MicroRNA Regulation of the Autotaxin-Lysophosphatidic Acid Signaling Axis
Abstract
1. Introduction to MiRNAs
2. Overview of Lysophosphatidic Acid Signaling and Autotaxin in Cancer
2.1. Lysophosphatidic Acid Signaling in Cancer
2.2. Autotaxin in Cancers
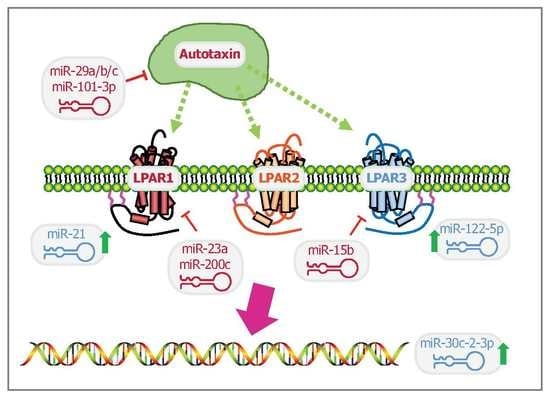
3. MiRNA as a Puppet Master of Lysophosphatidic Acid Signaling Events
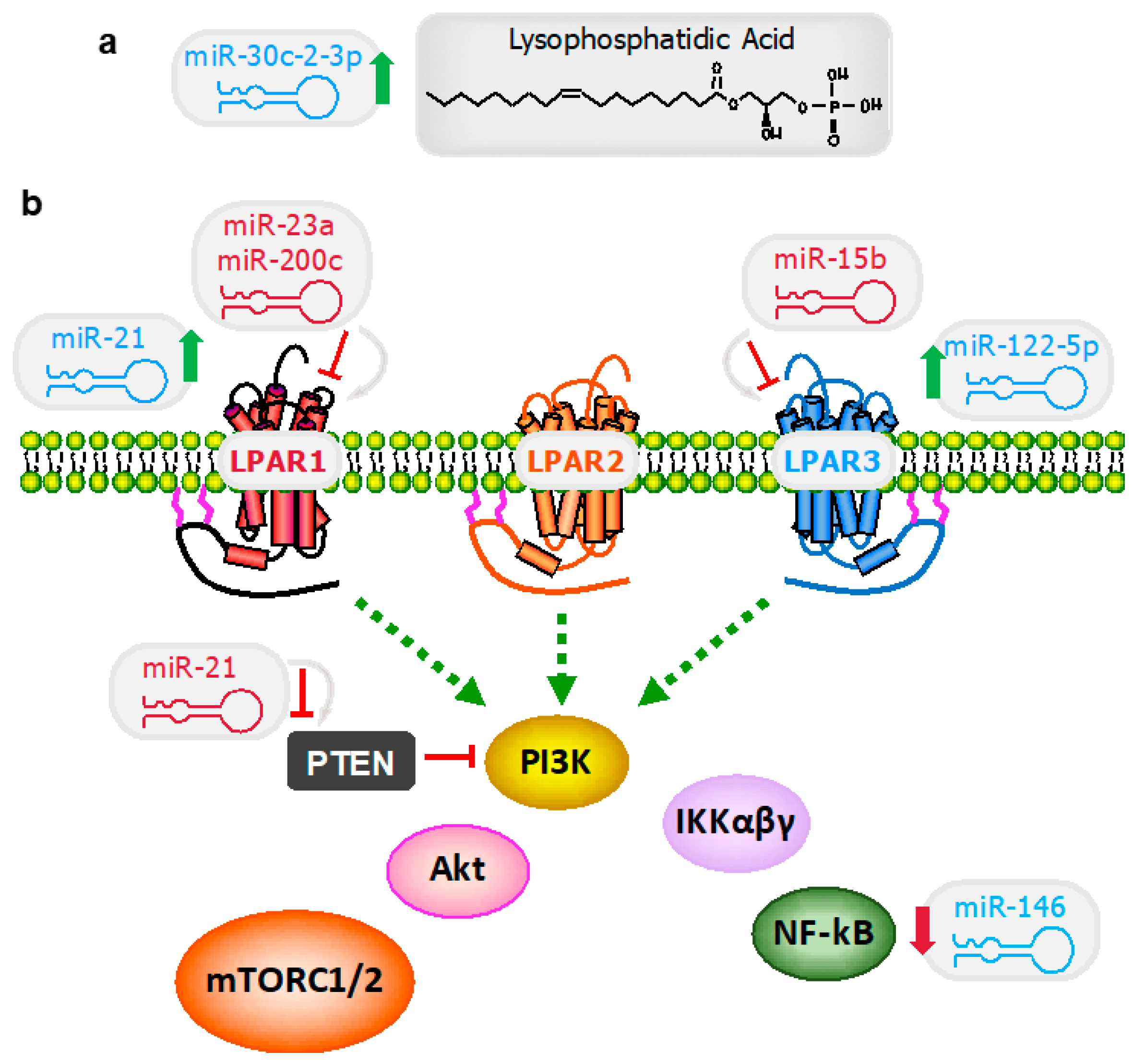
3.1. Lysophosphatidic Acid Signaling Regulation
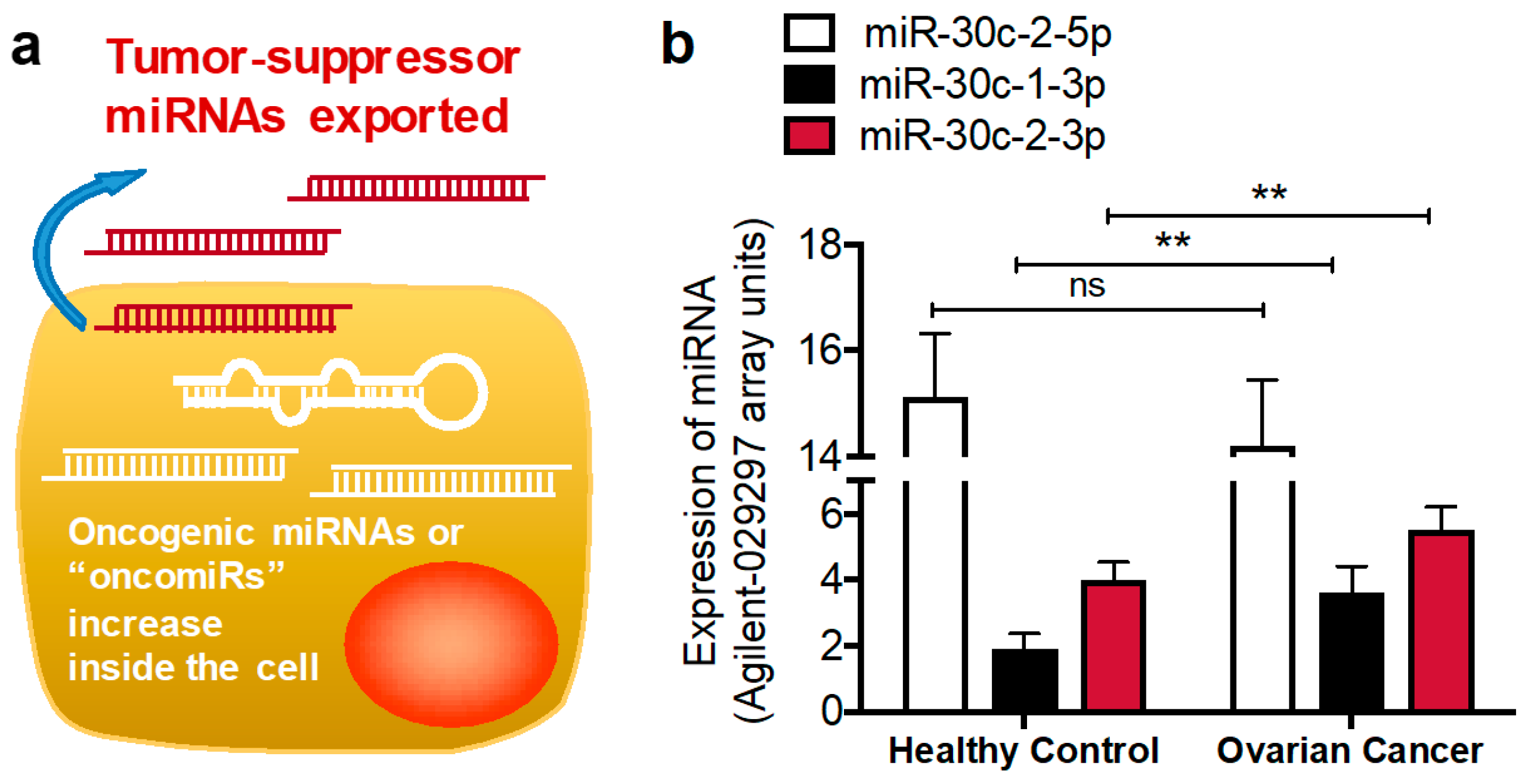
3.1.1. MiR-30c-2-3p
3.1.2. MiR-21
3.1.3. MiR-146a
3.2. Lysophosphatidic Acid Receptors
3.2.1. MiR-122
3.2.2. MiR-15b
3.2.3. MiR-200c
3.2.4. miR-23a
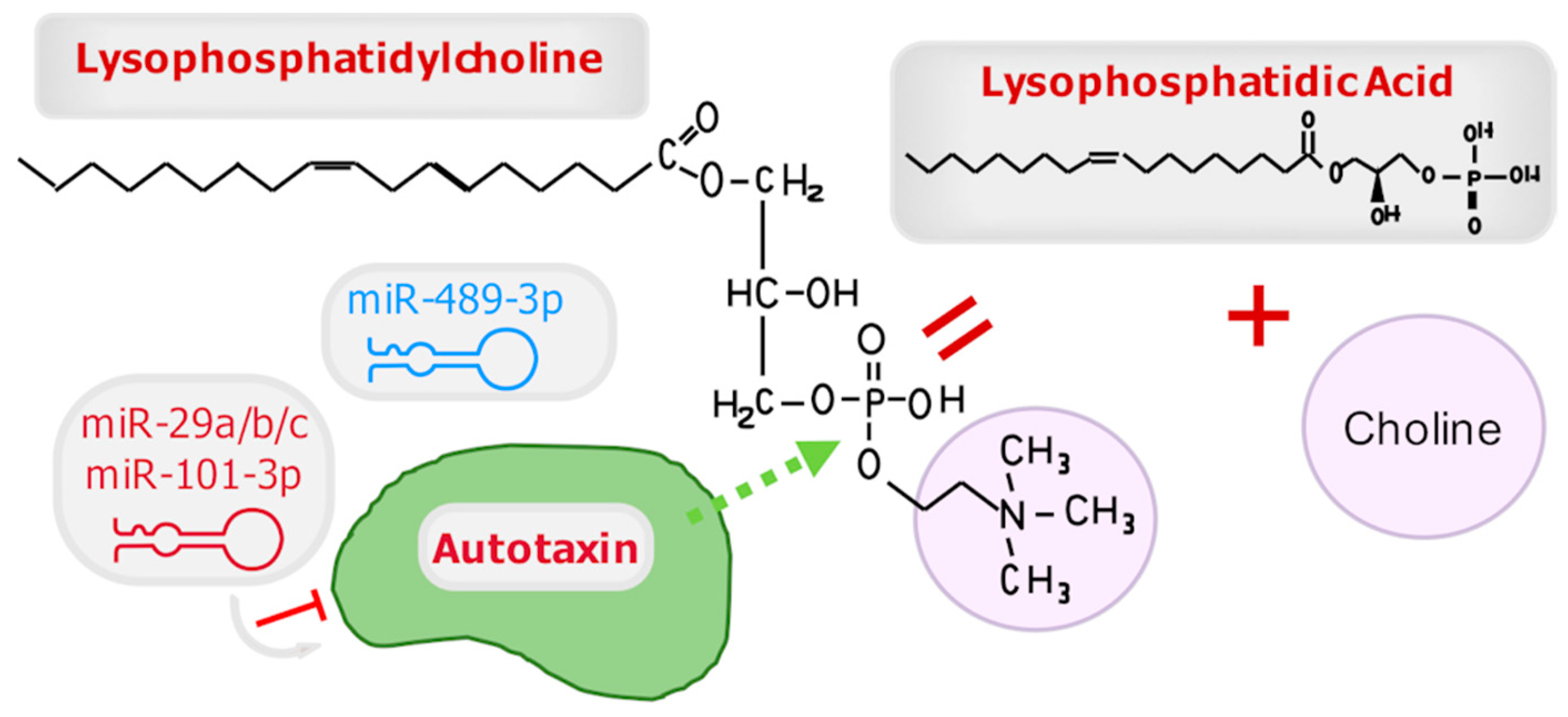
3.3. Autotaxin
3.3.1. MiR-29a/b/c
3.3.2. MiR-489-3p
3.3.3. MiR-101-3p
4. MiRNAs as Therapeutics
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Akiyama, Y.; Yuasa, Y. Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS ONE 2013, 8, e62589. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Sevignani, C.; Calin, G.A.; Siracusa, L.D.; Croce, C.M. Mammalian microRNAs: A small world for fine-tuning gene expression. Mamm. Genome 2006, 17, 189–202. [Google Scholar] [CrossRef]
- Chen, P.Y.; Weinmann, L.; Gaidatzis, D.; Pei, Y.; Zavolan, M.; Tuschl, T.; Meister, G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA 2008, 14, 263–274. [Google Scholar] [CrossRef]
- Chalfie, M.; Horvitz, H.R.; Sulston, J.E. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 1981, 24, 59–69. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Mills, G.B.; Moolenaar, W.H. The emerging role of lysophosphatidic acid in cancer. Nat. Rev. Cancer 2003, 3, 582–591. [Google Scholar] [CrossRef]
- Westermann, A.M.; Havik, E.; Postma, F.R.; Beijnen, J.H.; Dalesio, O.; Moolenaar, W.H.; Rodenhuis, S. Malignant effusions contain lysophosphatidic acid (LPA)-like activity. Ann. Oncol. 1998, 9, 437–442. [Google Scholar] [CrossRef]
- Eichholtz, T.; Jalink, K.; Fahrenfort, I.; Moolenaar, W.H. The bioactive phospholipid lysophosphatidic acid is released from activated platelets. Biochem. J. 1993, 291, 677–680. [Google Scholar] [CrossRef]
- Xu, Y.; Gaudette, D.C.; Boynton, J.D.; Frankel, A.; Fang, X.J.; Sharma, A.; Hurteau, J.; Casey, G.; Goodbody, A.; Mellors, A.; et al. Characterization of an ovarian cancer activating factor in ascites from ovarian cancer patients. Clin. Cancer Res. 1995, 1, 1223–1232. [Google Scholar]
- Frankel, A.; Mills, G.B. Peptide and lipid growth factors decrease cis-diamminedichloroplatinum-induced cell death in human ovarian cancer cells. Clin. Cancer Res. 1996, 2, 1307–1313. [Google Scholar]
