Characterization of JAK1 Pseudokinase Domain in Cytokine Signaling
Abstract
1. Introduction
2. Results
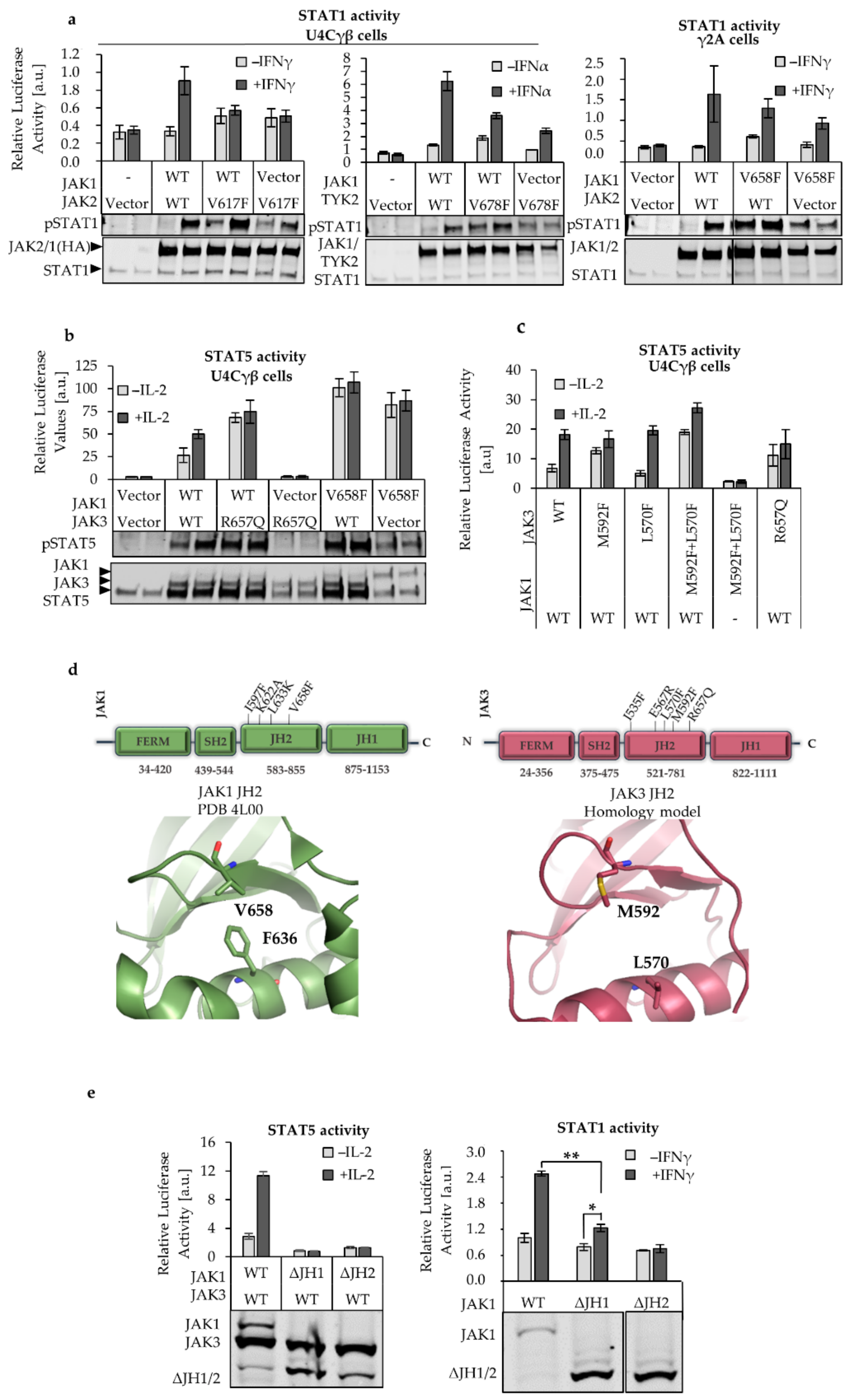
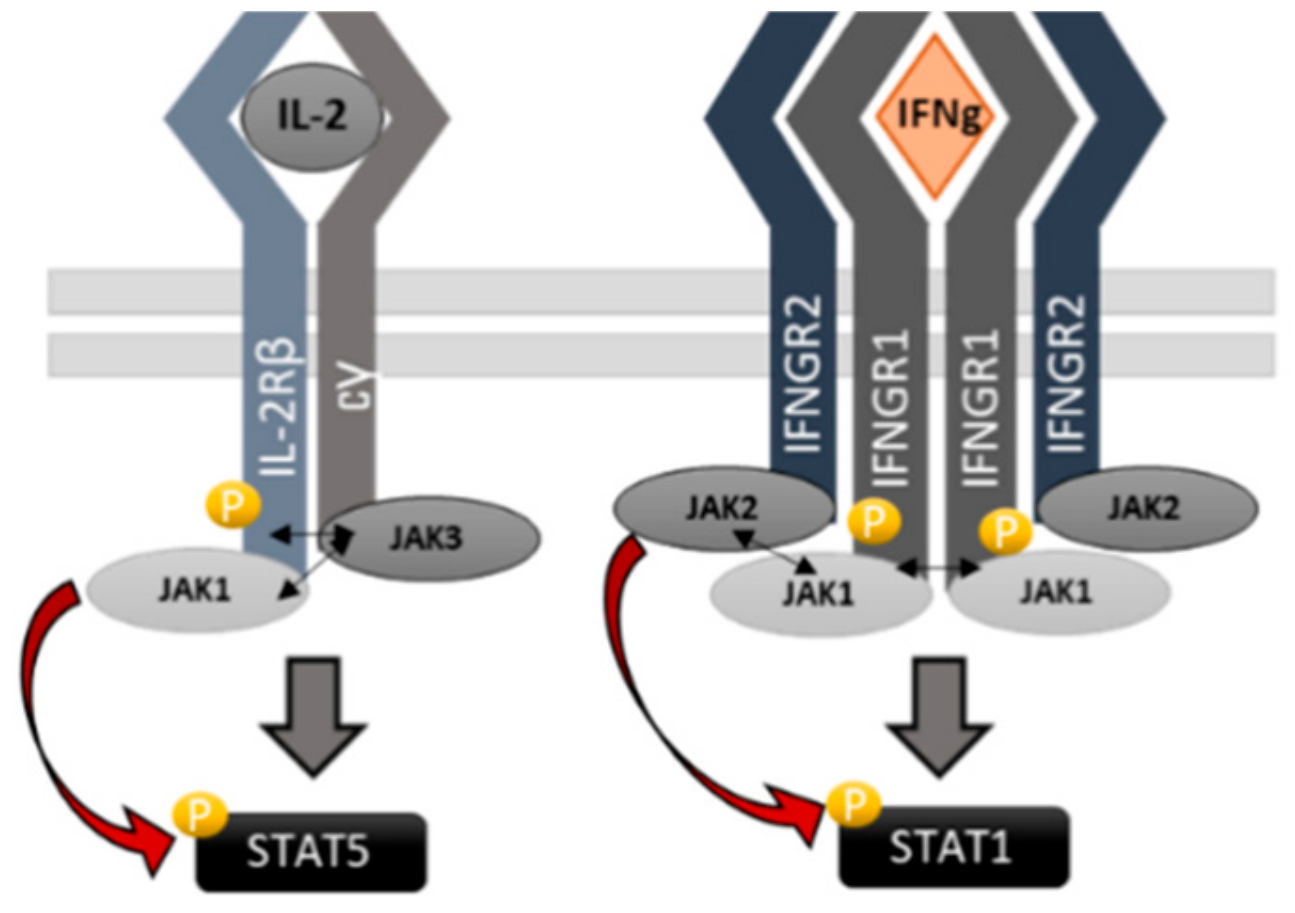
2.1. JAK1 Is Dominant STAT Activator in IL-2, but not in IFNγ and IFNα Signaling
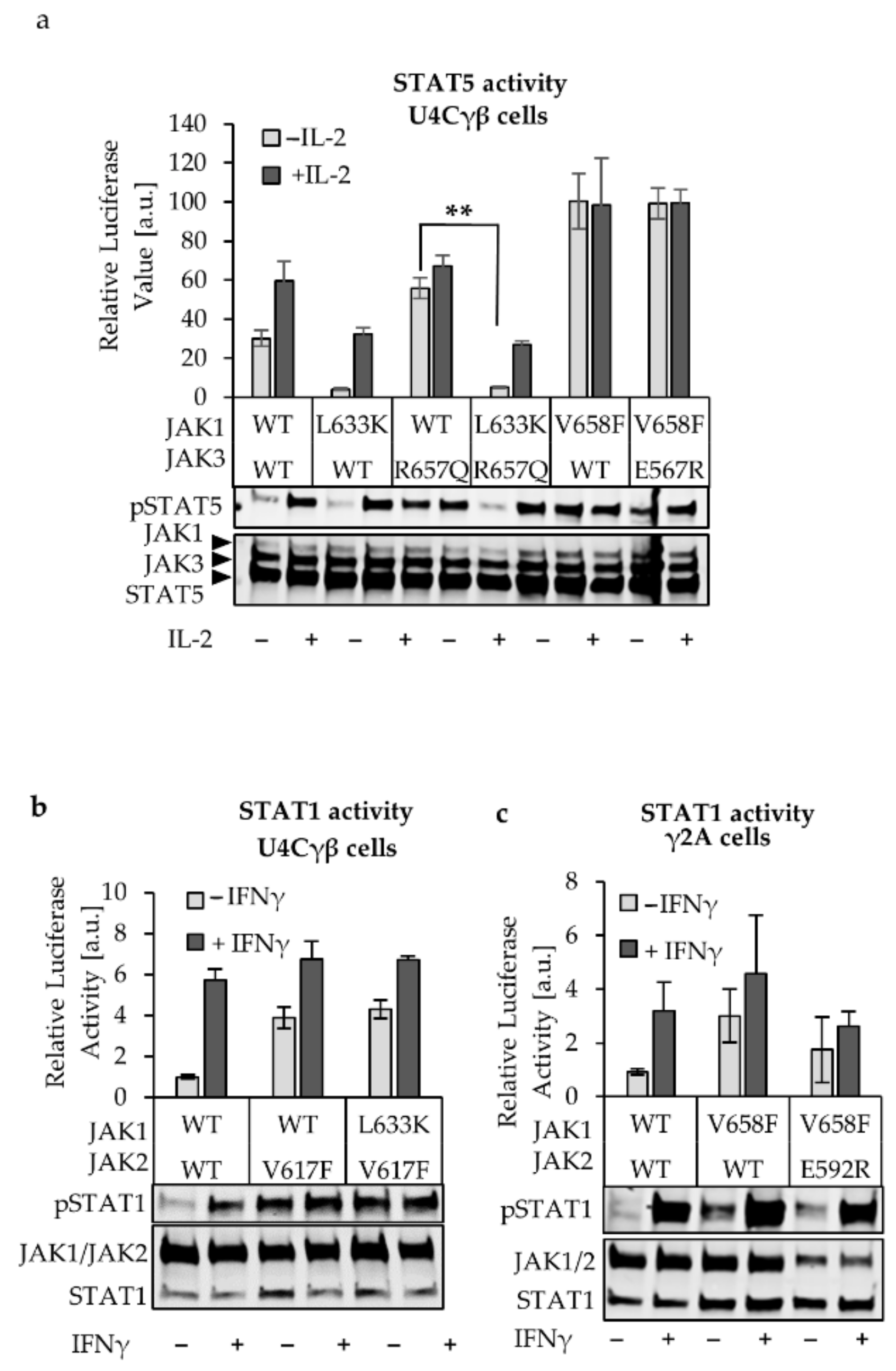
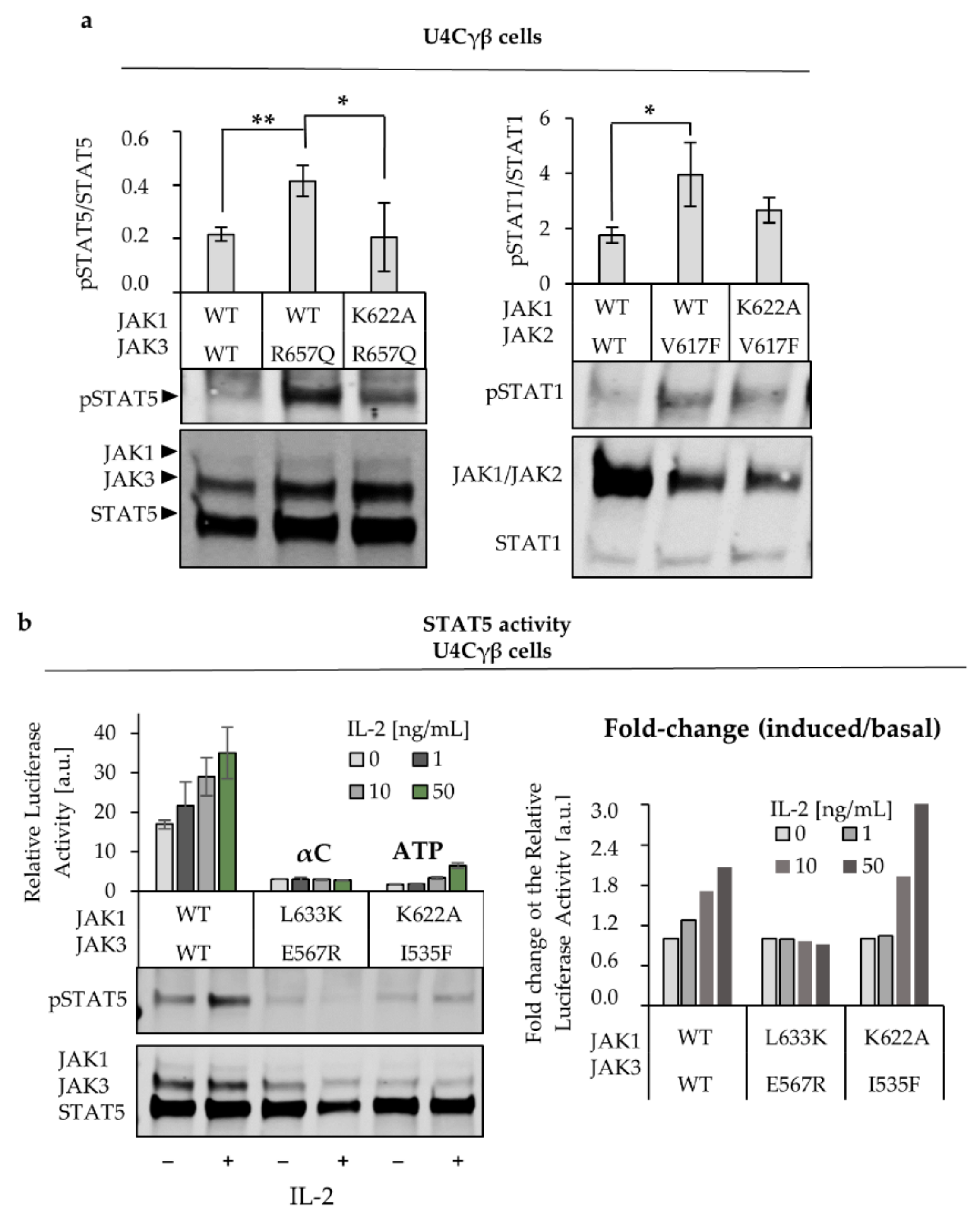
2.2. L633K in the Outer Face of JAK1 JH2 aC-helix Inhibits WT and Hyperactive IL-2, IFNγ, and IFNα Signaling at Variable Degrees
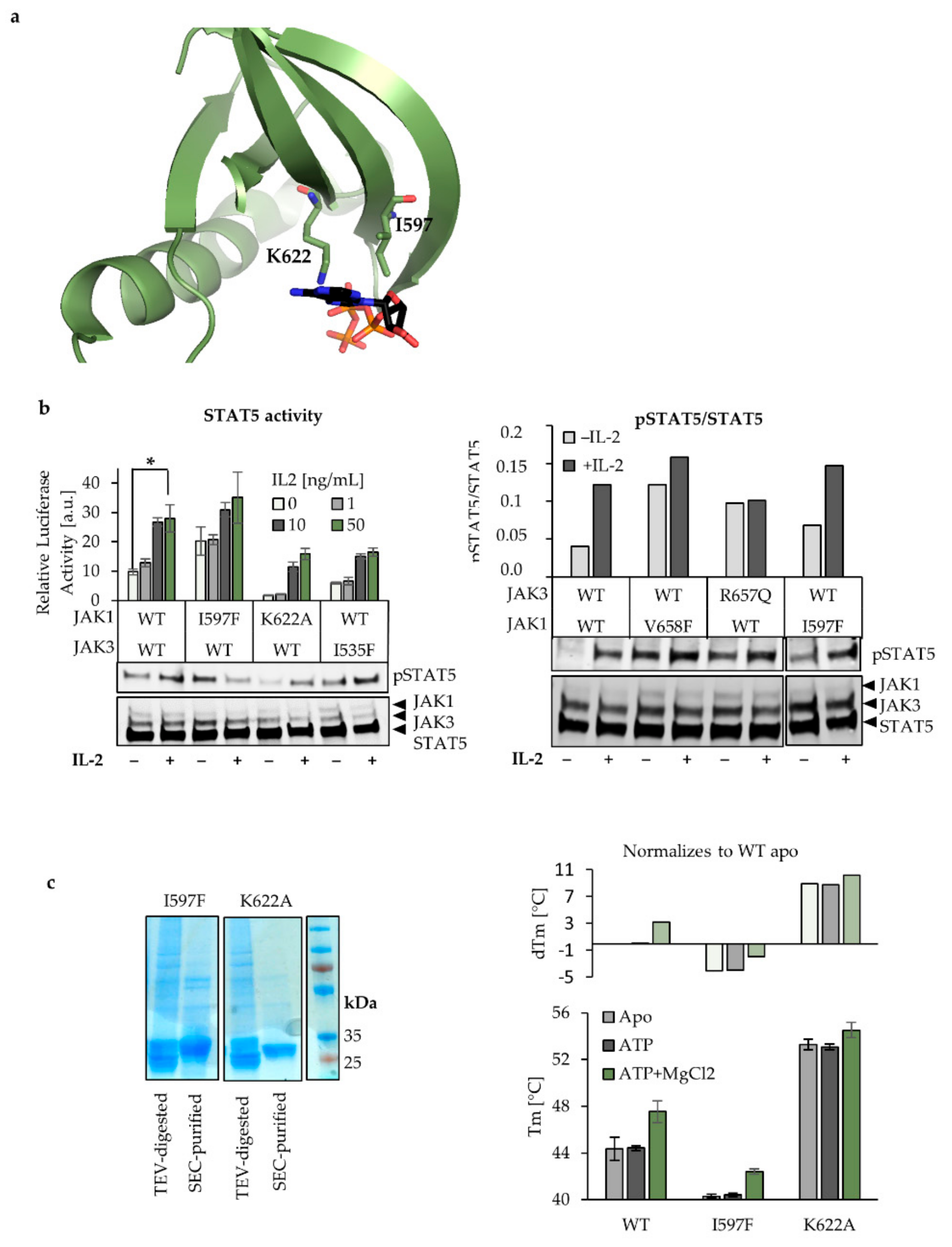
2.3. Characterization of ATP Binding to JAK1 JH2
2.4. JAK1 L633K Inhibits Hyperactive JAK3 but Does not Inhibit Hyperactive JAK2 and TYK2
2.5. JH2 Mutations in JAK Heterodimer Partners Show a Cumulative Inhibitory Effect
3. Discussion
4. Materials and Methods
4.1. Plasmid Constructs and Mutagenesis
4.2. Mammalian Cell Culture
4.3. Cell Transfection and Immunolabeling
4.4. Luciferase and Dual Luciferase Assays
4.5. Recombinant Protein Production and Purification
4.6. Differential Scanning Fluorometry (DSF)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schwartz, D.M.; Bonelli, M.; Gadina, M.; O’Shea, J.J. Type I/II cytokines, JAKs, and new strategies for treating autoimmune diseases. Nat. Rev. Rheumatol. 2016, 12, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Saharinen, P.; Silvennoinen, O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem. 2002, 277, 47954–47963. [Google Scholar] [CrossRef] [PubMed]
- Saharinen, P.; Takaluoma, K.; Silvennoinen, O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol. Cell. Biol. 2000, 20, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Sanz Sanz, A.; Niranjan, Y.; Hammarén, H.; Ungureanu, D.; Ruijtenbeek, R.; Touw, I.P.; Silvennoinen, O.; Hilhorst, R. The JH2 domain and SH2-JH2 linker regulate JAK2 activity: A detailed kinetic analysis of wild type and V617F mutant kinase domains. Biochim. Biophys. Acta BBA Proteins Proteom. 2014, 1844, 1835–1841. [Google Scholar] [CrossRef]
- Lupardus, P.J.; Ultsch, M.; Wallweber, H.; Bir Kohli, P.; Johnson, A.R.; Eigenbrot, C. Structure of the Pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 8025–8030. [Google Scholar] [CrossRef]
