A Mouse Model to Assess STAT3 and STAT5A/B Combined Inhibition in Health and Disease Conditions
Abstract
1. Introduction
2. Results and Discussion
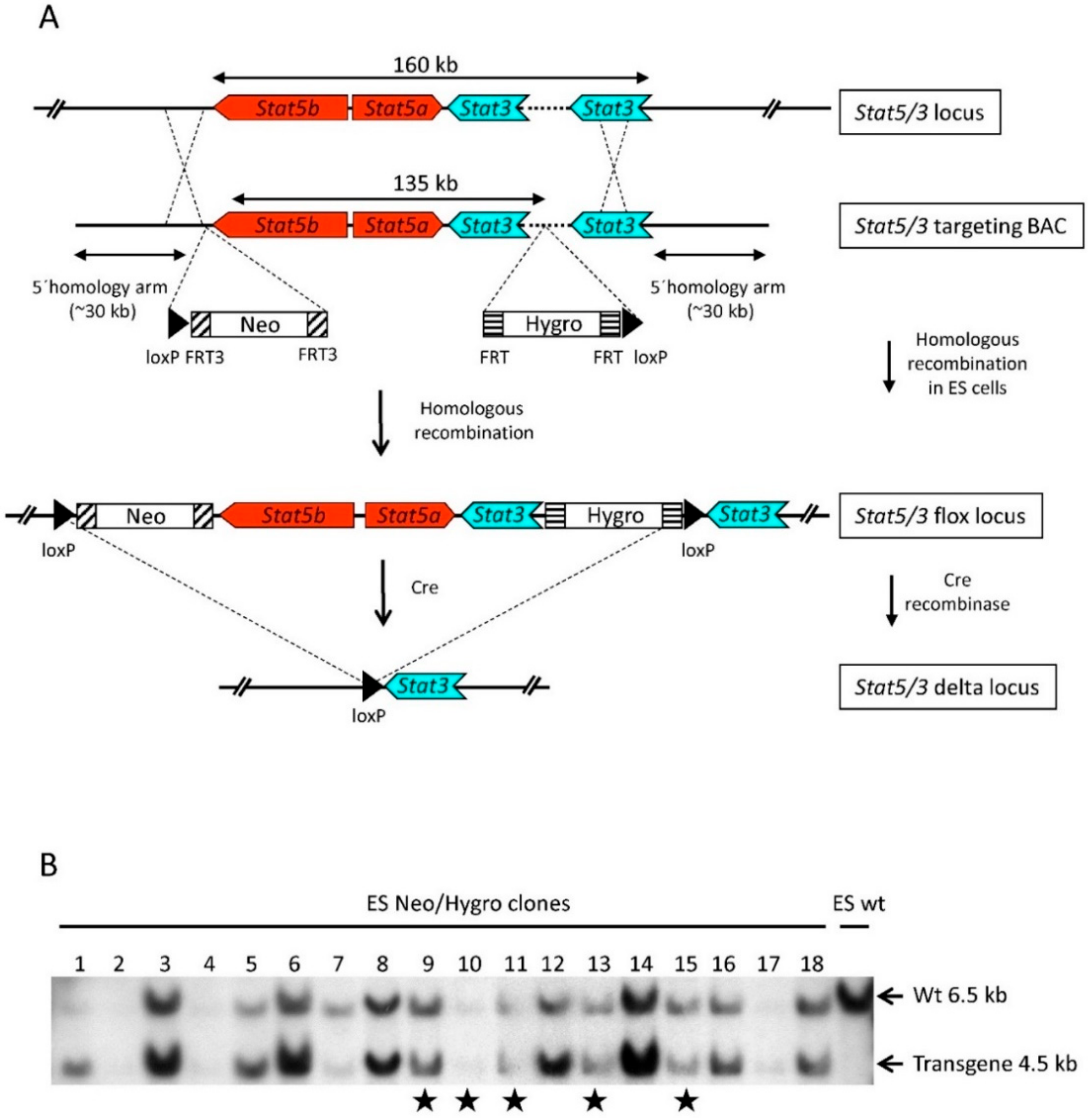
2.1. Generation of Stat3 and Stat5a/b Flox (Stat5/3loxP/loxP) Mice
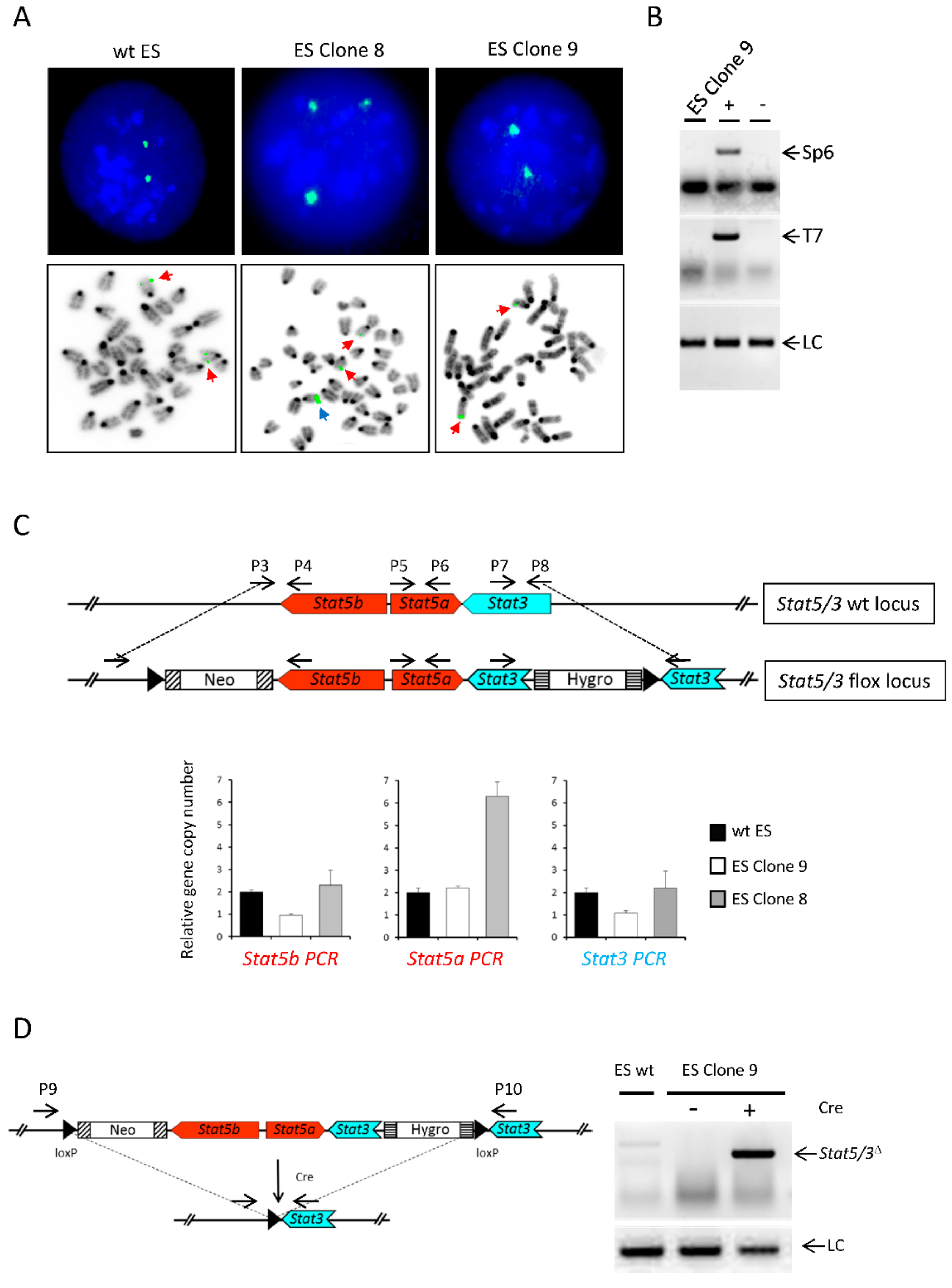
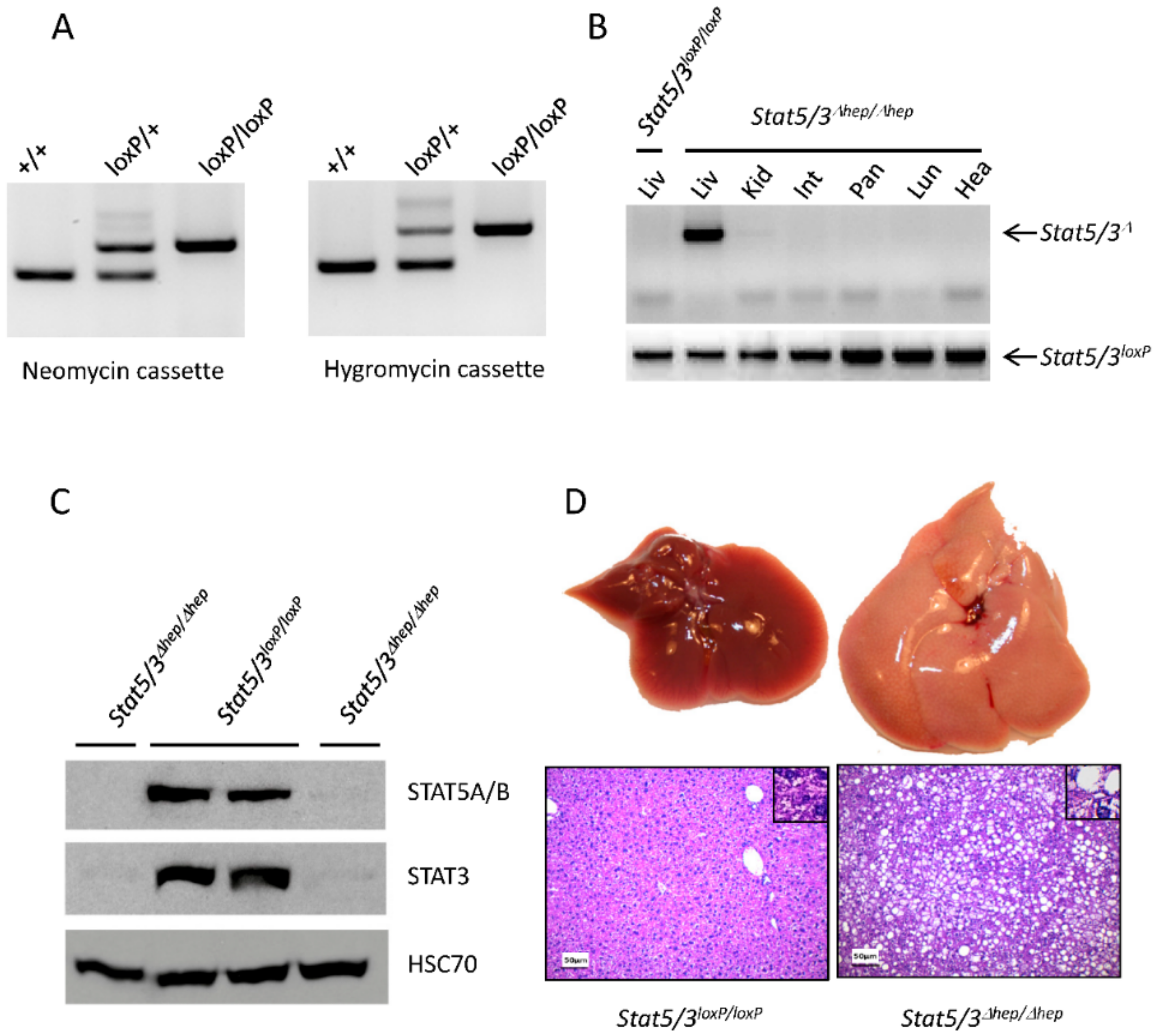
2.2. Functional Validation of Stat5/3loxP/loxP Mice
3. Material and Methods
3.1. BAC Recombineering
3.2. ES Cell Targeting
3.3. FISH analysis of ES Clones
3.4. Quantitative Genomic PCR
3.5. Animals
3.6. Western Blot Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, H. Genetically engineered mouse models for drug development and preclinical trials. Biomol. Ther. 2014, 22, 267–274. [Google Scholar] [CrossRef]
- Palais, G.; Nguyen Dinh Cat, A.; Friedman, H.; Panek-Huet, N.; Millet, A.; Tronche, F.; Gellen, B.; Mercadier, J.J.; Peterson, A.; Jaisser, F. Targeted transgenesis at the hprt locus: An efficient strategy to achieve tightly controlled in vivo conditional expression with the tet system. Physiol. Genom. 2009, 37, 140–146. [Google Scholar] [CrossRef]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The jak/stat signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef]
- Schindler, C.; Levy, D.E.; Decker, T. Jak-stat signaling: From interferons to cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Schwartz, D.M.; Villarino, A.V.; Gadina, M.; McInnes, I.B.; Laurence, A. The jak-stat pathway: Impact on human disease and therapeutic intervention. Annu. Rev. Med. 2015, 66, 311–328. [Google Scholar] [CrossRef]
- Loh, C.Y.; Arya, A.; Naema, A.F.; Wong, W.F.; Sethi, G.; Looi, C.Y. Signal transducer and activator of transcription (stats) proteins in cancer and inflammation: Functions and therapeutic implication. Front. Oncol. 2019, 9, 48. [Google Scholar] [CrossRef]
