Should Tumor Infiltrating Lymphocytes, Androgen Receptor, and FOXA1 Expression Predict the Clinical Outcome in Triple Negative Breast Cancer Patients?
Abstract
1. Introduction
2. Results
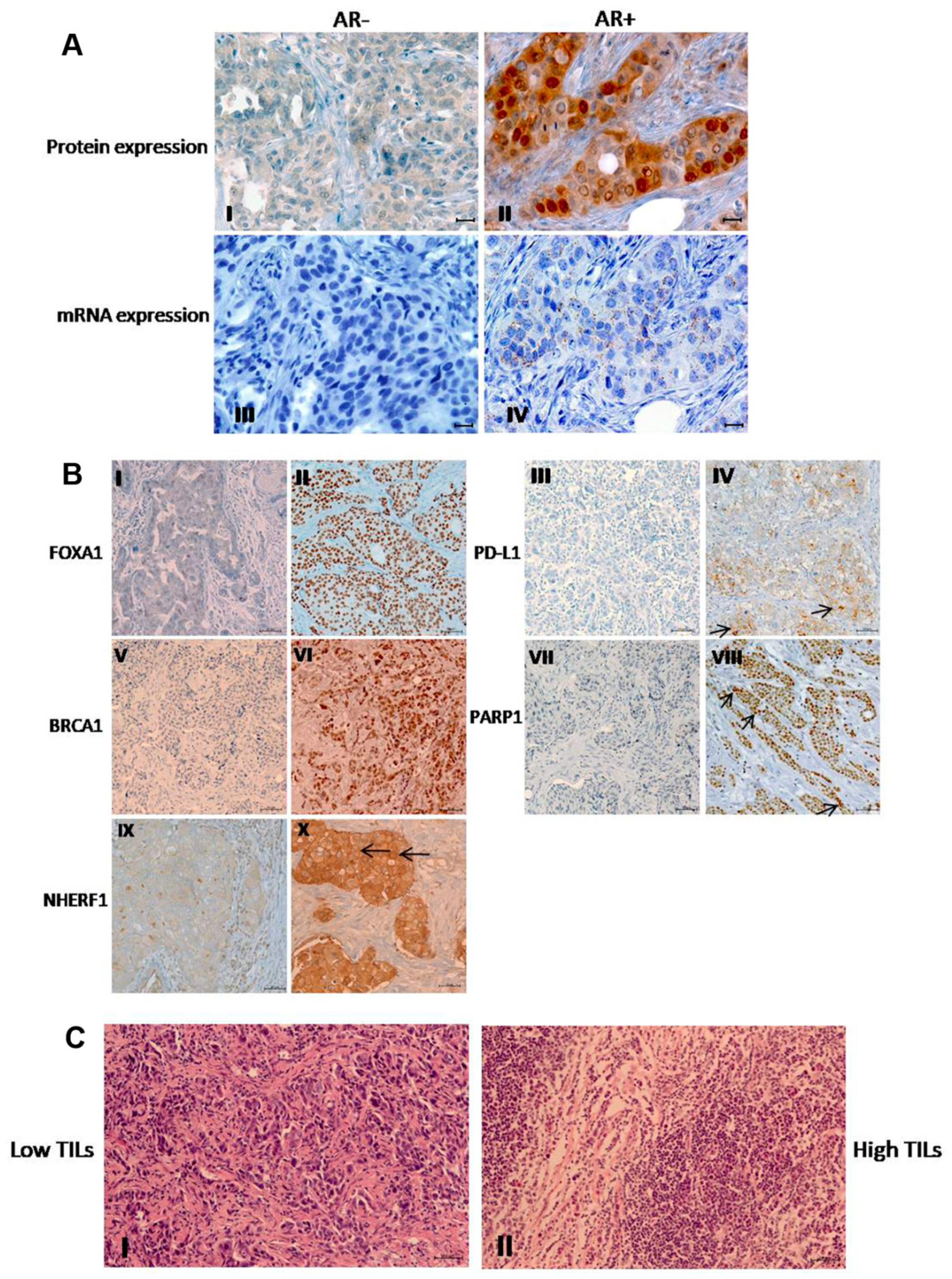
2.1. Protein Expression Profiling of AR, FOXA1, PD-L1, BRCA1, PARP1, and NHERF1
2.2. Relationship Between Tumor Markers Expression and Clinicopathological Features
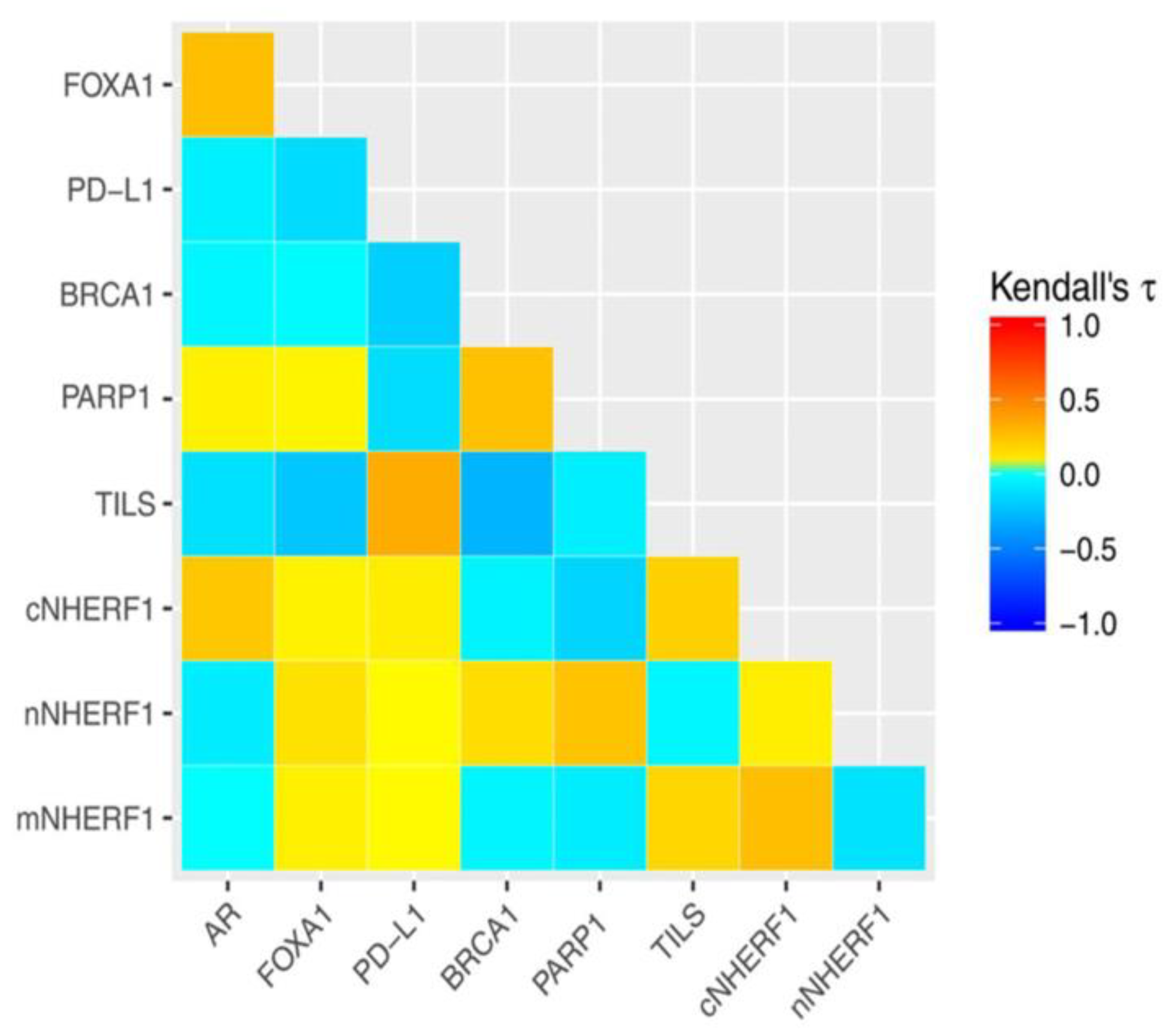
2.3. Association Between Protein Expressions Analyzed
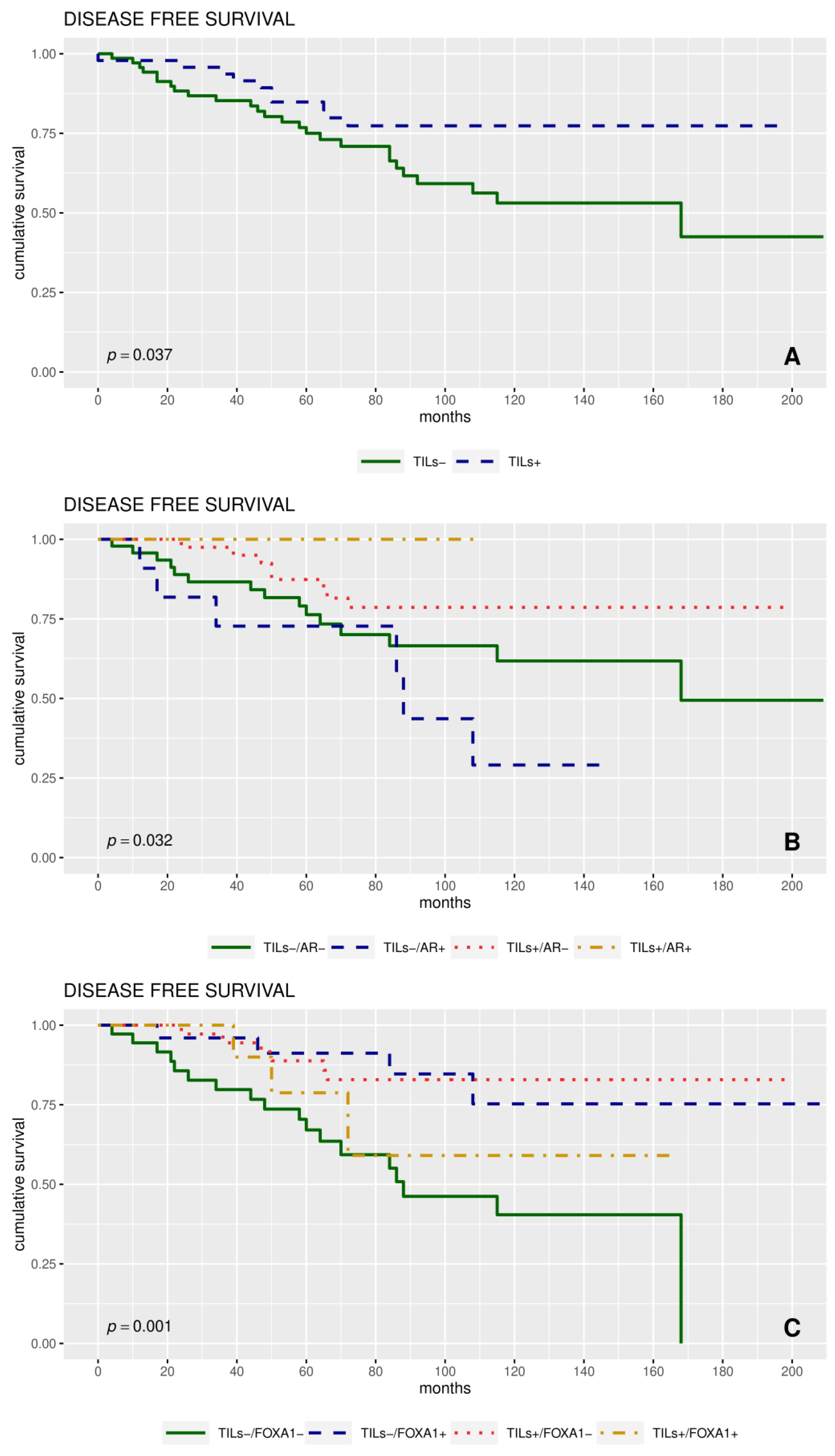
2.4. Expression of Proteins and Patient Clinical Outcome
3. Discussion
4. Materials and Methods
4.1. Patients and Clinicopathological Characteristics
4.2. TMA and Immunohistochemistry
4.3. Immunohistochemical Assessment
4.4. RNA Scope
4.5. Follow Up and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sharma, P. Biology and management of patients with Triple-Negative Breast Cancer. Oncologist 2016, 21, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Altekruse, S.F.; Li, C.I.; Chen, V.W.; Clarke, C.A.; Ries, L.A.; Cronin, K.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 2014, 106, pii: dju055. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [PubMed]
- Wahba, H.A.; El-Hadaad, H.A. Current approaches in treatment of triple-negative breast cancer. Cancer Biol. Med. 2015, 12, 106–116. [Google Scholar] [PubMed]
- Early Breast Cancer Trialists’Collaborative Group (EBCTCG). Comparisons between different polychemotherapy regimens for early breast cancer: Meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet 2012, 379, 432–444. [Google Scholar] [CrossRef]
- Pruneri, G.; Gray, K.P.; Vingiani, A.; Viale, G.; Curigliano, G.; Criscitiello, C.; Láng, I.; Ruhstaller, T.; Gianni, L.; Goldhirsch, A.; et al. Tumor-infiltrating lymphocytes (TILs) are a powerful prognostic marker in patients with triple-negative breast cancer enrolled in the IBCSG phase III randomized clinical trial 22-00. Breast Cancer Res. Treat. 2016, 158, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Müller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef]
- Hida, A.I.; Watanabe, T.; Sagara, Y.; Kashiwaba, M.; Sagara, Y.; Aogi, K.; Ohi, Y.; Tanimoto, A. Diffuse distribution of tumor-infiltrating lymphocytes is a marker for better prognosis and chemotherapeutic effect in triple-negative breast cancer. Breast Cancer Res. Treat. 2019, 1–12. [Google Scholar] [CrossRef]
- Stanton, S.E.; Adams, S.; Disis, M.L. Variation in the incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes: A systematic review. JAMA Oncol. 2016, 2, 1354–1360. [Google Scholar] [CrossRef]