- Ungureanu, D.; Wu, J.; Pekkala, T.; Niranjan, Y.; Young, C.; Jensen, O.N.; Xu, C.F.; Neubert, T.A.; Skoda, R.C.; Hubbard, S.R.; et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat. Struct. Mol. Biol. 2011, 18, 971–976. [Google Scholar] [CrossRef]
- Shan, Y.; Gnanasambandan, K.; Ungureanu, D.; Kim, E.T.; Hammarén, H.; Yamashita, K.; Silvennoinen, O.; Shaw, D.E.; Hubbard, S.R. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 Tyrosine kinase. Nat. Struct. Mol. Biol. 2014, 21, 579–584. [Google Scholar] [CrossRef]
- Hammarén, H.M.; Ungureanu, D.; Grisouard, J.; Skoda, R.C.; Hubbard, S.R.; Silvennoinen, O. ATP Binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. USA. 2015, 112, 4642–4647. [Google Scholar] [CrossRef]
- Raivola, J.; Hammarén, H.M.; Virtanen, A.T.; Bulleeraz, V.; Ward, A.C.; Silvennoinen, O. Hyperactivation of oncogenic JAK3 mutants depend on ATP binding to the pseudokinase domain. Front. Oncol. 2018, 8, 560. [Google Scholar] [CrossRef]
- Hammarén, H.M.; Virtanen, A.T.; Raivola, J.; Silvennoinen, O. The regulation of JAKs in Cytokine signaling and its breakdown in disease. Cytokine 2019, 118, 48–63. [Google Scholar] [CrossRef]
- Candotti, F.; Oakes, S.A.; Johnston, J.A.; Giliani, S.; Schumacher, R.F.; Mella, P.; Fiorini, M.; Ugazio, A.G.; Badolato, R.; Notarangelo, L.D.; et al. Structural and functional basis for JAK3-deficient severe combined immunodeficiency. Blood 1997, 90, 3996–4003. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Husa, M.; Li, D.; Hofmann, S.R.; Watford, W.; Roberts, J.L.; Buckley, R.H.; Changelian, P.; Candotti, F. Jak3 and the pathogenesis of severe combined immunodeficiency. Mol. Immunol. 2004, 41, 727–737. [Google Scholar] [CrossRef]
- Levine, R.L.; Loriaux, M.; Huntly, B.J.; Loh, M.L.; Beran, M.; Stoffregen, E.; Berger, R.; Clark, J.J.; Willis, S.G.; Nguyen, K.T.; et al. The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood 2005, 106, 3377–3379. [Google Scholar] [CrossRef] [PubMed]
- Jelinek, J.; Oki, Y.; Gharibyan, V.; Bueso-Ramos, C.; Prchal, J.T.; Verstovsek, S.; Beran, M.; Estey, E.; Kantarjian, H.M.; Issa, J.P. JAK2 mutation 1849G>T is rare in acute leukemias but can be found in CMML, philadelphia chromosome-negative CML, and megakaryocytic leukemia. Blood 2005, 106, 3370–3373. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.M.; Scott, M.A.; Campbell, P.J.; Green, A.R. Progenitors homozygous for the V617F mutation occur in most patients with polycythemia vera, but not essential thrombocythemia. Blood 2006, 108, 2435–2437. [Google Scholar] [CrossRef] [PubMed]
