Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies
Abstract
1. Introduction
2. The Circulating Transcriptome
3. MiRNAs in Biological Fluids
4. PiRNAs in Biological Fluids
5. SnRNAs and SnoRNAs
6. LncRNAs
7. CircRNAs
8. Final Remarks and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Li, T.; Huang, Y.; Sun, J.; Zhu, Y.; Yang, Y.; Lu, Z.J. RNA Biomarkers: Frontier of Precision Medicine for Cancer. Noncoding RNA 2017, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Sole, C.; Arnaiz, E.; Manterola, L.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Tolani, B.; Nie, X.; Zhi, X.; Hu, M.; He, B. Review of the clinical applications and technological advances of circulating tumor DNA in cancer monitoring. Ther. Clin. Risk Manag. 2017, 13, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Matullo, G.; Naccarati, A.; Pardini, B. MicroRNA expression profiling in bladder cancer: The challenge of next-generation sequencing in tissues and biofluids. Int. J. Cancer 2016, 138, 2334–2345. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, C.; Dragomir, M.; Tanase, M.; Giza, D.; Purnichescu-Purtan, R.; Chen, M.; Yeung, S.-C.J.; Calin, G.A. Circulating miRNAs in sepsis—A network under attack: An in-silico prediction of the potential existence of miRNA sponges in sepsis. PLoS ONE 2017, 12, e0183334. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Alexander, R.P.; Fang, G.; Rozowsky, J.; Snyder, M.; Gerstein, M.B. Annotating non-coding regions of the genome. Nat. Rev. Genet. 2010, 11, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Izzotti, A.; Calin, G.A.; Steele, V.E.; Croce, C.M.; De Flora, S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2009, 23, 3243–3250. [Google Scholar] [CrossRef] [PubMed]
- Ferdin, J.; Nishida, N.; Wu, X.; Nicoloso, M.S.; Shah, M.Y.; Devlin, C.; Ling, H.; Shimizu, M.; Kumar, K.; Cortez, M.A.; et al. HINCUTs in cancer: Hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ. 2013, 20, 1675–1687. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Rigoutsos, I.; Lee, S.K.; Nam, S.Y.; Anfossi, S.; Pasculli, B.; Pichler, M.; Jing, Y.; Rodriguez-Aguayo, C.; Telonis, A.G.; Rossi, S.; et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol. 2017, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.W.; Lo, Y.M.; Leung, S.F.; Tsang, Y.S.; Chan, L.Y.; Johnson, P.J.; Hjelm, N.M.; Lee, J.C.; Huang, D.P. Analysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin. Chem. 1999, 45, 1292–1294. [Google Scholar]
- Kopreski, M.S.; Benko, F.A.; Kwak, L.W.; Gocke, C.D. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1999, 5, 1961–1965. [Google Scholar]
- Holford, N.C.; Sandhu, H.S.; Thakkar, H.; Butt, A.N.; Swaminathan, R. Stability of β-Actin mRNA in Plasma. Ann. N. Y. Acad. Sci. 2008, 1137, 108–111. [Google Scholar] [CrossRef]
- Fernando, M.R.; Norton, S.E.; Luna, K.K.; Lechner, J.M.; Qin, J. Stabilization of cell-free RNA in blood samples using a new collection device. Clin. Biochem. 2012, 45, 1497–1502. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Pardini, B.; Calin, G.A. MicroRNAs and Long Non-Coding RNAs and Their Hormone-Like Activities in Cancer. Cancers (Basel) 2019, 11, 378. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Kohli, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.; Patel, T.; et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci. Rep. 2016, 6, 19413. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, G.; Cordero, F.; Tarallo, S.; Arigoni, M.; Riccardo, F.; Gallo, G.; Ronco, G.; Allasia, M.; Kulkarni, N.; Matullo, G.; et al. Small non-coding RNA profiling in human biofluids and surrogate tissues from healthy individuals: Description of the diverse and most represented species. Oncotarget 2018, 9, 3097–3111. [Google Scholar] [CrossRef] [PubMed]
- Yeri, A.; Courtright, A.; Reiman, R.; Carlson, E.; Beecroft, T.; Janss, A.; Siniard, A.; Richholt, R.; Balak, C.; Rozowsky, J.; et al. Total Extracellular Small RNA Profiles from Plasma, Saliva, and Urine of Healthy Subjects. Sci. Rep. 2017, 7, 44061. [Google Scholar] [CrossRef]
- Freedman, J.E.; Gerstein, M.; Mick, E.; Rozowsky, J.; Levy, D.; Kitchen, R.; Das, S.; Shah, R.; Danielson, K.; Beaulieu, L.; et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat. Commun. 2016, 7, 11106. [Google Scholar] [CrossRef]
- Ben-Dov, I.Z.; Whalen, V.M.; Goilav, B.; Max, K.E.; Tuschl, T. Cell and Microvesicle Urine microRNA Deep Sequencing Profiles from Healthy Individuals: Observations with Potential Impact on Biomarker Studies. PLoS ONE 2016, 11, e0147249. [Google Scholar] [CrossRef]