- Buchert, M.; Burns, C.J.; Ernst, M. Targeting jak kinase in solid tumors: Emerging opportunities and challenges. Oncogene 2016, 35, 939–951. [Google Scholar] [CrossRef]
- Lee, C.K.; Raz, R.; Gimeno, R.; Gertner, R.; Wistinghausen, B.; Takeshita, K.; DePinho, R.A.; Levy, D.E. Stat3 is a negative regulator of granulopoiesis but is not required for g-csf-dependent differentiation. Immunity 2002, 17, 63–72. [Google Scholar] [CrossRef]
- Welte, T.; Zhang, S.S.; Wang, T.; Zhang, Z.; Hesslein, D.G.; Yin, Z.; Kano, A.; Iwamoto, Y.; Li, E.; Craft, J.E.; et al. Stat3 deletion during hematopoiesis causes crohn’s disease-like pathogenesis and lethality: A critical role of stat3 in innate immunity. Proc. Natl. Acad. Sci. USA 2003, 100, 1879–1884. [Google Scholar] [CrossRef]
- Cernkovich, E.R.; Deng, J.; Bond, M.C.; Combs, T.P.; Harp, J.B. Adipose-specific disruption of signal transducer and activator of transcription 3 increases body weight and adiposity. Endocrinology 2008, 149, 1581–1590. [Google Scholar] [CrossRef]
- Alonzi, T.; Maritano, D.; Gorgoni, B.; Rizzuto, G.; Libert, C.; Poli, V. Essential role of stat3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol. Cell. Biol. 2001, 21, 1621–1632. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, J.J.; Kalinowski, A.; Liu, M.G.; Zhang, S.S.; Gao, Q.; Chai, G.X.; Ji, L.; Iwamoto, Y.; Li, E.; Schneider, M.; et al. Cardiomyocyte-restricted knockout of stat3 results in higher sensitivity to inflammation, cardiac fibrosis, and heart failure with advanced age. Proc. Natl. Acad. Sci. USA 2003, 100, 12929–12934. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Kaisho, T.; Yoshida, N.; Takeda, J.; Kishimoto, T.; Akira, S. Stat3 activation is responsible for il-6-dependent t cell proliferation through preventing apoptosis: Generation and characterization of t cell-specific stat3-deficient mice. J. Immunol. 1998, 161, 4652–4660. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Riedlinger, G.; Miyoshi, K.; Tang, W.; Li, C.; Deng, C.X.; Robinson, G.W.; Hennighausen, L. Inactivation of stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 2004, 24, 8037–8047. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed]