- Bandaranayake, R.M.; Ungureanu, D.; Shan, Y.; Shaw, D.E.; Silvennoinen, O.; Hubbard, S.R. Crystal structures of the JAK2 Pseudokinase domain and the pathogenic mutant V617F. Nat. Struct. Mol. Biol. 2012, 19, 754–759. [Google Scholar] [CrossRef]
- Hammarén, H.M.; Virtanen, A.T.; Abraham, B.G.; Peussa, H.; Hubbard, S.R.; Silvennoinen, O. Janus kinase 2 activation mechanisms revealed by analysis of suppressing mutations. J. Allergy Clin. Immunol. 2019, 143, 1549–1559. [Google Scholar] [CrossRef]
- Leroy, E.; Balligand, T.; Pecquet, C.; Mouton, C.; Colau, D.; Shiau, A.K.; Dusa, A.; Constantinescu, S.N. Differential effect of inhibitory strategies of the V617 mutant of JAK2 on cytokine receptor signaling. J. Allergy Clin. Immunol. 2019, 144, 224–235. [Google Scholar] [CrossRef]
- Kornev, A.P.; Taylor, S.S. Dynamics-driven allostery in protein kinases. Trends Biochem. Sci. 2015, 40, 628–647. [Google Scholar] [CrossRef]
- Jeong, E.G.; Kim, M.S.; Nam, H.K.; Min, C.K.; Lee, S.; Chung, Y.J.; Yoo, N.J.; Lee, S.H. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin. Cancer Res. 2008, 14, 3716–3721. [Google Scholar] [CrossRef]
- Canté-Barrett, K.; Uitdehaag, J.C.M.; Meijerink, J.P.P. Structural modeling of JAK1 mutations in T-Cell acute lymphoblastic leukemia reveals a second contact site between pseudokinase and kinase domains. Haematologica 2016, 101, 189. [Google Scholar] [CrossRef] [PubMed]
- Flex, E.; Petrangeli, V.; Stella, L.; Chiaretti, S.; Hornakova, T.; Knoops, L.; Ariola, C.; Fodale, V.; Clappier, E.; Paoloni, F.; et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J. Exp. Med. 2008, 205, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Haan, C.; Behrmann, I.; Haan, S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of janus kinases. J. Cell. Mol. Med. 2010, 14, 504–527. [Google Scholar] [CrossRef] [PubMed]