- Umu, S.U.; Langseth, H.; Bucher-Johannessen, C.; Fromm, B.; Keller, A.; Meese, E.; Lauritzen, M.; Leithaug, M.; Lyle, R.; Rounge, T.B. A comprehensive profile of circulating RNAs in human serum. RNA Biol. 2018, 15, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Yao, J.; Wu, D.C.; Nottingham, R.M.; Mohr, S.; Hunicke-Smith, S.; Lambowitz, A.M. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 2016, 22, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Danielson, K.M.; Rubio, R.; Abderazzaq, F.; Das, S.; Wang, Y.E. High Throughput Sequencing of Extracellular RNA from Human Plasma. PLoS ONE 2017, 12, e0164644. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Vickers, K.; Xiong, Y.; Zhao, S.; Sheng, Q.; Zhang, P.; Zhou, W.; Flynn, C.R. Comprehensive evaluation of extracellular small RNA isolation methods from serum in high throughput sequencing. BMC Genom. 2017, 18, 50. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Lobb, R.J.; Hastie, M.L.; Norris, E.L.; van Amerongen, R.; Gorman, J.J.; Moller, A. Oncogenic transformation of lung cells results in distinct exosome protein profile similar to the cell of origin. Proteomics 2017, 17. [Google Scholar] [CrossRef]
- Ma, P.; Pan, Y.; Li, W.; Sun, C.; Liu, J.; Xu, T.; Shu, Y. Extracellular vesicles-mediated noncoding RNAs transfer in cancer. J. Hematol. Oncol. 2017, 10, 57. [Google Scholar] [CrossRef]
- Yu, Z.; Zhao, S.; Ren, L.; Wang, L.; Chen, Z.; Hoffman, R.M.; Zhou, J. Pancreatic cancer-derived exosomes promote tumor metastasis and liver pre-metastatic niche formation. Oncotarget 2017, 8, 63461–63483. [Google Scholar] [CrossRef]
- Barile, L.; Vassalli, G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017, 174, 63–78. [Google Scholar] [CrossRef]
- Takahashi, K.; Yan, I.K.; Wood, J.; Haga, H.; Patel, T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol. Cancer Res. MCR 2014, 12, 1377–1387. [Google Scholar] [CrossRef]
- Takahashi, K.; Yan, I.K.; Kogure, T.; Haga, H.; Patel, T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 2014, 4, 458–467. [Google Scholar] [CrossRef]
- Bullock, M.D.; Silva, A.M.; Kanlikilicer-Unaldi, P.; Filant, J.; Rashed, M.H.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. Exosomal Non-Coding RNAs: Diagnostic, Prognostic and Therapeutic Applications in Cancer. Noncoding RNA 2015, 1, 53–68. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006, 20, 2202–2207. [Google Scholar] [CrossRef]
- Awad, H.; Bratasz, A.; Nuovo, G.; Burry, R.; Meng, X.; Kelani, H.; Brown, M.; Ramadan, M.E.; Williams, J.; Bouhliqah, L.; et al. MiR-155 deletion reduces ischemia-induced paralysis in an aortic aneurysm repair mouse model: Utility of immunohistochemistry and histopathology in understanding etiology of spinal cord paralysis. Ann. Diagn. Pathol. 2018, 36, 12–20. [Google Scholar] [CrossRef]
- Fabbri, M.; Garzon, R.; Andreeff, M.; Kantarjian, H.M.; Garcia-Manero, G.; Calin, G.A. MicroRNAs and noncoding RNAs in hematological malignancies: Molecular, clinical and therapeutic implications. Leukemia 2008, 22, 1095–1105. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin. Oncol. 2006, 33, 167–173. [Google Scholar] [CrossRef]
- Sevignani, C.; Calin, G.A.; Nnadi, S.C.; Shimizu, M.; Davuluri, R.V.; Hyslop, T.; Demant, P.; Croce, C.M.; Siracusa, L.D. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc. Natl. Acad. Sci. USA 2007, 104, 8017–8022. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Munker, R.; Calin, G.A. MicroRNA profiling in cancer. Clin. Sci. (Lond.) 2011, 121, 141–158. [Google Scholar] [CrossRef]
- Cogswell, J.P.; Ward, J.; Taylor, I.A.; Waters, M.; Shi, Y.; Cannon, B.; Kelnar, K.; Kemppainen, J.; Brown, D.; Chen, C.; et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimer’s Dis. JAD 2008, 14, 27–41. [Google Scholar] [CrossRef]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef]
- Pardini, B.; Cordero, F.; Naccarati, A.; Viberti, C.; Birolo, G.; Oderda, M.; Di Gaetano, C.; Arigoni, M.; Martina, F.; Calogero, R.A.; et al. microRNA profiles in urine by next-generation sequencing can stratify bladder cancer subtypes. Oncotarget 2018, 9, 20658–20669. [Google Scholar] [CrossRef]
- Da Silveira, J.C.; Veeramachaneni, D.N.; Winger, Q.A.; Carnevale, E.M.; Bouma, G.J. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: A possible new form of cell communication within the ovarian follicle. Biol. Reprod. 2012, 86, 71. [Google Scholar] [CrossRef]
- Iftikhar, H.; Carney, G.E. Evidence and potential in vivo functions for biofluid miRNAs: From expression profiling to functional testing: Potential roles of extracellular miRNAs as indicators of physiological change and as agents of intercellular information exchange. Bioessays 2016, 38, 367–378. [Google Scholar] [CrossRef]
- Anfossi, S.; Fu, X.; Nagvekar, R.; Calin, G.A. MicroRNAs, Regulatory Messengers Inside and Outside Cancer Cells. Adv. Exp. Med. Biol. 2018, 1056, 87–108. [Google Scholar] [CrossRef]
- Bullrich, F.; Fujii, H.; Calin, G.; Mabuchi, H.; Negrini, M.; Pekarsky, Y.; Rassenti, L.; Alder, H.; Reed, J.C.; Keating, M.J.; et al. Characterization of the 13q14 tumor suppressor locus in CLL: Identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res. 2001, 61, 6640–6648. [Google Scholar]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Katakowski, M.; Buller, B.; Wang, X.; Rogers, T.; Chopp, M. Functional microRNA is transferred between glioma cells. Cancer Res. 2010, 70, 8259–8263. [Google Scholar] [CrossRef]