- Blaas, L.; Kornfeld, J.W.; Schramek, D.; Musteanu, M.; Zollner, G.; Gumhold, J.; van Zijl, F.; Schneller, D.; Esterbauer, H.; Egger, G.; et al. Disruption of the growth hormone—Signal transducer and activator of transcription 5—Insulinlike growth factor 1 axis severely aggravates liver fibrosis in a mouse model of cholestasis. Hepatology 2010, 51, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Mair, M.; Zollner, G.; Schneller, D.; Musteanu, M.; Fickert, P.; Gumhold, J.; Schuster, C.; Fuchsbichler, A.; Bilban, M.; Tauber, S.; et al. Signal transducer and activator of transcription 3 protects from liver injury and fibrosis in a mouse model of sclerosing cholangitis. Gastroenterology 2010, 138, 2499–2508. [Google Scholar] [CrossRef]
- Deng, Y.R.; Ma, H.D.; Tsuneyama, K.; Yang, W.; Wang, Y.H.; Lu, F.T.; Liu, C.H.; Liu, P.; He, X.S.; Diehl, A.M.; et al. Stat3-mediated attenuation of ccl4-induced mouse liver fibrosis by the protein kinase inhibitor sorafenib. J. Autoimmun. 2013, 46, 25–34. [Google Scholar] [CrossRef]
- Hosui, A.; Kimura, A.; Yamaji, D.; Zhu, B.M.; Na, R.; Hennighausen, L. Loss of stat5 causes liver fibrosis and cancer development through increased tgf-{beta} and stat3 activation. J. Exp. Med. 2009, 206, 819–831. [Google Scholar] [CrossRef]
- Gleixner, K.V.; Schneeweiss, M.; Eisenwort, G.; Berger, D.; Herrmann, H.; Blatt, K.; Greiner, G.; Byrgazov, K.; Hoermann, G.; Konopleva, M.; et al. Combined targeting of stat3 and stat5: A novel approach to overcome drug resistance in chronic myeloid leukemia. Haematologica 2017, 102, 1519–1529. [Google Scholar] [CrossRef][Green Version]