- Xu, Y.; Shen, Z.; Wiper, D.W.; Wu, M.; Morton, R.E.; Elson, P.; Kennedy, A.W.; Belinson, J.; Markman, M.; Casey, G. Lysophosphatidic acid as a potential biomarker for ovarian and other gynecologic cancers. Jama 1998, 280, 719–723. [Google Scholar] [CrossRef]
- Murph, M.M.; Liu, W.; Yu, S.; Lu, Y.; Hall, H.; Hennessy, B.T.; Lahad, J.; Schaner, M.; Helland, A.; Kristensen, G.; et al. Lysophosphatidic acid-induced transcriptional profile represents serous epithelial ovarian carcinoma and worsened prognosis. PLoS ONE 2009, 4, e5583. [Google Scholar] [CrossRef]
- Hecht, J.H.; Weiner, J.A.; Post, S.R.; Chun, J. Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J. Cell Biol. 1996, 135, 1071–1083. [Google Scholar] [CrossRef]
- Bandoh, K.; Aoki, J.; Hosono, H.; Kobayashi, S.; Kobayashi, T.; Murakami-Murofushi, K.; Tsujimoto, M.; Arai, H.; Inoue, K. Molecular cloning and characterization of a novel human G-protein-coupled receptor, EDG7, for lysophosphatidic acid. J. Biol. Chem. 1999, 274, 27776–27785. [Google Scholar] [CrossRef]
- Noguchi, K.; Ishii, S.; Shimizu, T. Identification of p2y9/GPR23 as a novel G protein-coupled receptor for lysophosphatidic acid, structurally distant from the Edg family. J. Biol. Chem. 2003, 278, 25600–25606. [Google Scholar] [CrossRef]
- Lee, C.W.; Rivera, R.; Gardell, S.; Dubin, A.E.; Chun, J. GPR92 as a new G12/13- and Gq-coupled lysophosphatidic acid receptor that increases cAMP, LPA5. J. Biol. Chem. 2006, 281, 23589–23597. [Google Scholar] [CrossRef]
- Kotarsky, K.; Boketoft, A.; Bristulf, J.; Nilsson, N.E.; Norberg, A.; Hansson, S.; Owman, C.; Sillard, R.; Leeb-Lundberg, L.M.; Olde, B. Lysophosphatidic acid binds to and activates GPR92, a G protein-coupled receptor highly expressed in gastrointestinal lymphocytes. J. Pharmacol. Exp. Ther. 2006, 318, 619–628. [Google Scholar] [CrossRef]
- Pasternack, S.M.; von Kugelgen, I.; Aboud, K.A.; Lee, Y.A.; Ruschendorf, F.; Voss, K.; Hillmer, A.M.; Molderings, G.J.; Franz, T.; Ramirez, A.; et al. G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat. Genet. 2008, 40, 329–334. [Google Scholar] [CrossRef]
- Tabata, K.; Baba, K.; Shiraishi, A.; Ito, M.; Fujita, N. The orphan GPCR GPR87 was deorphanized and shown to be a lysophosphatidic acid receptor. Biochem. Biophys. Res. Commun. 2007, 363, 861–866. [Google Scholar] [CrossRef]
- McIntyre, T.M.; Pontsler, A.V.; Silva, A.R.; St Hilaire, A.; Xu, Y.; Hinshaw, J.C.; Zimmerman, G.A.; Hama, K.; Aoki, J.; Arai, H.; et al. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc. Natl. Acad. Sci. USA 2003, 100, 131–136. [Google Scholar] [CrossRef]