- Redis, R.S.; Calin, S.; Yang, Y.; You, M.J.; Calin, G.A. Cell-to-cell miRNA transfer: From body homeostasis to therapy. Pharmacol. Ther. 2012, 136, 169–174. [Google Scholar] [CrossRef]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidus, J.; Chang, B.H.; Kurre, P. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci. Rep. 2015, 5, 11295. [Google Scholar] [CrossRef]
- Larrea, E.; Sole, C.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.M.; Araujo, A.M.; Araiz, M.; Fernandez-Mercado, M.; et al. New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies. Int. J. Mol. Sci. 2016, 17, 627. [Google Scholar] [CrossRef]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef]
- Sohn, W.; Kim, J.; Kang, S.H.; Yang, S.R.; Cho, J.Y.; Cho, H.C.; Shim, S.G.; Paik, Y.H. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 2015, 47, e184. [Google Scholar] [CrossRef]
- Xu, J.; Cao, Z.; Liu, W.; You, L.; Zhou, L.; Wang, C.; Lou, W.; Sun, B.; Miao, Y.; Liu, X.; et al. Plasma miRNAs Effectively Distinguish Patients With Pancreatic Cancer From Controls: A Multicenter Study. Ann. Surg. 2016, 263, 1173–1179. [Google Scholar] [CrossRef]
- Cao, Z.; Liu, C.; Xu, J.; You, L.; Wang, C.; Lou, W.; Sun, B.; Miao, Y.; Liu, X.; Wang, X.; et al. Plasma microRNA panels to diagnose pancreatic cancer: Results from a multicenter study. Oncotarget 2016, 7, 41575–41583. [Google Scholar] [CrossRef]
- Komatsu, S.; Ichikawa, D.; Miyamae, M.; Kawaguchi, T.; Morimura, R.; Hirajima, S.; Okajima, W.; Ohashi, T.; Imamura, T.; Konishi, H.; et al. Malignant potential in pancreatic neoplasm; new insights provided by circulating miR-223 in plasma. Expert Opin. Biol. Ther. 2015, 15, 773–785. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, L.; Xiao, Y.; Zhu, J.; Li, Z. Circulating microRNA-182 in plasma and its potential diagnostic and prognostic value for pancreatic cancer. Med. Oncol. 2014, 31, 225. [Google Scholar] [CrossRef]
- Cote, G.A.; Gore, A.J.; McElyea, S.D.; Heathers, L.E.; Xu, H.; Sherman, S.; Korc, M. A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am. J. Gastroenterol. 2014, 109, 1942–1952. [Google Scholar] [CrossRef]
- Li, A.; Yu, J.; Kim, H.; Wolfgang, C.L.; Canto, M.I.; Hruban, R.H.; Goggins, M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 3600–3610. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, J.; Zhang, S.; Yu, D.; Chen, Y.; Liu, Q.; Shi, M.; Ni, C.; Zhu, M. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol. Rep. 2013, 30, 276–284. [Google Scholar] [CrossRef]
- Bhat, S.A.; Majid, S.; Rehman, M.U. Scenario and future prospects of microRNAs in gastric cancer: A review. Iran. J. Basic Med. Sci. 2019, 22, 345–352. [Google Scholar] [CrossRef]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef]
- Liu, X.; Pan, B.; Sun, L.; Chen, X.; Zeng, K.; Hu, X.; Xu, T.; Xu, M.; Wang, S. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2018, 27, 746–754. [Google Scholar] [CrossRef]
- Kral, J.; Korenkova, V.; Novosadova, V.; Langerova, L.; Schneiderova, M.; Liska, V.; Levy, M.; Veskrnova, V.; Spicak, J.; Opattova, A.; et al. Expression profile of miR-17/92 cluster is predictive of treatment response in rectal cancer. Carcinogenesis 2018, 39, 1359–1367. [Google Scholar] [CrossRef]
- Tsukamoto, M.; Iinuma, H.; Yagi, T.; Matsuda, K.; Hashiguchi, Y. Circulating Exosomal MicroRNA-21 as a Biomarker in Each Tumor Stage of Colorectal Cancer. Oncology 2017, 92, 360–370. [Google Scholar] [CrossRef]
- Ren, D.; Lin, B.; Zhang, X.; Peng, Y.; Ye, Z.; Ma, Y.; Liang, Y.; Cao, L.; Li, X.; Li, R.; et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget 2017, 8, 49807–49823. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, Y.; Guo, X.; Zhou, L.; Jia, Z.; Peng, Z.; Tang, Y.; Liu, W.; Zhu, B.; Wang, L.; et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J. Cell. Mol. Med. 2017, 21, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Nadal, E.; Truini, A.; Nakata, A.; Lin, J.; Reddy, R.M.; Chang, A.C.; Ramnath, N.; Gotoh, N.; Beer, D.G.; Chen, G. A Novel Serum 4-microRNA Signature for Lung Cancer Detection. Sci. Rep. 2015, 5, 12464. [Google Scholar] [CrossRef] [PubMed]
- Moltzahn, F.; Olshen, A.B.; Baehner, L.; Peek, A.; Fong, L.; Stoppler, H.; Simko, J.; Hilton, J.F.; Carroll, P.; Blelloch, R. Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res. 2011, 71, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Lodes, M.J.; Caraballo, M.; Suciu, D.; Munro, S.; Kumar, A.; Anderson, B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS ONE 2009, 4, e6229. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal miRNAs as Biomarkers for Prostate Cancer. Front. Genet. 2013, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Samsonov, R.; Shtam, T.; Burdakov, V.; Glotov, A.; Tsyrlina, E.; Berstein, L.; Nosov, A.; Evtushenko, V.; Filatov, M.; Malek, A. Lectin-induced agglutination method of urinary exosomes isolation followed by mi-RNA analysis: Application for prostate cancer diagnostic. Prostate 2016, 76, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, A.; Brennan, S.; Kennedy, P.J.; Hutvagner, G.; Tran, N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci. Rep. 2016, 6, 24922. [Google Scholar] [CrossRef]