- Testa, G.; Zhang, Y.; Vintersten, K.; Benes, V.; Pijnappel, W.W.; Chambers, I.; Smith, A.J.; Smith, A.G.; Stewart, A.F. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat. Biotechnol. 2003, 21, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Seed, B. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat. Biotechnol. 2003, 21, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Muyrers, J.P.; Zhang, Y.; Testa, G.; Stewart, A.F. Rapid modification of bacterial artificial chromosomes by et-recombination. Nucleic Acids Res. 1999, 27, 1555–1557. [Google Scholar] [CrossRef] [PubMed]
- Magin, T.M.; McWhir, J.; Melton, D.W. A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 1992, 20, 3795–3796. [Google Scholar] [CrossRef] [PubMed]
- Rebuzzini, P.; Zuccotti, M.; Redi, C.A.; Garagna, S. Chromosomal abnormalities in embryonic and somatic stem cells. Cytogenet. Genome Res. 2015, 147, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shimshek, D.R.; Kim, J.; Hubner, M.R.; Spergel, D.J.; Buchholz, F.; Casanova, E.; Stewart, A.F.; Seeburg, P.H.; Sprengel, R. Codon-improved cre recombinase (icre) expression in the mouse. Genesis 2002, 32, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Lewandoski, M.; Meyers, E.N.; Martin, G.R. Analysis of fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 1997, 62, 159–168. [Google Scholar] [PubMed]
- Takeda, K.; Noguchi, K.; Shi, W.; Tanaka, T.; Matsumoto, M.; Yoshida, N.; Kishimoto, T.; Akira, S. Targeted disruption of the mouse stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 1997, 94, 3801–3804. [Google Scholar] [CrossRef]
- Cui, Y.; Hosui, A.; Sun, R.; Shen, K.; Gavrilova, O.; Chen, W.; Cam, M.C.; Gao, B.; Robinson, G.W.; Hennighausen, L. Loss of signal transducer and activator of transcription 5 leads to hepatosteatosis and impaired liver regeneration. Hepatology 2007, 46, 504–513. [Google Scholar] [CrossRef]
- Saam, J.R.; Gordon, J.I. Inducible gene knockouts in the small intestinal and colonic epithelium. J. Biol. Chem. 1999, 274, 38071–38082. [Google Scholar] [CrossRef]