- Basile, D.; Garattini, S.K.; Bonotto, M.; Ongaro, E.; Casagrande, M.; Cattaneo, M.; Fanotto, V.; Carlo, E.D.; Loupakis, F.; Urbano, F.; et al. Immunotherapy for colorectal cancer: Where are we heading? Expert Opin. Biol. Ther. 2017, 17, 709–721. [Google Scholar] [CrossRef]
- Gardini, A.C.; Scarpi, E.; Faloppi, L.; Scartozzi, M.; Silvestris, N.; Santini, D.; de Stefano, G.; Marisi, G.; Negri, F.V.; Foschi, F.G.; et al. Immune inflammation indicators and implication for immune modulation strategies in advanced hepatocellular carcinoma patients receiving sorafenib. Oncotarget 2016, 7, 67142. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Criscitiello, C.; Goubar, A.; Viale, G.; Conte, P.; Guarneri, V.; Ficarra, G.; Mathieu, M.C.; Delaloge, S.; Curigliano, G.; et al. Prognostic value of tumor-infiltrating lymphocytes on residual disease after primary chemotherapy for triple-negative breast cancer: A retrospective multicenter study. Ann. Oncol. 2014, 25, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, M.; Jiang, Z.; Wang, X.A. Comprehensive Immunologic Portrait of Triple-Negative Breast. Cancer Transl. Oncol. 2018, 11, 311–329. [Google Scholar] [CrossRef] [PubMed]
- Saleh, R.; Taha, R.Z.; Sasidharan Nair, V.; Alajez, N.M.; Elkord, E. PD-L1 Blockade by Atezolizumab Downregulates Signaling Pathways Associated with Tumor Growth, Metastasis, and Hypoxia in Human Triple Negative Breast Cancer. Cancers 2019, 11, 1050. [Google Scholar] [CrossRef] [PubMed]
- Raninga, P.V.; Lee, A.C.; Sinha, D.; Shih, Y.Y.; Mittal, D.; Makhale, A.; Bain, A.L.; Nanayakarra, D.; Tonissen, K.F.; Kalimutho, M.; et al. Therapeutic cooperation between auranofin, a thioredoxin reductase inhibitor and anti-PD-L1 antibody for treatment of triple-negative breast cancer. Int. J. Cancer. 2019. [Google Scholar] [CrossRef] [PubMed]
- Guiu, S.; Mollevi, C.; Charon-Barra, C.; Boissière, F.; Crapez, E.; Chartron, E.; Lamy, P.J.; Gutowski, M.; Bourgier, C.; Romieu, G.; et al. Prognostic value of androgen receptor and FOXA1 co-expression in non-metastatic triple negative breast cancer and correlation with other biomarkers. Br. J. Cancer. 2018, 119, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Simental, J.A.; Sar, M.; Lane, M.V.; French, F.S.; Wilson, E.M. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 1991, 266, 510–518. [Google Scholar]