- Mello-Grand, M.; Gregnanin, I.; Sacchetto, L.; Ostano, P.; Zitella, A.; Bottoni, G.; Oderda, M.; Marra, G.; Munegato, S.; Pardini, B.; et al. Circulating microRNAs combined with PSA for accurate and non-invasive prostate cancer detection. Carcinogenesis 2019, 40, 246–253. [Google Scholar] [CrossRef]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef]
- Petrescu, G.E.D.; Sabo, A.A.; Torsin, L.I.; Calin, G.A.; Dragomir, M.P. MicroRNA based theranostics for brain cancer: Basic principles. J. Exp. Clin. Cancer Res. CR 2019, 38, 231. [Google Scholar] [CrossRef]
- Giannopoulou, L.; Kasimir-Bauer, S.; Lianidou, E.S. Liquid biopsy in ovarian cancer: Recent advances on circulating tumor cells and circulating tumor DNA. Clin. Chem. Lab. Med. 2018, 56, 186–197. [Google Scholar] [CrossRef]
- Andreu, Z.; Otta Oshiro, R.; Redruello, A.; Lopez-Martin, S.; Gutierrez-Vazquez, C.; Morato, E.; Marina, A.I.; Olivier Gomez, C.; Yanez-Mo, M. Extracellular vesicles as a source for non-invasive biomarkers in bladder cancer progression. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2017, 98, 70–79. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Fujita, K.; Jingushi, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Ueda, Y.; Tanigawa, G.; Yoshioka, I.; Ueda, K.; et al. MiR-21-5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget 2017, 8, 24668–24678. [Google Scholar] [CrossRef]
- Foj, L.; Ferrer, F.; Serra, M.; Arevalo, A.; Gavagnach, M.; Gimenez, N.; Filella, X. Exosomal and Non-Exosomal Urinary miRNAs in Prostate Cancer Detection and Prognosis. Prostate 2017, 77, 573–583. [Google Scholar] [CrossRef]
- Rodriguez, M.; Bajo-Santos, C.; Hessvik, N.P.; Lorenz, S.; Fromm, B.; Berge, V.; Sandvig, K.; Line, A.; Llorente, A. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 2017, 16, 156. [Google Scholar] [CrossRef]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef]
- Shen, M.M.; Abate-Shen, C. Molecular genetics of prostate cancer: New prospects for old challenges. Genes Dev. 2010, 24, 1967–2000. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Chen, A.; Wang, L.; Li, B.Y.; Sherman, J.; Ryu, J.E.; Hamamura, K.; Liu, Y.; Nakshatri, H.; Yokota, H. Reduction in Migratory Phenotype in a Metastasized Breast Cancer Cell Line via Downregulation of S100A4 and GRM3. Sci. Rep. 2017, 7, 3459. [Google Scholar] [CrossRef]
- Giannopoulou, L.; Zavridou, M.; Kasimir-Bauer, S.; Lianidou, E.S. Liquid biopsy in ovarian cancer: The potential of circulating miRNAs and exosomes. Transl. Res. J. Lab. Clin. Med. 2019, 205, 77–91. [Google Scholar] [CrossRef]
- Xie, B.; Ding, Q.; Han, H.; Wu, D. miRCancer: A microRNA–cancer association database constructed by text mining on literature. Bioinformatics 2013, 29, 638–644. [Google Scholar] [CrossRef]
- Big Data Analytics in Bioinformatics and Healthcare; Baoying, W., Ruowang, L., William, P., Eds.; IGI Global: Hershey, PA, USA, 2015; pp. 1–528. [Google Scholar]
- Xie, B.; Hochberg, R.; Ding, Q.; Wu, D. miRSAT & miRCDB: An Integrated MicroRNA Sequesce Analysis Tool and a Cancer-Associated MicroRNA Database. In Proceedings of the ISCA 2nd International Conference on Bioinformatics and Computational Biology, BICoB-2010, Honolulu, HI, USA, 24–26 March 2010; pp. 159–164. [Google Scholar]
- Choudhury, S.N.; Li, Y. miR-21 and let-7 in the Ras and NF-kappaB pathways. Microrna 2012, 1, 65–69. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 2009, 106, 7113–7118. [Google Scholar] [CrossRef]
- Fernandez-Mercado, M.; Manterola, L.; Lawrie, C.H. MicroRNAs in Lymphoma: Regulatory Role and Biomarker Potential. Curr. Genom. 2015, 16, 349–358. [Google Scholar] [CrossRef]
- Zhi, F.; Shao, N.; Wang, R.; Deng, D.; Xue, L.; Wang, Q.; Zhang, Y.; Shi, Y.; Xia, X.; Wang, S.; et al. Identification of 9 serum microRNAs as potential noninvasive biomarkers of human astrocytoma. Neuro-oncology 2015, 17, 383–391. [Google Scholar] [CrossRef]
- Goze, C.; Reynes, C.; Forestier, L.; Sabatier, R.; Duffau, H. Pilot Study of Whole Blood MicroRNAs as Potential Tools for Diffuse Low-Grade Gliomas Detection. Cell. Mol. Neurobiol. 2018, 38, 715–725. [Google Scholar] [CrossRef]
- Yang, C.; Wang, C.; Chen, X.; Chen, S.; Zhang, Y.; Zhi, F.; Wang, J.; Li, L.; Zhou, X.; Li, N.; et al. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int. J. Cancer 2013, 132, 116–127. [Google Scholar] [CrossRef]
- Witwer, K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef]
- Mo, M.H.; Chen, L.; Fu, Y.; Wang, W.; Fu, S.W. Cell-free Circulating miRNA Biomarkers in Cancer. J. Cancer 2012, 3, 432–448. [Google Scholar] [CrossRef]