- Murakami, M.; Shiraishi, A.; Tabata, K.; Fujita, N. Identification of the orphan GPCR, P2Y(10) receptor as the sphingosine-1-phosphate and lysophosphatidic acid receptor. Biochem. Biophys. Res. Commun. 2008, 371, 707–712. [Google Scholar] [CrossRef]
- Rai, V.; Toure, F.; Chitayat, S.; Pei, R.; Song, F.; Li, Q.; Zhang, J.; Rosario, R.; Ramasamy, R.; Chazin, W.J.; et al. Lysophosphatidic acid targets vascular and oncogenic pathways via RAGE signaling. J. Exp. Med. 2012, 209, 2339–2350. [Google Scholar] [CrossRef]
- Lee, B.H.; Kim, J.; Lee, R.M.; Choi, S.H.; Kim, H.J.; Hwang, S.H.; Lee, M.K.; Bae, C.S.; Kim, H.C.; Rhim, H.; et al. Gintonin enhances performance of mice in rotarod test: Involvement of lysophosphatidic acid receptors and catecholamine release. Neurosci. Lett. 2016, 612, 256–260. [Google Scholar] [CrossRef]
- Yu, S.; Murph, M.M.; Lu, Y.; Liu, S.; Hall, H.S.; Liu, J.; Stephens, C.; Fang, X.; Mills, G.B. Lysophosphatidic acid receptors determine tumorigenicity and aggressiveness of ovarian cancer cells. J. Natl. Cancer Inst. 2008, 100, 1630–1642. [Google Scholar] [CrossRef]
- Murph, M.M.; Nguyen, G.H.; Radhakrishna, H.; Mills, G.B. Sharpening the edges of understanding the structure/function of the LPA1 receptor: Expression in cancer and mechanisms of regulation. Biochim. Biophys. Acta 2008, 1781, 547–557. [Google Scholar] [CrossRef][Green Version]
- Hennessy, B.T.; Murph, M.; Nanjundan, M.; Carey, M.; Auersperg, N.; Almeida, J.; Coombes, K.R.; Liu, J.; Lu, Y.; Gray, J.W.; et al. Ovarian cancer: Linking genomics to new target discovery and molecular markers--the way ahead. Adv. Exp. Med. Biol. 2008, 617, 23–40. [Google Scholar]
- Liu, S.; Murph, M.; Panupinthu, N.; Mills, G.B. ATX-LPA receptor axis in inflammation and cancer. Cell Cycle 2009, 8, 3695–3701. [Google Scholar] [CrossRef]
- Liu, S.; Umezu-Goto, M.; Murph, M.; Lu, Y.; Liu, W.; Zhang, F.; Yu, S.; Stephens, L.C.; Cui, X.; Murrow, G.; et al. Expression of autotaxin and lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell 2009, 15, 539–550. [Google Scholar] [CrossRef]
- Stracke, M.L.; Krutzsch, H.C.; Unsworth, E.J.; Arestad, A.; Cioce, V.; Schiffmann, E.; Liotta, L.A. Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J. Biol. Chem. 1992, 267, 2524–2529. [Google Scholar]
- Brindley, D.N.; Benesch, M.G.K.; Murph, M.M. Melanoma—Current Clinical Management and Future Therapeutics; InTech Open: London, UK, 2015; pp. 297–324. [Google Scholar]
- Umezu-Goto, M.; Kishi, Y.; Taira, A.; Hama, K.; Dohmae, N.; Takio, K.; Yamori, T.; Mills, G.B.; Inoue, K.; Aoki, J.; et al. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J. Cell Biol. 2002, 158, 227–233. [Google Scholar] [CrossRef]