- Moslin, R.; Gardner, D.; Santella, J.; Zhang, Y.; Duncia, J.V.; Liu, C.; Lin, J.; Tokarski, J.S.; Strnad, J.; Pedicord, D.; et al. Identification of imidazo[1,2-B] pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling. Med. Chem. Commun. 2017, 8, 700–712. [Google Scholar] [CrossRef]
- Taylor, S.S.; Kornev, A.P. Protein Kinases: Evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011, 36, 65–77. [Google Scholar] [CrossRef]
- Haan, C.; Rolvering, C.; Raulf, F.; Kapp, M.; Drückes, P.; Thoma, G.; Behrmann, I.; Zerwes, H. Jak1 has a dominant role over Jak3 in signal transduction through Γc-containing cytokine receptors. Chem. Biol. 2011, 18, 314–323. [Google Scholar] [CrossRef]
- Ferrao, R.D.; Wallweber, H.J.; Lupardus, P.J. Receptor-mediated dimerization of JAK2 FERM domains is required for JAK2 activation. eLife 2018, 7, 38089. [Google Scholar] [CrossRef]
- Dusa, A.; Mouton, C.; Pecquet, C.; Herman, M.; Constantinescu, S.N. JAK2 V617F constitutive activation requires JH2 residue F595: A pseudokinase domain target for specific inhibitors. PLoS ONE 2010, 5, e11157. [Google Scholar] [CrossRef]
- Gnanasambandan, K.; Magis, A.; Sayeski, P.P. The Constitutive activation of Jak2-V617F is mediated by a Pi stacking mechanism involving phenylalanines 595 and 617. Biochemistry 2010, 49, 9972–9984. [Google Scholar] [CrossRef]
- Toms, A.; Deshpande, A.; McNally, R.; Jeong, Y.; Rogers, J.; Un Kim, C.; Gruner, S.; Ficarro, S.; Marto, J.A.; Sattler, M.; et al. Structure of a pseudokinase domain switch that controls oncogenic activation of Jak kinases. Nat. Struct. Mol. Biol. 2013, 20, 1221. [Google Scholar] [CrossRef]
- Eletto, D.; Burns, S.O.; Angulo, I.; Plagnol, V.; Gilmour, K.C.; Henriquez, F.; Curtis, J.; Gaspar, M.; Nowak, K.; Daza-Cajigal, V.; et al. Biallelic JAK1 mutations in immunodeficient patient with mycobacterial infection. Nat. Commun. 2016, 7, 13992. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2018, 47, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Hammarén, H.M.; Virtanen, A.T.; Silvennoinen, O. Nucleotide-binding mechanisms in pseudokinases. Biosci. Rep. 2016, 36. [Google Scholar] [CrossRef] [PubMed]