- Munagala, R.; Aqil, F.; Gupta, R.C. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumour Biol. J. Int. Soc. Oncodeve. Biol. Med. 2016, 37, 10703–10714. [Google Scholar] [CrossRef]
- Shi, R.; Wang, P.Y.; Li, X.Y.; Chen, J.X.; Li, Y.; Zhang, X.Z.; Zhang, C.G.; Jiang, T.; Li, W.B.; Ding, W.; et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget 2015, 6, 26971–26981. [Google Scholar] [CrossRef]
- Chiam, K.; Wang, T.; Watson, D.I.; Mayne, G.C.; Irvine, T.S.; Bright, T.; Smith, L.; White, I.A.; Bowen, J.M.; Keefe, D.; et al. Circulating Serum Exosomal miRNAs As Potential Biomarkers for Esophageal Adenocarcinoma. J. Gastrointest. Surg. Off. J. Soc. Surg. Aliment. Tract 2015, 19, 1208–1215. [Google Scholar] [CrossRef]
- Warnecke-Eberz, U.; Chon, S.H.; Holscher, A.H.; Drebber, U.; Bollschweiler, E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: Comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. J. Int. Soc. Oncodeve. Biol. Med. 2015, 36, 4643–4653. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Duechler, M.; Czyz, M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014, 347, 29–37. [Google Scholar] [CrossRef]
- Aushev, V.N.; Zborovskaya, I.B.; Laktionov, K.K.; Girard, N.; Cros, M.P.; Herceg, Z.; Krutovskikh, V. Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS ONE 2013, 8, e78649. [Google Scholar] [CrossRef]
- Camacho, L.; Guerrero, P.; Marchetti, D. MicroRNA and protein profiling of brain metastasis competent cell-derived exosomes. PLoS ONE 2013, 8, e73790. [Google Scholar] [CrossRef]
- Chugh, P.E.; Sin, S.H.; Ozgur, S.; Henry, D.H.; Menezes, P.; Griffith, J.; Eron, J.J.; Damania, B.; Dittmer, D.P. Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog. 2013, 9, e1003484. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Valentino, A.; Reclusa, P.; Sirera, R.; Giallombardo, M.; Camps, C.; Pauwels, P.; Crispi, S.; Rolfo, C. Exosomal microRNAs in liquid biopsies: Future biomarkers for prostate cancer. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Nat. Cancer Inst. Mex. 2017, 19, 651–657. [Google Scholar] [CrossRef]
- Fabris, L.; Ceder, Y.; Chinnaiyan, A.M.; Jenster, G.W.; Sorensen, K.D.; Tomlins, S.; Visakorpi, T.; Calin, G.A. The Potential of MicroRNAs as Prostate Cancer Biomarkers. Eur. Urol. 2016, 70, 312–322. [Google Scholar] [CrossRef]
- Meng, Y.; Sun, J.; Wang, X.; Hu, T.; Ma, Y.; Kong, C.; Piao, H.; Yu, T.; Zhang, G. Exosomes: A Promising Avenue for the Diagnosis of Breast Cancer. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef]
- Joyce, D.P.; Kerin, M.J.; Dwyer, R.M. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int. J. Cancer 2016, 139, 1443–1448. [Google Scholar] [CrossRef]
- Nassar, F.J.; Nasr, R.; Talhouk, R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol. Ther. 2017, 172, 34–49. [Google Scholar] [CrossRef]
- Drokow, E.K.; Sun, K.; Ahmed, H.A.W.; Akpabla, G.S.; Song, J.; Shi, M. Circulating microRNA as diagnostic biomarkers for haematological cancers: A systematic review and meta-analysis. Cancer Manag. Res. 2019, 11, 4313–4326. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, Z.; Pang, Y.; Cui, L.; Qian, T.; Quan, L.; Zhao, H.; Shi, J.; Ke, X.; Fu, L. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 2019, 12, 51. [Google Scholar] [CrossRef]
- Mirzaei, H.; Fathullahzadeh, S.; Khanmohammadi, R.; Darijani, M.; Momeni, F.; Masoudifar, A.; Goodarzi, M.; Mardanshah, O.; Stenvang, J.; Jaafari, M.R.; et al. State of the art in microRNA as diagnostic and therapeutic biomarkers in chronic lymphocytic leukemia. J. Cell. Physiol. 2018, 233, 888–900. [Google Scholar] [CrossRef]
- Ciccone, M.; Calin, G.A. MicroRNAs in Chronic Lymphocytic Leukemia: An Old Disease with New Genetic Insights. Microrna 2016, 5, 106–112. [Google Scholar] [CrossRef]
- Litwinska, Z.; Machalinski, B. miRNAs in chronic myeloid leukemia: Small molecules, essential function. Leuk Lymphoma 2017, 58, 1297–1305. [Google Scholar] [CrossRef]
- Carvalho de Oliveira, J.; Molinari Roberto, G.; Baroni, M.; Bezerra Salomao, K.; Alejandra Pezuk, J.; Sol Brassesco, M. MiRNA Dysregulation in Childhood Hematological Cancer. Int. J. Mol. Sci. 2018, 19, 2688. [Google Scholar] [CrossRef]
- Zhuo, X.; Zhou, W.; Li, D.; Chang, A.; Wang, Y.; Wu, Y.; Zhou, Q. Plasma microRNA expression signature involving miR-548q, miR-630 and miR-940 as biomarkers for nasopharyngeal carcinoma detection. Cancer Biomark. 2018, 23, 579–587. [Google Scholar] [CrossRef]
- Liu, X.; Luo, H.N.; Tian, W.D.; Lu, J.; Li, G.; Wang, L.; Zhang, B.; Liang, B.J.; Peng, X.H.; Lin, S.X.; et al. Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol. Ther. 2013, 14, 1133–1142. [Google Scholar] [CrossRef]
- Zheng, X.H.; Cui, C.; Ruan, H.L.; Xue, W.Q.; Zhang, S.D.; Hu, Y.Z.; Zhou, X.X.; Jia, W.H. Plasma microRNA profiling in nasopharyngeal carcinoma patients reveals miR-548q and miR-483-5p as potential biomarkers. Chin. J. Cancer 2014, 33, 330–338. [Google Scholar] [CrossRef]