- Nam, S.W.; Clair, T.; Kim, Y.S.; McMarlin, A.; Schiffmann, E.; Liotta, L.A.; Stracke, M.L. Autotaxin (NPP-2), a metastasis-enhancing motogen, is an angiogenic factor. Cancer Res. 2001, 61, 6938–6944. [Google Scholar]
- Xu, A.; Khan, M.A.K.; Chen, F.; Zhong, Z.; Chen, H.C.; Song, Y. Overexpression of autotaxin is associated with human renal cell carcinoma and bladder carcinoma and their progression. Med. Oncol. 2016, 33, e131. [Google Scholar] [CrossRef]
- Kehlen, A.; Englert, N.; Seifert, A.; Klonisch, T.; Dralle, H.; Langner, J.; Hoang-Vu, C. Expression, regulation and function of autotaxin in thyroid carcinomas. Int. J. Cancer 2004, 109, 833–838. [Google Scholar] [CrossRef]
- Kishi, Y.; Okudaira, S.; Tanaka, M.; Hama, K.; Shida, D.; Kitayama, J.; Yamori, T.; Aoki, J.; Fujimaki, T.; Arai, H. Autotaxin is overexpressed in glioblastoma multiforme and contributes to cell motility of glioblastoma by converting lysophosphatidylcholine to lysophosphatidic acid. J. Biol. Chem. 2006, 281, 17492–17500. [Google Scholar] [CrossRef]
- Kuppa, S.S.; Jia, W.; Liu, S.; Nguyen, H.; Smyth, S.S.; Mills, G.B.; Dobbin, K.K.; Hardman, W.J.; Murph, M.M. Autotaxin exacerbates tumor progression by enhancing MEK1 and overriding the function of miR-489-3p. Cancer Lett. 2018, 432, 84–92. [Google Scholar] [CrossRef]
- Masuda, A.; Nakamura, K.; Izutsu, K.; Igarashi, K.; Ohkawa, R.; Jona, M.; Higashi, K.; Yokota, H.; Okudaira, S.; Kishimoto, T.; et al. Serum autotaxin measurement in haematological malignancies: A promising marker for follicular lymphoma. Br. J. Haematol. 2008, 143, 60–70. [Google Scholar] [CrossRef]
- Murph, M.M.; Jiang, G.W.; Altman, M.K.; Jia, W.; Nguyen, D.T.; Fambrough, J.M.; Hardman, W.J.; Nguyen, H.T.; Tran, S.K.; Alshamrani, A.A.; et al. Vinyl sulfone analogs of lysophosphatidylcholine irreversibly inhibit autotaxin and prevent angiogenesis in melanoma. Bioorg. Med. Chem. 2015, 23, 5999–6013. [Google Scholar] [CrossRef]
- van Meeteren, L.A.; Ruurs, P.; Stortelers, C.; Bouwman, P.; van Rooijen, M.A.; Pradere, J.P.; Pettit, T.R.; Wakelam, M.J.; Saulnier-Blache, J.S.; Mummery, C.L.; et al. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol. Cell Biol. 2006, 26, 5015–5022. [Google Scholar] [CrossRef]
- Tanaka, M.; Okudaira, S.; Kishi, Y.; Ohkawa, R.; Iseki, S.; Ota, M.; Noji, S.; Yatomi, Y.; Aoki, J.; Arai, H. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J. Biol. Chem. 2006, 281, 25822–25830. [Google Scholar] [CrossRef]
- Altman, M.K.; Gopal, V.; Jia, W.; Yu, S.; Hall, H.; Mills, G.B.; McGinnis, A.C.; Bartlett, M.G.; Jiang, G.; Madan, D.; et al. Targeting melanoma growth and viability reveals dualistic functionality of the phosphonothionate analogue of carba cyclic phosphatidic acid. Mol. Cancer 2010, 9, e140. [Google Scholar] [CrossRef]
