Oncogenic Role of an Epigenetic Reader of m6A RNA Modification: YTHDF1 in Merkel Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples and MCC Cell Lines
2.2. Immunohistochemistry (IHC) and Western Blot
2.3. Gene Expression Analysis and Copy Number Analysis
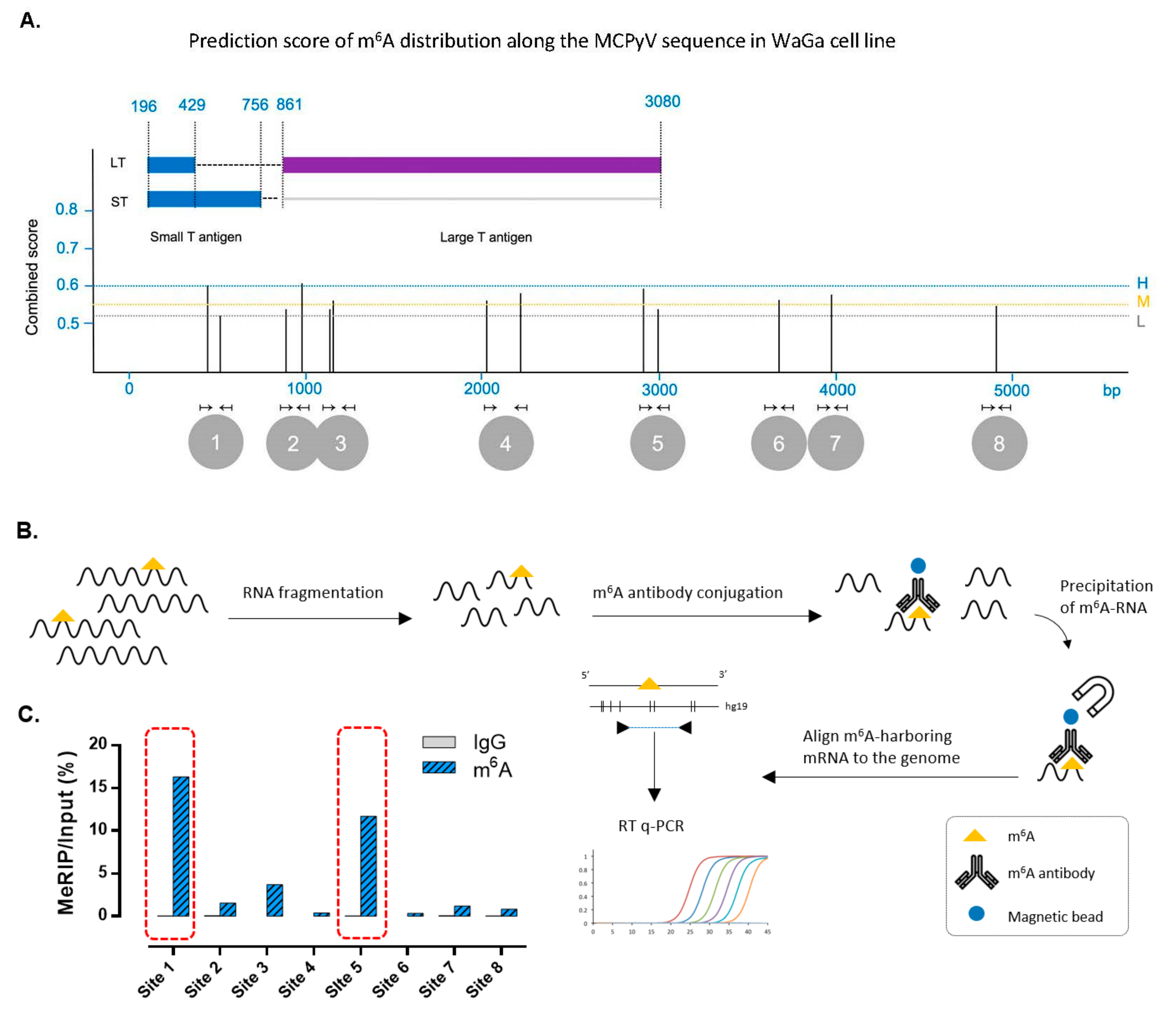
2.4. m6A Distribution Prediction
2.5. m6A Methylated RNA Immunoprecipitation (meRIP)
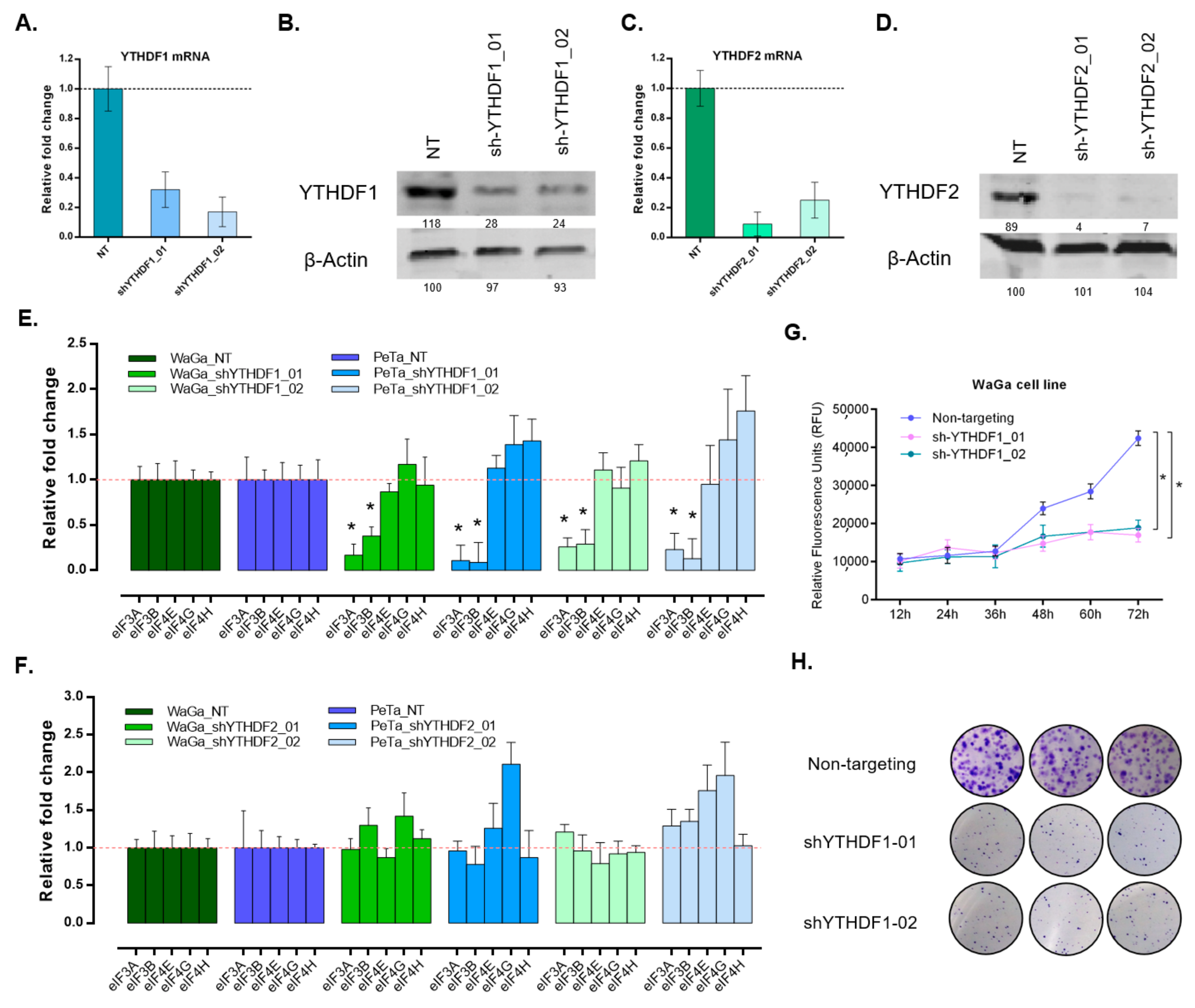
2.6. shRNA-Mediated Knockdown
2.7. Proliferation and Colony Formation Assay
2.8. Quantitative RT-PCR
3. Results
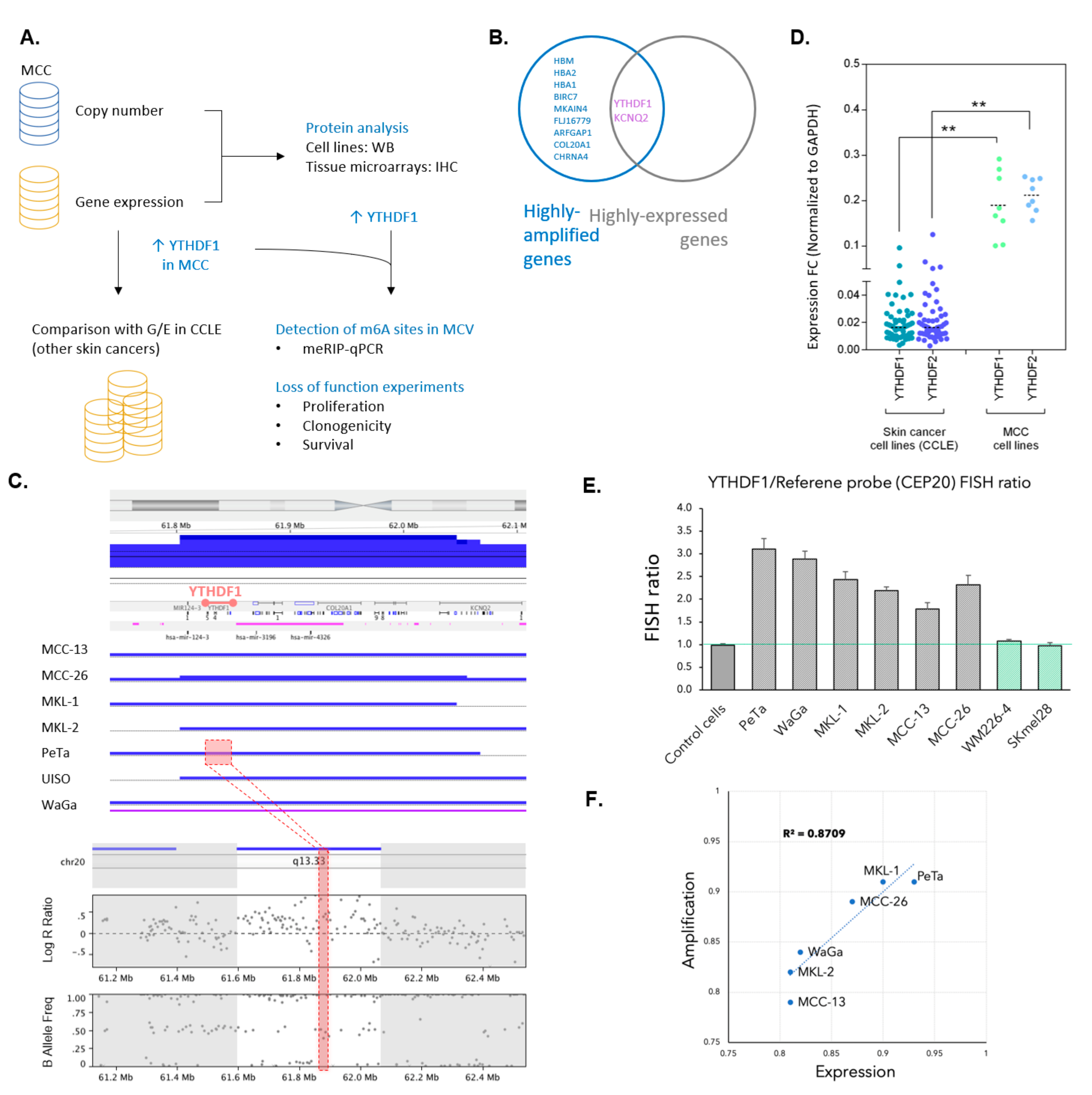
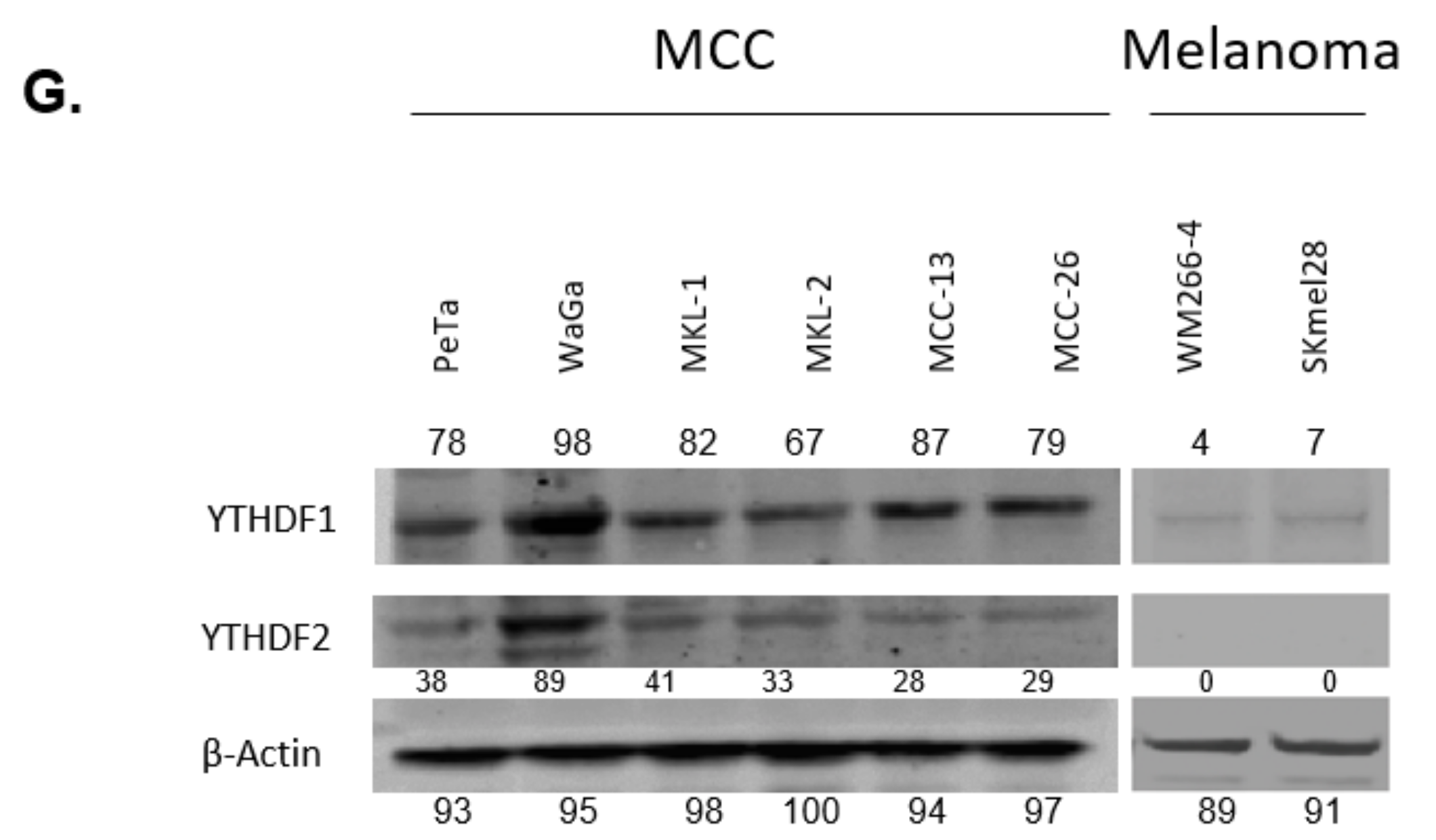
3.1. YTHDF1 Is Highly Amplified and Expressed in Merkel Cell Carcinoma Cell Lines
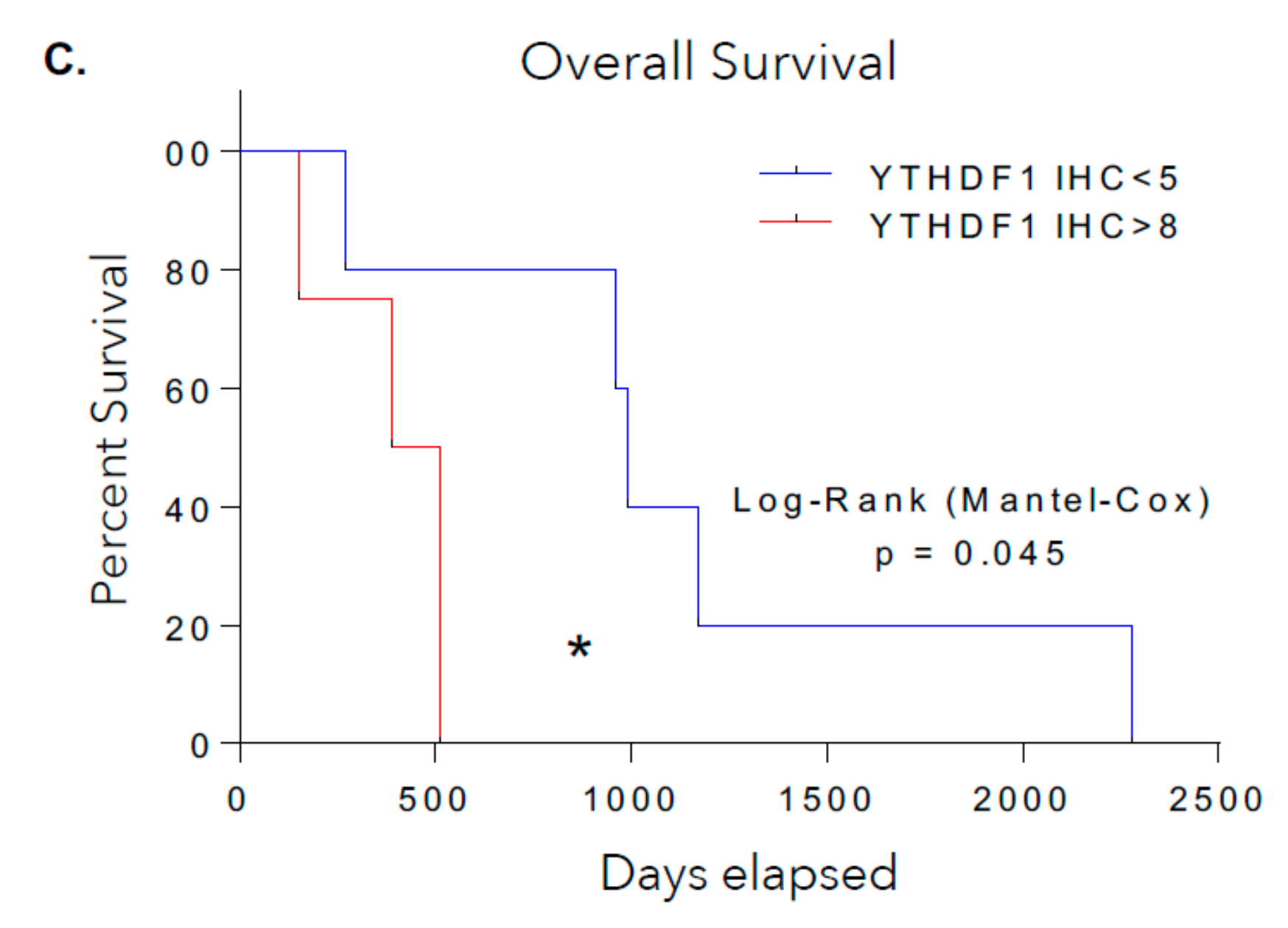
3.2. YTHDF1 Is Highly Amplified and Expressed in Merkel Cell Carcinoma Tumor Biopsies
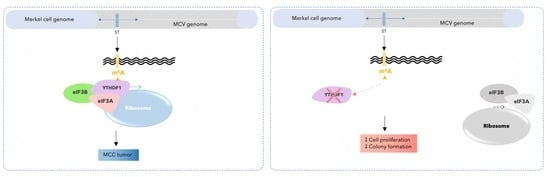
3.3. m6A RNA Modification Is Present in the Sequence of Merkel Cell Polyomavirus Transcripts in MCC Cell Lines