- Wang, L.; Yu, B.; Cen, J.; Peng, X.; Liu, Y.; Zeng, F.; Liu, X. Clinical significance of plasma miR-24 dysregulation in nasopharyngeal carcinoma. Nan Fang Yi Ke Da Xue Xue Bao 2015, 35, 743–747. [Google Scholar]
- Zeng, X.; Xiang, J.; Wu, M.; Xiong, W.; Tang, H.; Deng, M.; Li, X.; Liao, Q.; Su, B.; Luo, Z.; et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS ONE 2012, 7, e46367. [Google Scholar] [CrossRef]
- Plieskatt, J.L.; Rinaldi, G.; Feng, Y.; Levine, P.H.; Easley, S.; Martinez, E.; Hashmi, S.; Sadeghi, N.; Brindley, P.J.; Bethony, J.M.; et al. Methods and matrices: Approaches to identifying miRNAs for nasopharyngeal carcinoma. J. Transl. Med. 2014, 12, 3. [Google Scholar] [CrossRef]
- Wen, W.; Mai, S.J.; Lin, H.X.; Zhang, M.Y.; Huang, J.L.; Hua, X.; Lin, C.; Long, Z.Q.; Lu, Z.J.; Sun, X.Q.; et al. Identification of two microRNA signatures in whole blood as novel biomarkers for diagnosis of nasopharyngeal carcinoma. J. Transl. Med. 2019, 17, 186. [Google Scholar] [CrossRef]
- Yi, S.J.; Liu, P.; Chen, B.L.; Ou-Yang, L.; Xiong, W.M.; Su, J.P. Circulating miR-31-5p may be a potential diagnostic biomarker in nasopharyngeal carcinoma. Neoplasma 2019. [Google Scholar] [CrossRef]
- Wang, H.; Wei, X.; Wu, B.; Su, J.; Tan, W.; Yang, K. Tumor-educated platelet miR-34c-3p and miR-18a-5p as potential liquid biopsy biomarkers for nasopharyngeal carcinoma diagnosis. Cancer Manag. Res. 2019, 11, 3351–3360. [Google Scholar] [CrossRef]
- Loosen, S.H.; Lurje, G.; Wiltberger, G.; Vucur, M.; Koch, A.; Kather, J.N.; Paffenholz, P.; Tacke, F.; Ulmer, F.T.; Trautwein, C.; et al. Serum levels of miR-29, miR-122, miR-155 and miR-192 are elevated in patients with cholangiocarcinoma. PLoS ONE 2019, 14, e0210944. [Google Scholar] [CrossRef]
- Bernuzzi, F.; Marabita, F.; Lleo, A.; Carbone, M.; Mirolo, M.; Marzioni, M.; Alpini, G.; Alvaro, D.; Boberg, K.M.; Locati, M.; et al. Serum microRNAs as novel biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Clin. Exp. Immunol. 2016, 185, 61–71. [Google Scholar] [CrossRef]
- Wu, X.; Xia, M.; Chen, D.; Wu, F.; Lv, Z.; Zhan, Q.; Jiao, Y.; Wang, W.; Chen, G.; An, F. Profiling of downregulated blood-circulating miR-150-5p as a novel tumor marker for cholangiocarcinoma. Tumour Biol. 2016, 37, 15019–15029. [Google Scholar] [CrossRef]
- Wang, L.J.; Zhang, K.L.; Zhang, N.; Ma, X.W.; Yan, S.W.; Cao, D.H.; Shi, S.J. Serum miR-26a as a diagnostic and prognostic biomarker in cholangiocarcinoma. Oncotarget 2015, 6, 18631–18640. [Google Scholar] [CrossRef]
- Cheng, Q.; Feng, F.; Zhu, L.; Zheng, Y.; Luo, X.; Liu, C.; Yi, B.; Jiang, X. Circulating miR-106a is a Novel Prognostic and Lymph Node Metastasis Indicator for Cholangiocarcinoma. Sci. Rep. 2015, 5, 16103. [Google Scholar] [CrossRef]
- Voigtlander, T.; Gupta, S.K.; Thum, S.; Fendrich, J.; Manns, M.P.; Lankisch, T.O.; Thum, T. MicroRNAs in Serum and Bile of Patients with Primary Sclerosing Cholangitis and/or Cholangiocarcinoma. PLoS ONE 2015, 10, e0139305. [Google Scholar] [CrossRef]
- Correa-Gallego, C.; Maddalo, D.; Doussot, A.; Kemeny, N.; Kingham, T.P.; Allen, P.J.; D’Angelica, M.I.; DeMatteo, R.P.; Betel, D.; Klimstra, D.; et al. Circulating Plasma Levels of MicroRNA-21 and MicroRNA-221 Are Potential Diagnostic Markers for Primary Intrahepatic Cholangiocarcinoma. PLoS ONE 2016, 11, e0163699. [Google Scholar] [CrossRef]
- Wang, S.; Yin, J.; Li, T.; Yuan, L.; Wang, D.; He, J.; Du, X.; Lu, J. Upregulated circulating miR-150 is associated with the risk of intrahepatic cholangiocarcinoma. Oncol. Rep. 2015, 33, 819–825. [Google Scholar] [CrossRef]
- Shigehara, K.; Yokomuro, S.; Ishibashi, O.; Mizuguchi, Y.; Arima, Y.; Kawahigashi, Y.; Kanda, T.; Akagi, I.; Tajiri, T.; Yoshida, H.; et al. Real-Time PCR-Based Analysis of the Human Bile MicroRNAome Identifies miR-9 as a Potential Diagnostic Biomarker for Biliary Tract Cancer. PLoS ONE 2011, 6, e23584. [Google Scholar] [CrossRef]
- Kojima, M.; Sudo, H.; Kawauchi, J.; Takizawa, S.; Kondou, S.; Nobumasa, H.; Ochiai, A. MicroRNA markers for the diagnosis of pancreatic and biliary-tract cancers. PLoS ONE 2015, 10, e0118220. [Google Scholar] [CrossRef]
- Baraniskin, A.; Nopel-Dunnebacke, S.; Schumacher, B.; Gerges, C.; Bracht, T.; Sitek, B.; Meyer, H.E.; Gerken, G.; Dechene, A.; Schlaak, J.F.; et al. Analysis of U2 small nuclear RNA fragments in the bile differentiates cholangiocarcinoma from primary sclerosing cholangitis and other benign biliary disorders. Dig. Dis. Sci. 2014, 59, 1436–1441. [Google Scholar] [CrossRef]
- Li, G.; Pu, Y. MicroRNA signatures in total peripheral blood of gallbladder cancer patients. Tumour Biol. 2015, 36, 6985–6990. [Google Scholar] [CrossRef]
- Kitagawa, T.; Taniuchi, K.; Tsuboi, M.; Sakaguchi, M.; Kohsaki, T.; Okabayashi, T.; Saibara, T. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol. Oncol. 2019, 13, 212–227. [Google Scholar] [CrossRef]
