex Karyotype. Cancers. 2020; 12(1):88. https://doi.org/10.3390/cancers12010088
Chicago/Turabian StyleFenwarth, Laurène, Nicolas Duployez, Xavier Thomas, Nicolas Boissel, Sandrine Geffroy, Alice Marceau-Renaut, Denis Caillot, Emmanuel Raffoux, Emilie Lemasle, Jean-Pierre Marolleau, and et al. 2020. "Clofarabine Improves Relapse-Free Survival of Acute Myeloid Leukemia in Younger Adults with Micro-Complex Karyotype" Cancers 12, no. 1: 88. https://doi.org/10.3390/cancers12010088
APA StyleFenwarth, L., Duployez, N., Thomas, X., Boissel, N., Geffroy, S., Marceau-Renaut, A., Caillot, D., Raffoux, E., Lemasle, E., Marolleau, J.-P., Berthon, C., Cheok, M. H., Peyrouze, P., Pigneux, A., Vey, N., Celli-Lebras, K., Terré, C., Preudhomme, C., & Dombret, H. (2020). Clofarabine Improves Relapse-Free Survival of Acute Myeloid Leukemia in Younger Adults with Micro-Complex Karyotype. Cancers, 12(1), 88. https://doi.org/10.3390/cancers12010088