- Jia, W.; Tran, S.K.; Ruddick, C.A.; Murph, M.M. The Src homology 3 binding domain is required for lysophosphatidic acid 3 receptor-mediated cellular viability in melanoma cells. Cancer Lett. 2015, 356, 589–596. [Google Scholar] [CrossRef]
- Baker, D.L.; Fujiwara, Y.; Pigg, K.R.; Tsukahara, R.; Kobayashi, S.; Murofushi, H.; Uchiyama, A.; Murakami-Murofushi, K.; Koh, E.; Bandle, R.W.; et al. Carba analogs of cyclic phosphatidic acid are selective inhibitors of autotaxin and cancer cell invasion and metastasis. J. Biol. Chem. 2006, 281, 22786–22793. [Google Scholar] [CrossRef]
- Murph, M.M. Research on Melanoma: A Glimpse into Current Directions and Future Trends; BoD–Books on Demand: Norderstedt, Germany, 2011. [Google Scholar]
- Mazereeuw-Hautier, J.; Gres, S.; Fanguin, M.; Cariven, C.; Fauvel, J.; Perret, B.; Chap, H.; Salles, J.P.; Saulnier-Blache, J.S. Production of lysophosphatidic acid in blister fluid: Involvement of a lysophospholipase D activity. J. Investig. Dermatol. 2005, 125, 421–427. [Google Scholar] [CrossRef]
- Dvinge, H.; Git, A.; Graf, S.; Salmon-Divon, M.; Curtis, C.; Sottoriva, A.; Zhao, Y.; Hirst, M.; Armisen, J.; Miska, E.A.; et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013, 497, e378. [Google Scholar] [CrossRef]
- Byrnes, C.C.; Jia, W.; Alshamrani, A.A.; Kuppa, S.S.; Murph, M.M. miR-122-5p expression and secretion in melanoma cells is amplified by the LPAR3 SH3-binding domain to regulate Wnt1. Mol. Cancer Res. 2019, 17, 299–309. [Google Scholar] [CrossRef]
- Jia, W.; Eneh, J.O.; Ratnaparkhe, S.; Altman, M.K.; Murph, M.M. MicroRNA-30c-2* expressed in ovarian cancer cells suppresses growth factor-induced cellular proliferation and downregulates the oncogene BCL9. Mol. Cancer Res. 2011, 9, 1732–1745. [Google Scholar] [CrossRef]
- Byrd, A.E.; Aragon, I.V.; Brewer, J.W. MicroRNA-30c-2* limits expression of proadaptive factor XBP1 in the unfolded protein response. J. Cell Biol. 2012, 196, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Mathew, L.K.; Lee, S.S.; Skuli, N.; Rao, S.; Keith, B.; Nathanson, K.L.; Lal, P.; Simon, M.C. Restricted expression of miR-30c-2-3p and miR-30a-3p in clear cell renal cell carcinomas enhances HIF2alpha activity. Cancer Discov. 2014, 4, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Shukla, K.; Sharma, A.K.; Ward, A.; Will, R.; Hielscher, T.; Balwierz, A.; Breunig, C.; Munstermann, E.; Konig, R.; Keklikoglou, I.; et al. MicroRNA-30c-2-3p negatively regulates NF-kappaB signaling and cell cycle progression through downregulation of TRADD and CCNE1 in breast cancer. Mol. Oncol. 