- Kistner, A.; Gossen, M.; Zimmermann, F.; Jerecic, J.; Ullmer, C.; Lubbert, H.; Bujard, H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc. Natl. Acad. Sci. USA 1996, 93, 10933–10938. [Google Scholar] [CrossRef] [PubMed]
- Singireddy, A.V.; Inglis, M.A.; Zuure, W.A.; Kim, J.S.; Anderson, G.M. Neither signal transducer and activator of transcription 3 (stat3) or stat5 signaling pathways are required for leptin’s effects on fertility in mice. Endocrinology 2013, 154, 2434–2445. [Google Scholar] [CrossRef] [PubMed]
- Baier, B.; Hunt, P.; Broman, K.W.; Hassold, T. Variation in genome-wide levels of meiotic recombination is established at the onset of prophase in mammalian males. PLoS Genet. 2014, 10, e1004125. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Anastassiadis, K.; Schnutgen, F.; von Melchner, H.; Stewart, A.F. Gene targeting and site-specific recombination in mouse es cells. Methods Enzymol. 2013, 533, 133–155. [Google Scholar] [PubMed]
- Vanden Berghe, T.; Hulpiau, P.; Martens, L.; Vandenbroucke, R.E.; Van Wonterghem, E.; Perry, S.W.; Bruggeman, I.; Divert, T.; Choi, S.M.; Vuylsteke, M.; et al. Passenger mutations confound interpretation of all genetically modified congenic mice. Immunity 2015, 43, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Pettitt, S.J.; Liang, Q.; Rairdan, X.Y.; Moran, J.L.; Prosser, H.M.; Beier, D.R.; Lloyd, K.C.; Bradley, A.; Skarnes, W.C. Agouti c57bl/6n embryonic stem cells for mouse genetic resources. Nat. Methods 2009, 6, 493–495. [Google Scholar] [CrossRef]
- Zheng, B.; Sage, M.; Sheppeard, E.A.; Jurecic, V.; Bradley, A. Engineering mouse chromosomes with cre-loxp: Range, efficiency, and somatic applications. Mol. Cell. Biol. 2000, 20, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Kellendonk, C.; Opherk, C.; Anlag, K.; Schutz, G.; Tronche, F. Hepatocyte-specific expression of cre recombinase. Genesis 2000, 26, 151–153. [Google Scholar] [CrossRef]