- Rodig, S.J.; Meraz, M.A.; White, J.M.; Lampe, P.A.; Riley, J.K.; Arthur, C.D.; King, K.L.; Sheehan, K.C.; Yin, L.; Pennica, D.; et al. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the jaks in cytokine-induced biologic responses. Cell 1998, 93, 373–383. [Google Scholar] [CrossRef]
- Vainchenker, W.; Constantinescu, S.N. JAK/STAT signaling in hematological malignancies. Oncogene 2012, 32, 2601. [Google Scholar] [CrossRef]
- Kleppe, M.; Spitzer, M.H.; Li, S.; Hill, C.E.; Dong, L.; Papalexi, E.; De Groote, S.; Bowman, R.L.; Keller, M.; Koppikar, P.; et al. Jak1 integrates cytokine sensing to regulate hematopoietic stem cell function and stress hematopoiesis. Cell Stem Cell 2017, 21, 489–501. [Google Scholar] [CrossRef]
- Briscoe, J.; Rogers, N.C.; Witthuhn, B.A.; Watling, D.; Harpur, A.G.; Wilks, A.F.; Stark, G.R.; Ihle, J.N.; Kerr, I.M. Kinase-negative mutants of JAK1 can sustain interferon-gamma-inducible gene expression but not an antiviral state. EMBO J. 1996, 15, 799–809. [Google Scholar] [CrossRef]
- Haan, S.; Margue, C.; Engrand, A.; Rolvering, C.; Schmitz-Van, d.L.; Heinrich, P.C.; Behrmann, I.; Haan, C. Dual role of the Jak1 FERM and kinase domains in cytokine receptor binding and in stimulation-dependent jak activation. J. Immunol. 2008, 180, 998. [Google Scholar] [CrossRef]
- Greenlund, A.C.; Farrar, M.A.; Viviano, B.L.; Schreiber, R.D. Ligand-Induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91). EMBO J. 1994, 13, 1591–1600. [Google Scholar] [CrossRef]
- Keil, E.; Finkenstädt, D.; Wufka, C.; Trilling, M.; Liebfried, P.; Strobl, B.; Müller, M.; Pfeffer, K. Important scaffold function of the janus kinase 2 uncovered by a novel mouse model harboring a Jak2 activation-loop mutation. Blood 2014, 123, 520. [Google Scholar] [CrossRef]
- Wallweber, H.J.; Tam, C.; Franke, Y.; Starovasnik, M.A.; Lupardus, P.J. Structural Basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat. Struct. Mol. Biol. 2014, 21, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Ferrao, R.; Wallweber, H.J.; Ho, H.; Tam, C.; Franke, Y.; Quinn, J.; Lupardus, P.J. The structural basis for class II cytokine receptor recognition by JAK1. Structure 2016, 24, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wlodawer, A.; Lubkowski, J. Crystal Structure of a complex of the intracellular domain of interferon λ receptor 1 (IFNLR1) and the FERM/SH2 domains of human JAK1. J. Mol. Biol. 2016, 428, 4651–4668. [Google Scholar] [CrossRef] [PubMed]