- Desai, K.V.; Michalowska, A.M.; Kondaiah, P.; Ward, J.M.; Shih, J.H.; Green, J.E. Gene expression profiling identifies a unique androgen-mediated inflammatory/immune signature and a PTEN (phosphatase and tensin homolog deleted on chromosome 10)-mediated apoptotic response specific to the rat ventral prostate. Mol. Endocrinol. 2004, 18, 2895–2907. [Google Scholar] [CrossRef]
- Liu, Y.X.; Zhang, K.J.; Tang, L.L. Clinical significance of androgen receptor expression in triple negative breast cancer-an immunohistochemistry study. Oncol. Lett. 2018, 15, 10008–10016. [Google Scholar] [CrossRef]
- Ni, M.; Chen, Y.; Lim, E.; Wimberly, H.; Bailey, S.T.; Imai, Y.; Rimm, D.L.; Liu, X.S.; Brown, M. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell 2011, 20, 119–131. [Google Scholar] [CrossRef]
- Barton, V.N.; Christenson, J.L.; Gordon, M.A.; Greene, L.I.; Rogers, T.J.; Butterfield, K.; Babbs, B.; Spoelstra, N.S.; D’Amato, N.C.; Elias, A.; et al. Androgen receptor supports an anchorage-independent, cancer stem cell-like population in triple-negative breast cancer. Cancer Res. 2017, 77, 3455–3466. [Google Scholar] [CrossRef] [PubMed]
- De Amicis, F.; Thirugnansampanthan, J.; Cui, Y.; Selever, J.; Beyer, A.; Parra, I.; Weigel, N.L.; Herynk, M.H.; Tsimelzon, A.; Lewis, M.T.; et al. Androgen receptor overexpression induces tamoxifen resistance in human breast cancer cells. Breast Cancer Res. Treat. 2010, 121, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.L.; Macarthur, S.; Ross-Innes, C.S.; Tilley, W.D.; Neal, D.E.; Mills, I.G.; Carroll, J.S. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011, 30, 3019–3027. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, A.; Holmes, K.A.; Ross-Innes, C.S.; Schmidt, D.; Carroll, J.S. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011, 43, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.J.; Jain, R.K.; Leung, S.; Choo, J.; Nielsen, T.; Huntsman, D.; Nakshatri, H.; Badve, S. FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Cancer Res. Treat. 2012, 131, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.S.; Liu, X.S.; Brodsky, A.S.; Li, W.; Meyer, C.A.; Szary, A.J.; Eeckhoute, J.; Shao, W.; Hestermann, E.V.; Geistlinger, T.R.; et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 2005, 122, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Laganiere, J.; Deblois, G.; Lefebvre, C.; Bataille, A.R.; Robert, F.; Giguère, V. From the cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. USA. 2005, 102, 11651–11656. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Fujino, K.; Monteiro, L.J.; Gomes, A.R.; Drost, R.; Davidson-Smith, H.; Takeda, S.; Khoo, U.S.; Jonkers, J.; Sproul, D.; et al. FOXA1 repression is associated with loss of BRCA1 and increased promoter methylation and chromatin silencing in breast cancer. Oncogene 2015, 34, 5012–5024. [Google Scholar] [CrossRef]