2015, 9, 1106–1119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cai, M.; Jiang, D.; Xu, L. Upregulated LncRNA-CCAT1 promotes hepatocellular carcinoma progression by functioning as miR-30c-2-3p sponge. Cell Biochem. Funct. 2019, 37, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Chen, P.; Sang, C.; Huang, D.; Geng, Q.; Wang, L. Modulation of apoptosis-related microRNAs following myocardial infarction in fat-1 transgenic mice vs. wild-type mice. J. Cell Mol. Med. 2018, 22, 5698–5707. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.T.; Liang, Q.; Yang, L.; Lin, X.L.; Wu, S.; Chen, Y.; Zhang, X.T.; Gao, Y.J.; Ge, Z.Z. RAB31 Targeted by MiR-30c-2-3p regulates the GLI1 signaling pathway, affecting gastric cancer cell proliferation and apoptosis. Front. Oncol. 2018, 8, e554. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Jia, W.; Beedle, A.M.; Kennedy, E.J.; Murph, M.M. Lysophosphatidic Acid Mediates Activating Transcription Factor 3 Expression Which Is a Target for Post-Transcriptional Silencing by miR-30c-2-3p. PLoS ONE 2015, 10, e0139489. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, J.D.; Baraniskin, A.; Hahn, S.A.; Mosel, F.; Bredemeier, M.; Wimberger, P.; Kimmig, R.; Kasimir-Bauer, S. Circulating U2 small nuclear RNA fragments as a novel diagnostic tool for patients with epithelial ovarian cancer. Clin. Chem. 2014, 60, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [PubMed]
- Sahay, D.; Leblanc, R.; Grunewald, T.G.; Ambatipudi, S.; Ribeiro, J.; Clezardin, P.; Peyruchaud, O. The LPA1/ZEB1/miR-21-activation pathway regulates metastasis in basal breast cancer. Oncotarget 2015, 6, 20604–20620. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Sun, C.; Hu, Z.; Yang, Y.; Zhu, Y.; Zheng, D.; Gu, L.; Lu, X. Lysophosphatidic acid induces MDA-MB-231 breast cancer cells migration through activation of PI3K/PAK1/ERK signaling. PLoS ONE 2010, 5, e15940. [Google Scholar] [CrossRef] [PubMed]
- Crone, S.G.; Jacobsen, A.; Federspiel, B.; Bardram, L.; Krogh, A.; Lund, A.H.; Friis-Hansen, L. MicroRNA-146a inhibits G protein-coupled receptor-mediated activation of NF-kappaB by targeting CARD10 and COPS8 in gastric cancer. Mol. Cancer 2012, 11, e71. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009, 183, 2150–2158. [Google Scholar] [CrossRef]