3.4. Silencing of YTHDF1 Impacts mRNA Translation Initiation and Reduces Proliferative and Clonogenic Capacity in MCC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schadendorf, D.; Lebbe, C.; Zur Hausen, A.; Avril, M.F.; Hariharan, S.; Bharmal, M.; Becker, J.C. Merkel cell carcinoma: Epidemiology, prognosis, therapy and unmet medical needs. Eur. J. Cancer 2017, 71, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Shuda, M.; Kwun, H.J.; Feng, H.; Chang, Y.; Moore, P.S. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J. Clin. Investig. 2011, 121, 3623–3634. [Google Scholar] [CrossRef] [PubMed]
- Canaani, D.; Kahana, C.; Lavi, S.; Groner, Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 1979, 6, 2879–2899. [Google Scholar] [CrossRef] [PubMed]
- Finkel, D.; Groner, Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology 1983, 131, 409–425. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef]
- Dimock, K.; Stoltzfus, C.M. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry 1977, 16, 471–478. [Google Scholar] [CrossRef]
- Krug, R.M.; Morgan, M.A.; Shatkin, A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5’-terminal 7-methylguanosine in cap structures. J. Virol. 1976, 20, 45–53. [Google Scholar] [CrossRef]
- Kane, S.E.; Beemon, K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Mol. Cell. Biol. 1985, 5, 2298–2306. [Google Scholar] [CrossRef]
- Sommer, S.; Salditt-Georgieff, M.; Bachenheimer, S.; Darnell, J.E.; Furuichi, Y.; Morgan, M.; Shatkin, A.J. The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res. 1976, 3, 749–765. [Google Scholar] [CrossRef] [PubMed]
- Lavi, S.; Shatkin, A.J. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc. Natl. Acad. Sci. USA 1975, 72, 2012–2016. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, N.S.; McIntyre, A.B.R.; McFadden, M.J.; Roder, A.E.; Kennedy, E.M.; Gandara, J.A.; Hopcraft, S.E.; Quicke, K.M.; Vazquez, C.; Willer, J.; et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 2016, 20, 654–665. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Simen Zhao, B.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Houben, R.; Grimm, J.; Willmes, C.; Weinkam, R.; Becker, J.C.; Schrama, D. Merkel cell carcinoma and Merkel cell polyomavirus: Evidence for hit-and-run oncogenesis. J. Investig. Dermatol. 2012, 132, 254–256. [Google Scholar] [CrossRef]