- Wu, P.E.; Shen, C.Y. ‘Hide-then-hit’ to explain the importance of genotypic polymorphism of DNA repair genes in determining susceptibility to cancer. J. Mol. Cell Biol. 2011, 3, 59–65. [Google Scholar] [CrossRef][Green Version]
- Saponaro, C.; Vagheggini, A.; Scarpi, E.; Centonze, M.; Catacchio, I.; Popescu, O.; Pastena, M.I.; Giotta, F.; Silvestris, N.; Mangia, A. NHERF1 and tumor microenvironment: A new scene in invasive breast carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 96. [Google Scholar] [CrossRef] [PubMed]
- Mangia, A.; Scarpi, E.; Partipilo, G.; Schirosi, L.; Opinto, G.; Giotta, F.; Simone, G. NHERF1 together with PARP1 and BRCA1 expression as a new potential biomarker to stratify breast cancer patients. Oncotarget 2017, 8, 65730–65742. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Tsvetkova, V.; Orvieto, E.; Piacentini, F.; Ficarra, G.; Griguolo, G.; Miglietta, F.; Giarratano, T.; Omarini, C.; Bonaguro, S.; et al. Immune characterization of breast cancer metastases: Prognostic implications. Breast Cancer Res. 2018, 20, 62. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, Z.; Wang, Z.; Wu, P.; Qiu, F.; Huang, J. Prognostic and predictive value of tumor-infiltrating lymphocytes in breast cancer: A systematic review and meta-analysis. Clin. Transl. Oncol. 2016, 18, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Qu, Q.; Chen, X.; Huang, O.; Wu, J.; Shen, K. The Prognostic Value of Tumor-Infiltrating Lymphocytes in Breast Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0152500. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Lai, Y.; Xu, J.; Huang, J. Prognostic value of FOXA1 in breast cancer: A systematic review and meta-analysis. Breast 2016, 27, 35–43. [Google Scholar] [CrossRef] [PubMed]
- McGhan, L.J.; McCullough, A.E.; Protheroe, C.A.; Dueck, A.C.; Lee, J.J.; Nunez-Nateras, R.; Castle, E.P.; Gray, R.J.; Wasif, N.; Goetz, M.P.; et al. Androgen receptor-positive triple negative breast cancer: A unique breast cancer subtype. Ann. Surg. Oncol. 2014, 21, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Pan, B.; Zhu, H.; Zhou, Y.; Mao, F.; Lin, Y.; Xu, Q.; Sun, Q. Prognostic value of androgen receptor in triple negative breast cancer: A meta-analysis. Oncotarget 2016, 7, 46482–46491. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.E.; Kang, S.H.; Lee, S.J.; Bae, Y.K. Androgen receptor expression predicts decreased survival in early stage triple-negative breast cancer. Ann. Surg. Oncol. 2015, 22, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Asano, Y.; Kashiwagi, S.; Goto, W.; Tanaka, S.; Morisaki, T.; Takashima, T.; Noda, S.; Onoda, N.; Ohsawa, M.; Hirakawa, K.; et al. Expression and clinical significance of androgen receptor in triple negative breast cancer. Cancers (Basel) 2017, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jae, E.; Yoon, M. Influence of Androgen Receptor Expression on the Survival Outcomes in Breast Cancer: A Meta-Analysis. J. Breast Cancer 2015, 18, 134–142. [Google Scholar] [CrossRef]
- Vera-Badillo, F.E.; Templeton, A.J.; de Gouveia, P.; Diaz-Padilla, I.; Bedard, P.L.; Al-Mubarak, M.; Seruga, B.; Tannock, I.F.; Ocana, A.; Amir, E. Androgen receptor expression and outcomes in early breast cancer: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2014, 106, djt319. [Google Scholar] [CrossRef] [PubMed]
- McNamara, K.M.; Yoda, T.; Miki, Y.; Chanplakorn, N.; Wongwaisayawan, S.; Incharoen, P.; Kongdan, Y.; Wang, L.; Takagi, K.; Mayu, T.; et al. Androgenic pathway in triple negative invasive ductal tumors: Its correlation with tumor cell proliferation. Cancer Sci. 2013, 104, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Paradiso, A.; Scarpi, E.; Malfettone, A.; Addati, T.; Giotta, F.; Simone, G.; Amadori, D.; Mangia, A. Nuclear NHERF1 expression as a prognostic marker in breast cancer. Cell Death Dis. 2013, 4, e904. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Yu, H.; Hao, C.; Martin, T.A.; Hargest, R.; He, J.; Cheng, S.; Jiang, W.G. NHERF1 regulates the progression of colorectal cancer through the interplay with VEGFR2 pathway. Oncotarget 2017, 8, 7753–7765. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, C.; Scarpi, E.; Zito, F.A.; Giotta, F.; Silvestris, N.; Mangia, A. Independent Negative Prognostic Role of TCF1 Expression within the Wnt/β-Catenin Signaling Pathway in Primary Breast Cancer Patients. Cancers (Basel) 2019, 11, 1035. [Google Scholar] [CrossRef] [PubMed]
- Massink, M.P.; Kooi, I.E.; van Mil, S.E.; Jordanova, E.S.; Ameziane, N.; Dorsman, J.C.; van Beek, D.M.; van der Voorn, J.P.; Sie, D.; Ylstra, B.; et al. Proper genomic profiling of (BRCA1-mutated) basal-like breast carcinomas requires prior removal of tumor infiltrating lymphocytes. Mol. Oncol. 2015, 9, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Mathot, L.; Stenninger, J. Behavior of seeds and soil in the mechanism of metastasis: A deeper understanding. Cancer Sci. 2012, 103, 626–631. [Google Scholar] [CrossRef]