- Holschneider, D.P.; Shih, J.C. Genotype to phenotype: Challenges and opportunities. Int. J. Dev. Neurosci. 2000, 18, 615–618. [Google Scholar] [CrossRef]
- Wang, J.; Sarov, M.; Rientjes, J.; Fu, J.; Hollak, H.; Kranz, H.; Xie, W.; Stewart, A.F.; Zhang, Y. An improved recombineering approach by adding reca to lambda red recombination. Mol. Biotechnol. 2006, 32, 43–53. [Google Scholar] [CrossRef]
- Casanova, E.; Fehsenfeld, S.; Greiner, E.; Stewart, A.F.; Schutz, G. Conditional mutagenesis of camkiv. Genesis 2002, 32, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Rostovskaya, M.; Fu, J.; Obst, M.; Baer, I.; Weidlich, S.; Wang, H.; Smith, A.J.; Anastassiadis, K.; Stewart, A.F. Transposon-mediated bac transgenesis in human es cells. Nucleic Acids Res. 2012, 40, e150. [Google Scholar] [CrossRef] [PubMed]
- Konig, M.; Reichel, M.; Marschalek, R.; Haas, O.A.; Strehl, S. A highly specific and sensitive fluorescence in situ hybridization assay for the detection of t(4;11)(q21;q23) and concurrent submicroscopic deletions in acute leukaemias. Br. J. Haematol. 2002, 116, 758–764. [Google Scholar] [CrossRef] [PubMed]
| % of Cell Interphases | ||||
|---|---|---|---|---|
| ES Clone | One Signal | Two Signals | Three Signals | Four Signals |
| wt | 1.5 | 73.0 | 20.5 | 5.0 |
| 8 | 0.0 | 9.0 | 85.0 | 6.0 |
| 9 | 4.5 | 93.5 | 2.0 | 0.0 |
| 10 | 0.0 | 3.0 | 27.5 | 69.5 |
| 11 | 0.0 | 4.0 | 83.5 | 12.5 |
| 13 | 0.0 | 11.0 | 80.0 | 9.0 |
| 15 | 0.0 | 1.0 | 29.0 | 70.0 |
| Genotype | ||||
|---|---|---|---|---|
| Stage | Stat5/3+/+ | Stat5/3Δ//+ | Stat5/3Δ/Δ | p Value |
| Blastocysts | 8 (9) | 23 (18) | 5 (9) | 0.1942 |
| Born mice | 22 (12) | 27 (25) | 0 (12) | <0.0001 |
| Primer | 5′-3′Sequence |
|---|---|
| P1 | CATCATACACTTCATTTTAGGACTGCC |
| P2 | TGGCCCAGTGTTCAGTGCTCTTCTTACC |
| P3 | GGGACTCTTAAAATGGAAATCTGG |