- Houben, R.; Shuda, M.; Weinkam, R.; Schrama, D.; Feng, H.; Chang, Y.; Moore, P.S.; Becker, J.C. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J. Virol. 2010, 84, 7064–7072. [Google Scholar] [CrossRef]
- Shuda, M.; Feng, H.; Kwun, H.J.; Rosen, S.T.; Gjoerup, O.; Moore, P.S.; Chang, Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. USA 2008, 105, 16272–16277. [Google Scholar] [CrossRef]
- Van Gele, M.; Kaghad, M.; Leonard, J.H.; Van Roy, N.; Naeyaert, J.M.; Geerts, M.L.; Van Belle, S.; Cocquyt, V.; Bridge, J.; Sciot, R.; et al. Mutation analysis of P73 and TP53 in Merkel cell carcinoma. Br. J. Cancer 2000, 82, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Leonard, J.H.; Dash, P.; Holland, P.; Kearsley, J.H.; Bell, J.R. Characterisation of four Merkel cell carcinoma adherent cell lines. Int. J. Cancer 1995, 60, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Ronan, S.G.; Green, A.D.; Shilkaitis, A.; Huang, T.S.; Das Gupta, T.K. Merkel cell carcinoma: In vitro and in vivo characteristics of a new cell line. J. Am. Acad. Dermatol. 