- Adams, S.; Loi, S.; Toppmeyer, D.; Cescon, D.W.; De Laurentiis, M.; Nanda, R.; Winer, E.P.; Mukai, H.; Tamura, K.; Armstrong, A.; et al. Phase 2 study of pembrolizumab as first-line therapy for PD-L1–positive metastatic triple-negative breast cancer (mTNBC): Preliminary data from KEYNOTE-086 cohort B. J. Clin. Oncol. 2017, 35, 1088. [Google Scholar] [CrossRef]
- Polk, A.; Svane, I.M.; Andersson, M.; Nielsen, D. Checkpoint inhibitors in breast cancer current status. Cancer Treat. Rev. 2018, 63, 122–134. [Google Scholar] [CrossRef]
- Mori, H.; Kubo, M.; Yamaguchi, R.; Nishimura, R.; Osako, T.; Arima, N.; Okumura, Y.; Okido, M.; Yamada, M.; et al. The combination of PD-L1 expression and decreased tumor-infiltrating lymphocytes is associated with a poor prognosis in triple-negative breast cancer. Oncotarget 2017, 8, 15584–15592. [Google Scholar] [CrossRef]
- Birner, P.; Oberhuber, G.; Stani, J.; Reithofer, C.; Samonigg, H.; Hausmaninger, H.; Kubista, E.; Kwasny, W.; Kandioler-Eckersberger, D.; Gnant, M.; et al. Evaluation of the United States Food and Drug Administration-approved scoring and test system of HER-2 protein expression in breast cancer. Clin. Cancer Res. 2001, 7, 1669–1675. [Google Scholar] [PubMed]
- Wolff, A.C.; Hammond, M.E.; Schwartz, J.N.; Hagerty, K.L.; Allred, D.C.; Cote, R.J.; Dowsett, M.; Fitzgibbons, P.L.; Hanna, W.M.; Langer, A.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 2007, 25, 118–145. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Thorat, M.A.; Marchio, C.; Morimiya, A.; Savage, K.; Nakshatri, H.; Reis-Filho, J.S.; Badve, S. Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J. Clin. Pathol. 2008, 61, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Fan, X.; Zhu, W.; Huang, C.; Zhuang, W.; Xu, H.; Lin, X.; Hu, D.; Huang, Y.; Jiang, K.; et al. Prognostic significance of PD-L1 expression and tumor infiltrating lymphocyte in surgically resectable non-small cell lung cancer. Oncotarget 2017, 8, 83986–83994. [Google Scholar] [CrossRef]
- Mazzotta, A.; Partipilo, G.; De Summa, S.; Giotta, F.; Simone, G.; Mangia, A. Nuclear PARP1 expression and its prognostic significance in breast cancer patients. Tumour Biol. 2016, 37, 6143–6153. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org (accessed on 5 January 2017).