- Baraniskin, A.; Nopel-Dunnebacke, S.; Ahrens, M.; Jensen, S.G.; Zollner, H.; Maghnouj, A.; Wos, A.; Mayerle, J.; Munding, J.; Kost, D.; et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int. J. Cancer 2013, 132, E48–E57. [Google Scholar] [CrossRef]
- Cui, L.; Lou, Y.; Zhang, X.; Zhou, H.; Deng, H.; Song, H.; Yu, X.; Xiao, B.; Wang, W.; Guo, J. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin. Biochem. 2011, 44, 1050–1057. [Google Scholar] [CrossRef]
- Yu, B.; Du, Q.; Li, H.; Liu, H.Y.; Ye, X.; Zhu, B.; Zhai, Q.; Li, X.X. Diagnostic potential of serum exosomal colorectal neoplasia differentially expressed long non-coding RNA (CRNDE-p) and microRNA-217 expression in colorectal carcinoma. Oncotarget 2017, 8, 83745–83753. [Google Scholar] [CrossRef]
- Vychytilova-Faltejskova, P.; Stitkovcova, K.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Kala, Z.; Svoboda, M.; Kiss, I.; Vyzula, R.; et al. Circulating PIWI-Interacting RNAs piR-5937 and piR-28876 Are Promising Diagnostic Biomarkers of Colon Cancer. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Pre. Oncol. 2018, 27, 1019–1028. [Google Scholar] [CrossRef]
- Qu, A.; Wang, W.; Yang, Y.; Zhang, X.; Dong, Y.; Zheng, G.; Wu, Q.; Zou, M.; Du, L.; Wang, Y.; et al. A serum piRNA signature as promising non-invasive diagnostic and prognostic biomarkers for colorectal cancer. Cancer Manag. Res. 2019, 11, 3703–3720. [Google Scholar] [CrossRef]
- Liao, J.; Yu, L.; Mei, Y.; Guarnera, M.; Shen, J.; Li, R.; Liu, Z.; Jiang, F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer 2010, 9, 198. [Google Scholar] [CrossRef]
- Su, J.; Liao, J.; Gao, L.; Shen, J.; Guarnera, M.A.; Zhan, M.; Fang, H.; Stass, S.A.; Jiang, F. Analysis of small nucleolar RNAs in sputum for lung cancer diagnosis. Oncotarget 2016, 7, 5131–5142. [Google Scholar] [CrossRef]
- Kohler, J.; Schuler, M.; Gauler, T.C.; Nopel-Dunnebacke, S.; Ahrens, M.; Hoffmann, A.C.; Kasper, S.; Nensa, F.; Gomez, B.; Hahnemann, M.; et al. Circulating U2 small nuclear RNA fragments as a diagnostic and prognostic biomarker in lung cancer patients. J. Cancer Res. Clin. Oncol. 2016, 142, 795–805. [Google Scholar] [CrossRef]
- Mazieres, J.; Catherinne, C.; Delfour, O.; Gouin, S.; Rouquette, I.; Delisle, M.B.; Prevot, G.; Escamilla, R.; Didier, A.; Persing, D.H.; et al. Alternative processing of the U2 small nuclear RNA produces a 19-22nt fragment with relevance for the detection of non-small cell lung cancer in human serum. PLoS ONE 2013, 8, e60134. [Google Scholar] [CrossRef]
- Arantes, L.; De Carvalho, A.C.; Melendez, M.E.; Lopes Carvalho, A. Serum, plasma and saliva biomarkers for head and neck cancer. Expert Rev. Mol. Diagn. 2018, 18, 85–112. [Google Scholar] [CrossRef]
- Wang, J.L.; Wang, X.; Yang, D.; Shi, W.J. The Expression of MicroRNA-155 in Plasma and Tissue Is Matched in Human Laryngeal Squamous Cell Carcinoma. Yonsei Med. J. 2016, 57, 298–305. [Google Scholar] [CrossRef]
- Ayaz, L.; Gorur, A.; Yaroglu, H.Y.; Ozcan, C.; Tamer, L. Differential expression of microRNAs in plasma of patients with laryngeal squamous cell carcinoma: Potential early-detection markers for laryngeal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2013, 139, 1499–1506. [Google Scholar] [CrossRef]
- Lucas Grzelczyk, W.; Szemraj, J.; Kwiatkowska, S.; Jozefowicz-Korczynska, M. Serum expression of selected miRNAs in patients with laryngeal squamous cell carcinoma (LSCC). Diagn. Pathol. 2019, 14, 49. [Google Scholar] [CrossRef]
- Zhou, E.; Luo, L.; Ma, L.J.; Yin, J.; Tan, Z.Q.; Miao, G.Y.; Liu, J.; Xiao, X.P. Diagnostic value of serum miRNA let-7a for laryngeal carcinoma and effects of let-7a on proliferation and apoptosis of laryngeal carcinoma cells. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 2019, 54, 534–539. [Google Scholar] [CrossRef]
- Xu, Y.; Lin, Y.P.; Yang, D.; Zhang, G.; Zhou, H.F. Expression of serum microRNA-378 and its clinical significance in laryngeal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 5137–5142. [Google Scholar]
- Yilmaz, S.S.; Guzel, E.; Karatas, O.F.; Yilmaz, M.; Creighton, C.J.; Ozen, M. MiR-221 as a pre- and postoperative plasma biomarker for larynx cancer patients. Laryngoscope 2015, 125, E377–E381. [Google Scholar] [CrossRef]
- Iliev, R.; Fedorko, M.; Machackova, T.; Mlcochova, H.; Svoboda, M.; Pacik, D.; Dolezel, J.; Stanik, M.; Slaby, O. Expression Levels of PIWI-interacting RNA, piR-823, Are Deregulated in Tumor Tissue, Blood Serum and Urine of Patients with Renal Cell Carcinoma. Anticancer Res. 2016, 36, 6419–6423. [Google Scholar] [CrossRef]
- Eissa, S.; Safwat, M.; Matboli, M.; Zaghloul, A.; El-Sawalhi, M.; Shaheen, A. Measurement of Urinary Level of a Specific Competing endogenous RNA network (FOS and RCAN mRNA/miR-324-5p, miR-4738-3p,/lncRNA miR-497-HG) Enables Diagnosis of Bladder Cancer. Urol. Oncol. 2019, 37, 292.e219–292.e227. [Google Scholar] [CrossRef]