1993, 29, 715–722. [Google Scholar] [CrossRef]
- Houben, R.; Dreher, C.; Angermeyer, S.; Borst, A.; Utikal, J.; Haferkamp, S.; Peitsch, W.K.; Schrama, D.; Hesbacher, S. Mechanisms of p53 restriction in Merkel cell carcinoma cells are independent of the Merkel cell polyoma virus T antigens. J. Investig. Dermatol. 2013, 133, 2453–2460. [Google Scholar] [CrossRef] [PubMed]
- Orouji, E.; Orouji, A.; Gaiser, T.; Larribere, L.; Gebhardt, C.; Utikal, J. MAP kinase pathway gene copy alterations in NRAS/BRAF wild-type advanced melanoma. Int. J. Cancer 2016, 138, 2257–2262. [Google Scholar] [CrossRef]
- Zhou, Y.; Zeng, P.; Li, Y.H.; Zhang, Z.; Cui, Q. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016, 44, e91. [Google Scholar] [CrossRef]
- Orouji, E.; Federico, A.; Larribere, L.; Novak, D.; Lipka, D.B.; Assenov, Y.; Sachindra, S.; Huser, L.; Granados, K.; Gebhardt, C.; et al. Histone methyltransferase SETDB1 contributes to melanoma tumorigenesis and serves as a new potential therapeutic target. Int. J. Cancer 2019, 145, 3462–3477. [Google Scholar] [CrossRef]