- Bhaumik, D.; Scott, G.K.; Schokrpur, S.; Patil, C.K.; Campisi, J.; Benz, C.C. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 2008, 27, 5643–5647. [Google Scholar] [CrossRef]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Maierthaler, M.; Benner, A.; Hoffmeister, M.; Surowy, H.; Jansen, L.; Knebel, P.; Chang-Claude, J.; Brenner, H.; Burwinkel, B. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int. J. Cancer 2017, 140, 176–187. [Google Scholar] [CrossRef]
- Ofir, M.; Hacohen, D.; Ginsberg, D. MiR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol. Cancer Res. 2011, 9, 440–447. [Google Scholar] [CrossRef]
- Xia, L.; Zhang, D.; Du, R.; Pan, Y.; Zhao, L.; Sun, S.; Hong, L.; Liu, J.; Fan, D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer 2008, 123, 372–379. [Google Scholar] [CrossRef] [PubMed]
- MacLean, J.A., 2nd; King, M.L.; Okuda, H.; Hayashi, K. WNT7A regulation by miR-15b in ovarian cancer. PLoS ONE 2016, 11, e0156109. [Google Scholar] [CrossRef] [PubMed]
- Li, G.C.; Qin, X.L.; Song, H.H.; Li, Y.N.; Qiu, Y.Y.; Cui, S.C.; Wang, Y.S.; Wang, H.; Gong, J.L. Upregulated microRNA-15b alleviates ovarian cancer through inhitbition of the PI3K/Akt pathway by targeting LPAR3. J. Cell Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Adam, L.; Zhong, M.; Choi, W.; Qi, W.; Nicoloso, M.; Arora, A.; Calin, G.; Wang, H.; Siefker-Radtke, A.; McConkey, D.; et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 2009, 15, 5060–5072. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef]
- Howe, E.N.; Cochrane, D.R.; Richer, J.K. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011, 13, e45. [Google Scholar] [CrossRef]
- Elson-Schwab, I.; Lorentzen, A.; Marshall, C.J. MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PLoS ONE 2010, 5, e13176. [Google Scholar] [CrossRef]
- Luna, C.; Li, G.; Huang, J.; Qiu, J.; Wu, J.; Yuan, F.; Epstein, D.L.; Gonzalez, P. Regulation of trabecular meshwork cell contraction and intraocular pressure by miR-200c. PLoS ONE 2012, 7, e51688. [Google Scholar] [CrossRef]
- Van Leeuwen, F.N.; Olivo, C.; Grivell, S.; Giepmans, B.N.; Collard, J.G.; Moolenaar, W.H. Rac activation by lysophosphatidic acid LPA1 receptors through the guanine nucleotide exchange factor Tiam1. J. Biol. Chem. 2003, 278, 400–406. [Google Scholar] [CrossRef]
- Yang, J.; Nie, Y.; Wang, F.; Hou, J.; Cong, X.; Hu, S.; Chen, X. Reciprocal regulation of miR-23a and lysophosphatidic acid receptor signaling in cardiomyocyte hypertrophy. Biochim. Biophys. Acta 2013, 1831, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Kolmert, J.; Yang, M.; Reinke, S.N.; Kamleh, M.A.; Snowden, S.; Heyder, T.; Levanen, B.; Erle, D.J.; Skold, C.M.; et al. Metabolomics analysis identifies sex-associated metabotypes of oxidative stress and the autotaxin-lysoPA axis in COPD. Eur. Respir. J. 2017, 49, e1602322. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.L.; Desiderio, D.M.; Miller, D.D.; Tolley, B.; Tigyi, G.J. Direct quantitative analysis of lysophosphatidic acid molecular species by stable isotope dilution electrospray ionization liquid chromatography-mass spectrometry. Anal. Biochem. 2001, 292, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lyu, L.; Zhang, X.; Zhang, J. Autotaxin is a novel target of microRNA-101-3p. FEBS Open Bio 2019, 9, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, e478. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Uhlmann, E.J.; Wong, A.H.; Karmali, P.; Basu, M.; Gabriely, G.; Jain, A.; Wang, Y.; Chiocca, E.A.; Stephens, R.; et al. MicroRNA-10b inhibition reduces E2F1-mediated transcription and miR-15/16 activity in glioblastoma. Oncotarget 2015, 6, 3770–3783. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Uhlmann, E.J.; Gabriely, G.; Volfovsky, N.; Wang, Y.; Teng, J.; Karmali, P.; Marcusson, E.; Peter, M.; Mohan, A.; et al. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: First steps toward the clinic. EMBO Mol. Med. 2016, 8, 268–287. [Google Scholar] [CrossRef] [PubMed]

| miRNA | Target | Alteration |
|---|---|---|
| miR-15b | BCL-2, LPAR3, WNT7A | LPAR3 mRNA and protein |
| miR-21 | PTEN | Upregulated by LPAR1 signaling through the PI3K pathway |
| miR-23a | LPAR1 | Reduced LPAR1 through LPAR3 in cardiomyocyte hypertrophy |
| miR-29a/b/c | ATX/ENPP2 | ATX mRNA and protein |
| miR-30c-2-3p | ATF3, BCL-9, CCNE1, HIF2A, RAB31, TRADD, XBP1 | Lysophosphatidic acid signaling, cell cycle and death |
| miR-101-3p | ATX/ENPP2 | ATX mRNA and protein |
| miR-122-5p | WNT1 | Lysophosphatidic acid signaling through LPAR3 |
| miR-146a | COPS8, CAR10 | Modest reduction of NF-kB activation and cytokine mRNA by 25 μM lysophosphatidic acid |
| miR-200c | LPAR1 | Cell contraction |
| miR-489-3p | MEK1 | Feedback loop with autotaxin signaling |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murph, M.M. MicroRNA Regulation of the Autotaxin-Lysophosphatidic Acid Signaling Axis. Cancers 2019, 11, 1369. https://doi.org/10.3390/cancers11091369
Murph MM. MicroRNA Regulation of the Autotaxin-Lysophosphatidic Acid Signaling Axis. Cancers. 2019; 11(9):1369. https://doi.org/10.3390/cancers11091369
Chicago/Turabian StyleMurph, Mandi M. 2019. "MicroRNA Regulation of the Autotaxin-Lysophosphatidic Acid Signaling Axis" Cancers 11, no. 9: 1369. https://doi.org/10.3390/cancers11091369
APA StyleMurph, M. M. (2019). MicroRNA Regulation of the Autotaxin-Lysophosphatidic Acid Signaling Axis. Cancers, 11(9), 1369. https://doi.org/10.3390/cancers11091369