| AR | FOXA1 | PD-L1 | BRCA1 | PARP1 | TILS | cNHERF1 | nNHERF1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| τ | p-value | τ | p-value | τ | p-value | τ | p-value | τ | p-value | τ | p-value | τ | p-value | τ | p-value | |
| FOXA1 | 0.354 | <0.001 | ||||||||||||||
| PD-L1 | −0.022 | 0.803 | −0.110 | 0.173 | ||||||||||||
| BRCA1 | −0.068 | 0.425 | −0.038 | 0.623 | −0.164 | 0.054 | ||||||||||
| PARP1 | 0.126 | 0.143 | 0.080 | 0.307 | −0.046 | 0.595 | 0.248 | 0.001 | ||||||||
| TILS | −0.116 | 0.145 | −0.124 | 0.085 | 0.259 | 0.001 | −0.172 | 0.020 | −0.003 | 0.966 | ||||||
| cNHERF1 | 0.241 | 0.002 | 0.129 | 0.073 | 0.138 | 0.079 | −0.060 | 0.418 | −0.051 | 0.496 | 0.199 | 0.004 | ||||
| nNHERF1 | −0.071 | 0.429 | 0.098 | 0.229 | 0.064 | 0.462 | 0.134 | 0.106 | 0.297 | <0.001 | −0.040 | 0.602 | −0.057 | 0.455 | ||
| mNHERF1 | 0.037 | 0.668 | 0.011 | 0.888 | −0.029 | 0.732 | −0.004 | 0.959 | −0.242 | 0.003 | 0.168 | 0.024 | 0.221 | 0.002 | −0.111 | 0.177 |
| Characteristics | pts. | DFS | OS | |||||
|---|---|---|---|---|---|---|---|---|
| Events | 5-year DFS (95% CI) a | HR (95% CI) b | p-Value c | Events | 5-year OS (95% CI) a | p-Value c | ||
| Overall | 124 | 36 | 80 (73–88) | 12 | 92 (87–97) | |||
| Age | ||||||||
| ≤51 | 65 | 22 | 79 (70–90) | 1.00 | 0.432 | 5 | 93 (87–100) | 0.460 |
| >51 | 59 | 14 | 81 (71−92) | 0.765 (0.39–1.50) | 7 | 89 (82-98) | ||
| Pre/post | ||||||||
| Post | 73 | 25 | 81 (73–90) | 1.00 | 0.810 | 7 | 94 (88–99) | 0.391 |
| Pre | 41 | 11 | 79 (66–93) | 1.09 (0.53–2.23) | 5 | 87 (77–98) | ||
| Size | ||||||||
| ≤2 cm | 59 | 15 | 87 (78–96) | 1.00 | 0.552 | 4 | 93 (86–100) | 0.431 |
| >2 cm | 64 | 21 | 74 (64–86) | 1.22 (0.63–2.38) | 8 | 90 (83–98) | ||
| Histological type | ||||||||
| IDC | 111 | 33 | 81 (74–89) | - | 0.947 | 9 | 93 (89–98) | 0.027 |
| ILC | 4 | 1 | 75 (43–100) | - | 0 | 100 (100–100) | ||
| Other | 9 | 2 | 74 (48–100) | - | 3 | 67 (42–100) | ||
| Lymph node status | ||||||||
| Negative | 65 | 16 | 83 (74–93) | 1.00 | 0.578 | 6 | 90 (83-98) | 0.894 |
| Positive | 55 | 17 | 79 (69–91) | 1.21 (0.61−2.40) | 6 | 92 (85–100) | ||
| Ki67 | ||||||||
| Negative (≤20%) | 12 | 4 | 82 (62–100) | 1.00 | 0.843 | 1 | 100 (100–100) | 0.945 |
| Positive (>20%) | 110 | 30 | 81 (73–89) | 0.90 (0.32–2.56) | 11 | 90 (85–96) | ||
| AR | ||||||||
| Negative (<10%) | 92 | 23 | 83 (75–91) | 1.00 | 0.219 | 7 | 93 (88–99) | 0.111 |
| Positive (≥10%) | 16 | 6 | 81 (64–100) | 1.75 (0.70–4.33) | 3 | 87 (71−100) | ||
| FOXA1 | ||||||||
| Negative | 76 | 24 | 79 (71−89) | 1.00 | 0.281 | 8 | 90 (84–97) | 0.990 |
| Positive | 37 | 7 | 88 (77–100) | 0.63 (0.27–1.47) | 4 | 91 (82–100) | ||
| PD-L1 | ||||||||
| Negative (<1%) | 83 | 24 | 83 (75–92) | 1.00 | 0.460 | 7 | 94 (88–99) | 0.432 |
| Positive (≥1%) | 22 | 7 | 79 (66–96) | 0.73 (0.31−1.69) | 4 | 86 (75–100) | ||
| BRCA1 | ||||||||
| Negative (<3%) | 46 | 11 | 84 (74–96) | 1.00 | 0.273 | 1 | 100 (100–100) | 0.030 |
| Positive (≥3%) | 60 | 18 | 82 (72–93) | 1.52 (0.72–3.23) | 8 | 88 (80–97) | ||
| PARP1 | ||||||||
| Negative (0–9) | 87 | 24 | 83 (75–91) | 1.00 | 0.523 | 6 | 95 (91−100) | 0.032 |
| Positive (10–18) | 20 | 6 | 85 (71−100) | 1.34 (0.55–3.28) | 4 | 80 (63–100) | ||
| TILs | ||||||||
| Negative (<50%) | 70 | 26 | 75 (65–87) | 1.00 | 0.037 | 10 | 88 (80–96) | 0.019 |
| Positive (≥50%) | 47 | 10 | 84 (75–96) | 0.47 (0.23–0.971) | 1 | 98 (94–100) | ||
| mNHERF1 | ||||||||
| Negative (0%) | 76 | 19 | 81 (73–91) | 1.00 | 0.478 | 8 | 89 (82–96) | 0.435 |