| P4 | CAGAATGTTCTAGAAGGTTTGG |
| P5 | GTGCATGCTTTGTAGGAATTCTATGG |
| P6 | GTGGTTCCTCTGGTTTGTTACGTAGG |
| P7 | ATCTTGGACACAAATGCAGAGCC |
| P8 | CTGGTTAAGAAAAAGCCATTCTACC |
| P9 | GCTTTGAAGCTTCATCCCTATCC |
| P10 | TAGCTTAGGATAATTTTCTTCATG |
| P11 | GGTTGGCGCCTACCGGTGGATGTGG |
| P12 | CAGTAGCCCAGTGTCCCAGCCAAACAG |
| P13 | AATCAGTAATAAGTGATGATAGAAGGG |
| P14 | CAGATGACCACTCCAGTCGGGGG |
| P15 | ACCAGCCAGCTATCAACTCG |
| P16 | TTACATTGGTCCAGCCACC |
| P17 | TCTGAGCATGGCCTCTAA |
| P18 | GCTGGAGTAAATTTCACAGTG |
| P19 | TCTCACTCGGAAGGACAT |
| Sp6 | GATTTAGGTGACACTATAG |
| T7 | GTAATACGACTCACTATAGGGC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moll, H.P.; Mohrherr, J.; Blaas, L.; Musteanu, M.; Stiedl, P.; Grabner, B.; Zboray, K.; König, M.; Stoiber, D.; Rülicke, T.; et al. A Mouse Model to Assess STAT3 and STAT5A/B Combined Inhibition in Health and Disease Conditions. Cancers 2019, 11, 1226. https://doi.org/10.3390/cancers11091226
Moll HP, Mohrherr J, Blaas L, Musteanu M, Stiedl P, Grabner B, Zboray K, König M, Stoiber D, Rülicke T, et al. A Mouse Model to Assess STAT3 and STAT5A/B Combined Inhibition in Health and Disease Conditions. Cancers. 2019; 11(9):1226. https://doi.org/10.3390/cancers11091226
Chicago/Turabian StyleMoll, Herwig P., Julian Mohrherr, Leander Blaas, Monica Musteanu, Patricia Stiedl, Beatrice Grabner, Katalin Zboray, Margit König, Dagmar Stoiber, Thomas Rülicke, and et al. 2019. "A Mouse Model to Assess STAT3 and STAT5A/B Combined Inhibition in Health and Disease Conditions" Cancers 11, no. 9: 1226. https://doi.org/10.3390/cancers11091226
APA StyleMoll, H. P., Mohrherr, J., Blaas, L., Musteanu, M., Stiedl, P., Grabner, B., Zboray, K., König, M., Stoiber, D., Rülicke, T., Strehl, S., Eferl, R., & Casanova, E. (2019). A Mouse Model to Assess STAT3 and STAT5A/B Combined Inhibition in Health and Disease Conditions. Cancers, 11(9), 1226. https://doi.org/10.3390/cancers11091226