- Ferrao, R.; Lupardus, P.J. The janus kinase (JAK) FERM and SH2 domains: Bringing specificity to JAK - receptor interactions. Front. Endocrinol. 2017, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.R. Mechanistic insights into regulation of JAK2 tyrosine kinase. Front. Endocrinol. 2018, 8, 361. [Google Scholar] [CrossRef] [PubMed]
- Lupardus, P.J.; Skiniotis, G.; Rice, A.J.; Thomas, C.; Fischer, S.; Walz, T.; Garcia, K.C. Structural snapshots of full-length Jak1, a transmembrane gp130/IL-6/IL-6Ralpha cytokine receptor complex, and the receptor-Jak1 holocomplex. Structure 2011, 19, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Koppikar, P.; Bhagwat, N.; Kilpivaara, O.; Manshouri, T.; Adli, M.; Hricik, T.; Liu, F.; Saunders, L.M.; Mullally, A.; Abdel-Wahab, O.; et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature 2012, 489, 155–159. [Google Scholar] [CrossRef]
- Meyer, S.C.; Keller, M.D.; Chiu, S.; Koppikar, P.; Guryanova, O.A.; Rapaport, F.; Xu, K.; Manova, K.; Pankov, D.; O’Reilly, R.J.; et al. CHZ868, a type II JAK2 inhibitor, reverses type I JAK inhibitor persistence and demonstrates efficacy in myeloproliferative neoplasms. Cancer. Cell. 2015, 28, 15–28. [Google Scholar] [CrossRef]
- Tvorogov, D.; Thomas, D.; Liau, N.P.D.; Dottore, M.; Barry, E.F.; Lathi, M.; Kan, W.L.; Hercus, T.R.; Stomski, F.; Hughes, T.P.; et al. Accumulation of JAK activation loop phosphorylation is linked to type I JAK inhibitor withdrawal syndrome in myelofibrosis. Sci. Adv. 2018, 4, 3834. [Google Scholar] [CrossRef]
- Leroy, E.; Dusa, A.; Colau, D.; Motamedi, A.; Cahu, X.; Mouton, C.; Huang, L.J.; Shiau, A.K.; Constantinescu, S.N. Uncoupling JAK2 V617F activation from cytokine-induced signalling by modulation of JH2 αC helix. Biochem. J. 2016, 473, 1579–1591. [Google Scholar] [CrossRef]
- Pine, R.; Canova, A.; Schindler, C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN alpha and IFN gamma, and is likely to autoregulate the p91 gene. EMBO J. 1994, 13, 158–167. [Google Scholar] [CrossRef] [PubMed]

| JAK | Mutation | Effect | Short Description. |
|---|---|---|---|