| Positive (>0%) | 42 | 14 | 78 (66–92) | 1.28 (0.64–2.57) | 3 | 95 (88–100) | ||
| cNHERF1 | ||||||||
| Negative (70%) | 57 | 14 | 81 (71−93) | 1.00 | 0.645 | 7 | 87 (78–96) | 0.202 |
| Positive (≥70%) | 61 | 19 | 79 (69–90) | 1.18 (0.59–2.35) | 4 | 95 (89–100) | ||
| nNHERF1 | ||||||||
| Negative (0%) | 98 | 17 | 78 (70–87) | 1.00 | 0.276 | 8 | 93 (87–98) | 0.321 |
| Positive (>0%) | 20 | 3 | 89 (76–100) | 0.52 (0.16–1.71) | 3 | 83 (68–100) | ||
| Characteristics | Categorical | Continuous | ||
|---|---|---|---|---|
| HR (95% CI) a | p-Value a | HR (95% CI) | p-Value a | |
| AR | 1.65 (0.54–5.02) | 0.382 | 1.00 (0.99–1.03) | 0.509 |
| FOXA1 | 0.38 (0.12–1.20) | 0.100 | 0.64 (0.16–2.48) | 0.517 |
| PD-L1 | 0.49 (0.13–1.76) | 0.272 | 1.00 (0.96–1.04) | 0.863 |
| BRCA1 | 1.47 (0.53–4.11) | 0.459 | 1.00 (0.99–1.02) | 0.386 |
| PARP1 | 1.40 (0.46–4.32) | 0.554 | 1.00 (0.99–1.02) | 0.583 |
| TILS | 0.34 (0.12–0.98) | 0.045 | 0.19 (0.02–1.51) | 0.116 |
| mNHERF1 | 2.28 (0.89–5.82) | 0.085 | 1.01 (1.00–1.02) | 0.104 |
| cNHERF1 | 0.92 (0.37–2.32) | 0.867 | 1.00 (0.99–1.02) | 0.805 |
| nNHERF1 | 0.62 (0.13–3.00) | 0.548 | 0.88 (0.70–1.10) | 0.263 |
| Characteristics | n | (%) |
|---|---|---|
| Patients Age: median value 51 (range 26–80) | ||
| ≤51 years | 65 | (52.4) |
| >51 years | 59 | (47.6) |
| Menopausal status | ||
| Pre | 83 | (66.9) |
| Post | 41 | (33.1) |
| Histological type | ||
| IDC | 111 | (89.5) |
| ILC | 4 | (3.2) |
| Other | 9 | (7.3) |
| Histological grade | ||
| G1 | 1 | (0.8) |
| G2 | 23 | (18.7) |
| G3 | 99 | (80.5) |
| Unknown | 1 | |
| Tumor size (cm) | ||
| ≤2 cm | 59 | (48.0) |
| >2 cm | 64 | (52.0) |
| Unknown | 1 | |
| Lymph node status | ||
| Negative | 65 | (54.2) |
| Positive | 55 | (45.8) |
| Unknown | 4 | |
| Ki67 | ||
| Negative (≤20%) | 12 | (9.8) |
| Positive (>20%) | 110 | (90.2) |
| Unknown | 2 | |
| TILs | ||
| Negative (<50%) | 70 | (59.8) |
| Positive (≥50%) | 47 | (40.2) |
| Unknown | 7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mangia, A.; Saponaro, C.; Vagheggini, A.; Opinto, G.; Centonze, M.; Vicenti, C.; Popescu, O.; Pastena, M.; Giotta, F.; Silvestris, N. Should Tumor Infiltrating Lymphocytes, Androgen Receptor, and FOXA1 Expression Predict the Clinical Outcome in Triple Negative Breast Cancer Patients? Cancers 2019, 11, 1393. https://doi.org/10.3390/cancers11091393
Mangia A, Saponaro C, Vagheggini A, Opinto G, Centonze M, Vicenti C, Popescu O, Pastena M, Giotta F, Silvestris N. Should Tumor Infiltrating Lymphocytes, Androgen Receptor, and FOXA1 Expression Predict the Clinical Outcome in Triple Negative Breast Cancer Patients? Cancers. 2019; 11(9):1393. https://doi.org/10.3390/cancers11091393
Chicago/Turabian StyleMangia, Anita, Concetta Saponaro, Alessandro Vagheggini, Giuseppina Opinto, Matteo Centonze, Chiara Vicenti, Ondina Popescu, Maria Pastena, Francesco Giotta, and Nicola Silvestris. 2019. "Should Tumor Infiltrating Lymphocytes, Androgen Receptor, and FOXA1 Expression Predict the Clinical Outcome in Triple Negative Breast Cancer Patients?" Cancers 11, no. 9: 1393. https://doi.org/10.3390/cancers11091393
APA StyleMangia, A., Saponaro, C., Vagheggini, A., Opinto, G., Centonze, M., Vicenti, C., Popescu, O., Pastena, M., Giotta, F., & Silvestris, N. (2019). Should Tumor Infiltrating Lymphocytes, Androgen Receptor, and FOXA1 Expression Predict the Clinical Outcome in Triple Negative Breast Cancer Patients? Cancers, 11(9), 1393. https://doi.org/10.3390/cancers11091393