- Tsai, K.; Courtney, D.G.; Cullen, B.R. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog. 2018, 14, e1006919. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bogler, O.; et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef]
- Verhaegen, M.E.; Mangelberger, D.; Harms, P.W.; Eberl, M.; Wilbert, D.M.; Meireles, J.; Bichakjian, C.K.; Saunders, T.L.; Wong, S.Y.; Dlugosz, A.A. Merkel Cell Polyomavirus Small T Antigen Initiates Merkel Cell Carcinoma-like Tumor Development in Mice. Cancer Res. 2017, 77, 3151–3157. [Google Scholar] [CrossRef] [PubMed]
- Verhaegen, M.E.; Mangelberger, D.; Harms, P.W.; Vozheiko, T.D.; Weick, J.W.; Wilbert, D.M.; Saunders, T.L.; Ermilov, A.N.; Bichakjian, C.K.; Johnson, T.M.; et al. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J. Investig. Dermatol. 2015, 135, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Fan, S.; Wu, M.; Zuo, Z.; Li, X.; Jiang, L.; Shen, Q.; Xu, P.; Zeng, L.; Zhou, Y.; et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 2019, 10, 4892. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orouji, E.; Peitsch, W.K.; Orouji, A.; Houben, R.; Utikal, J. Oncogenic Role of an Epigenetic Reader of m6A RNA Modification: YTHDF1 in Merkel Cell Carcinoma. Cancers 2020, 12, 202. https://doi.org/10.3390/cancers12010202
Orouji E, Peitsch WK, Orouji A, Houben R, Utikal J. Oncogenic Role of an Epigenetic Reader of m6A RNA Modification: YTHDF1 in Merkel Cell Carcinoma. Cancers. 2020; 12(1):202. https://doi.org/10.3390/cancers12010202
Chicago/Turabian StyleOrouji, Elias, Wiebke K. Peitsch, Azadeh Orouji, Roland Houben, and Jochen Utikal. 2020. "Oncogenic Role of an Epigenetic Reader of m6A RNA Modification: YTHDF1 in Merkel Cell Carcinoma" Cancers 12, no. 1: 202. https://doi.org/10.3390/cancers12010202
APA StyleOrouji, E., Peitsch, W. K., Orouji, A., Houben, R., & Utikal, J. (2020). Oncogenic Role of an Epigenetic Reader of m6A RNA Modification: YTHDF1 in Merkel Cell Carcinoma. Cancers, 12(1), 202. https://doi.org/10.3390/cancers12010202