| JAK1 | L633K | LOF | At the solvent exposed face of the JH2 αC-helix, homologous to the JAK2 E592R. |
| I597F | GOF/- | Residing JH2 ATP-binding site and designed to inhibit ATP binding. Homologous to JAK2 I559F. | |
| K622A | LOF | Removes conserved β3 lysine in JH2. Designed to inhibit ATP binding. Homologous mutations shown to inhibit hyperactivation in JAK2 and JAK3. | |
| V658F | GOF | Homologous to JAK2 V617F and TYK2 V678FF. Resides in the JH2 β4-β5 loop and potentially disturbs the SH2-JH2 linker and causes cytokine independent activation. Mutation in JAK1 or JAK2 cause ALL. | |
| JAK2 | E592R | LOF | At the solvent exposed face of the JH2 αC-helix, shown to inhibit JAK2 V617F-driven dimerization of EPOR [19]. |
| I559F | LOF | In the JH2 β2. Designed to sterically inhibit ATP binding and shown to inhibit ATP binding in recombinant JH2 [8]. | |
| V617F | GOF | Homologous to JAK1 V658F and TYK2 V678F. | |
| TYK2 | L653R | LOF | At the solvent exposed face of the JH2 αC-helix, homologous to the JAK2 E592R. |
| V603F | LOF | At the ATP, binding pocket of JH2, designed to inhibit ATP binding. Homologous to JAK2 I559F. | |
| V678F | GOF | Constitutive active mutation in JH2. Homologous to JAK1 V658F and JAK1 V617F. | |
| JAK3 | E567R | LOF | Resides at the solvent exposed face of the JH2 αC-helix. Homologous to the JAK2 E592R. |
| I535F | LOF | At the ATP binding pocket of JH2 and homologous to JAK2 I559F.Shown to inhibit constitutive JAK3 activation [9]. | |
| R657Q | GOF | Activating mutation found in ALL patient. Resides in the JH1-JH2 interface. | |
| M592F | GOF | Homologous to constitutively active JAK1 V658F, JAK2 V617F and TYK2 V678F. | |
| L570F | - | Mutation designed to create the WT state as in JAK1, JAK2 and TYK2 (F595 in JAK2). Stacks with the mutated JAK2 V617F and enables the hyperactivation via the FFV-triad formation (see text). | |
| M592F + L570F | GOF | Double mutant designed to create a complete FFV-triad into JAK3 (see above). |
| Fw | Rev | |
|---|---|---|
| JAK1ΔJH2 (583–855) | atcctcaagaaggatctgaaaccagcaactgaagtggacccc | cttcagttgctggtttcagatccttcttgaggatccgatcg |
| JAK1Δ JH1-HA (583–1153) | ctgaaaccagcaactgaagtgtacccatacgatgttccagattacgcttag | ctaagcgtaatctggaacatcgtatgggtattcagttgctggtttcagatccttctt |
| JAK1 L633K | cagggatatttccaaggccttcttcgaggc | gcctcgaagaaggccttggaaatatccctg |
| JAK1 I597F | gagaacacacttctattctgggaccctgatgg | cccagaatagaagtgtgttctcgtgcctctcc |
| JAK1 V658F | ctatggcgtctgtttccgcgacgtggag | ctccacgtcgcggaaacagacgccatag |
| JAK1 K622A | gaagataaaagtgatcctcgcagtcttagaccccagccacagg | cctgtggctggggtctaagactgcgaggatcacttttatcttc |
| JAK2 E592R | gcacacagaaactattcacggtctttctttgaagcagc | gctgcttcaaagaaagaccgtgaatagtttctgtgtgc |
| JAK2 V617F | atggagtatgtttctgtggagacgagaatattctgg | tcgtctccacagaaacatactccataatttaaaacc |
| JAK2 I559F | ggccaaggcacttttacaaagttttttaaaggcgtacgaagagaagtagg | cctacttctcttcgtacgcctttaaaaaactttgtaaaagtgccttggcc |
| TYK2 V603F | cacaaggaccaacttctatgagggccgcc | ggcggccctcatagaagttggtccttgtg |
| TYK2 L653R | ccatgacatcgcccgggccttctacgagacagccagcc | cgtagaaggcccgggcgatgtcatggtgactagggtcc |
| TYK2 V678F | gcatggcgtctgtttccgcggccctga | tcagggccgcggaaacagacgccatgc |
| JAK3 I535F | ggtccttcaccaagttttaccggggctgtcgc | gcgacagccccggtaaaacttggtgaaggacc |
| JAK3 L570F | ggagtcattctttgaagcagcgagcttgatgagcc | ctcgctgcttcaaagaatgactccatgcagttcttgtgc |
| JAK3 M592F | ggcgtgtgctttgctggagacagcaccatggtgcagg | gtctccagcaaagcacacgccgtggagcagcacgagatgccgg |
| JAK3 E567R | gcacaagaactgcatgcgttcattcctggaagc | gcttccaggaatgaacgcatgcagttcttgtgc |
| JAK3 R657Q | aaggtgctcctggctcaggagggggctgatggg | cccatcagccccctcctgagccaggagcacctt |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raivola, J.; Haikarainen, T.; Silvennoinen, O. Characterization of JAK1 Pseudokinase Domain in Cytokine Signaling. Cancers 2020, 12, 78. https://doi.org/10.3390/cancers12010078
Raivola J, Haikarainen T, Silvennoinen O. Characterization of JAK1 Pseudokinase Domain in Cytokine Signaling. Cancers. 2020; 12(1):78. https://doi.org/10.3390/cancers12010078
Chicago/Turabian StyleRaivola, Juuli, Teemu Haikarainen, and Olli Silvennoinen. 2020. "Characterization of JAK1 Pseudokinase Domain in Cytokine Signaling" Cancers 12, no. 1: 78. https://doi.org/10.3390/cancers12010078
APA StyleRaivola, J., Haikarainen, T., & Silvennoinen, O. (2020). Characterization of JAK1 Pseudokinase Domain in Cytokine Signaling. Cancers, 12(1), 78. https://doi.org/10.3390/cancers12010078