- Appaiah, H.N.; Goswami, C.P.; Mina, L.A.; Badve, S.; Sledge, G.W., Jr.; Liu, Y.; Nakshatri, H. Persistent upregulation of U6:SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Res. BCR 2011, 13, R86. [Google Scholar] [CrossRef]
- Kuhlmann, J.D.; Baraniskin, A.; Hahn, S.A.; Mosel, F.; Bredemeier, M.; Wimberger, P.; Kimmig, R.; Kasimir-Bauer, S. Circulating U2 small nuclear RNA fragments as a novel diagnostic tool for patients with epithelial ovarian cancer. Clin. Chem. 2014, 60, 206–213. [Google Scholar] [CrossRef]
- Muinelo-Romay, L.; Casas-Arozamena, C.; Abal, M. Liquid Biopsy in Endometrial Cancer: New Opportunities for Personalized Oncology. Int. J. Mol. Sci. 2018, 19, 2311. [Google Scholar] [CrossRef]
- Torres, A.; Torres, K.; Pesci, A.; Ceccaroni, M.; Paszkowski, T.; Cassandrini, P.; Zamboni, G.; Maciejewski, R. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int. J. Cancer 2013, 132, 1633–1645. [Google Scholar] [CrossRef]
- Montagnana, M.; Benati, M.; Danese, E.; Giudici, S.; Perfranceschi, M.; Ruzzenenete, O.; Salvagno, G.L.; Bassi, A.; Gelati, M.; Paviati, E.; et al. Aberrant MicroRNA Expression in Patients With Endometrial Cancer. Int. J. Gynecol. Cancer 2017, 27, 459–466. [Google Scholar] [CrossRef]
- Jia, W.; Wu, Y.; Zhang, Q.; Gao, G.; Zhang, C.; Xiang, Y. Identification of four serum microRNAs from a genome-wide serum microRNA expression profile as potential non-invasive biomarkers for endometrioid endometrial cancer. Oncol. Lett. 2013, 6, 261–267. [Google Scholar] [CrossRef]
- Benati, M.; Montagnana, M.; Danese, E.; Paviati, E.; Giudici, S.; Franchi, M.; Lippi, G. Evaluation of mir-203 Expression Levels and DNA Promoter Methylation Status in Serum of Patients with Endometrial Cancer. Clin. Lab. 2017, 63, 1675–1681. [Google Scholar] [CrossRef]
- Gao, Y.; Dai, M.; Liu, H.; He, W.; Lin, S.; Yuan, T.; Chen, H.; Dai, S. Diagnostic value of circulating miR-21: An update meta-analysis in various cancers and validation in endometrial cancer. Oncotarget 2016, 7, 68894–68908. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, N.; Yin, D.; Li, Y.K.; Guo, L.; Shi, L.P.; Huang, X. Changes in the Expression of Serum MiR-887-5p in Patients With Endometrial Cancer. Int. J. Gynecol. Cancer 2016, 26, 1143–1147. [Google Scholar] [CrossRef]
- Srivastava, A.; Moxley, K.; Ruskin, R.; Dhanasekaran, D.N.; Zhao, Y.D.; Ramesh, R. A Non-invasive Liquid Biopsy Screening of Urine-Derived Exosomes for miRNAs as Biomarkers in Endometrial Cancer Patients. AAPS J. 2018, 20, 82. [Google Scholar] [CrossRef]
- Nahand, J.S.; Taghizadeh-Boroujeni, S.; Karimzadeh, M.; Borran, S.; Pourhanifeh, M.H.; Moghoofei, M.; Bokharaei-Salim, F.; Karampoor, S.; Jafari, A.; Asemi, Z.; et al. microRNAs: New prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J. Cell. Physiol. 2019, 234, 17064–17099. [Google Scholar] [CrossRef]
- Okoye, J.O.; Ngokere, A.A.; Onyenekwe, C.C.; Erinle, C.A. Comparable expression of miR-let-7b, miR-21, miR-182, miR-145, and p53 in serum and cervical cells: Diagnostic implications for early detection of cervical lesions. Int. J. Health Sci. (Qassim) 2019, 13, 29–38. [Google Scholar]
- Xin, F.; Liu, P.; Ma, C.F. A circulating serum miRNA panel as early detection biomarkers of cervical intraepithelial neoplasia. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4846–4851. [Google Scholar]
- Jia, W.; Wu, Y.; Zhang, Q.; Gao, G.E.; Zhang, C.; Xiang, Y. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol. Clin. Oncol. 2015, 3, 851–858. [Google Scholar] [CrossRef]
- Kong, Q.; Tang, Z.; Xiang, F.; Jiang, J.; Yue, H.; Wu, R.; Kang, X. Diagnostic Value of Serum hsa-mir-92a in Patients with Cervical Cancer. Clin. Lab. 2017, 63, 335–340. [Google Scholar] [CrossRef]
- Wang, W.T.; Zhao, Y.N.; Yan, J.X.; Weng, M.Y.; Wang, Y.; Chen, Y.Q.; Hong, S.J. Differentially expressed microRNAs in the serum of cervical squamous cell carcinoma patients before and after surgery. J. Hematol. Oncol. 2014, 7, 6. [Google Scholar] [CrossRef]
- Ma, Q.; Wan, G.; Wang, S.; Yang, W.; Zhang, J.; Yao, X. Serum microRNA-205 as a novel biomarker for cervical cancer patients. Cancer Cell Int. 2014, 14, 81. [Google Scholar] [CrossRef]
- Khare, D.; Goldschmidt, N.; Bardugo, A.; Gur-Wahnon, D.; Ben-Dov, I.Z.; Avni, B. Plasma microRNA profiling: Exploring better biomarkers for lymphoma surveillance. PLoS ONE 2017, 12, e0187722. [Google Scholar] [CrossRef]
- Cordeiro, A.; Navarro, A.; Gaya, A.; Diaz-Beya, M.; Gonzalez-Farre, B.; Castellano, J.J.; Fuster, D.; Martinez, C.; Martinez, A.; Monzo, M. PiwiRNA-651 as marker of treatment response and survival in classical Hodgkin lymphoma. Oncotarget 2016, 7, 46002–46013. [Google Scholar] [CrossRef]
- Lopez-Santillan, M.; Larrabeiti-Etxebarria, A.; Arzuaga-Mendez, J.; Lopez-Lopez, E.; Garcia-Orad, A. Circulating miRNAs as biomarkers in diffuse large B-cell lymphoma: A systematic review. Oncotarget 2018, 9, 22850–22861. [Google Scholar] [CrossRef]
- Drusco, A.; Bottoni, A.; Lagana, A.; Acunzo, M.; Fassan, M.; Cascione, L.; Antenucci, A.; Kumchala, P.; Vicentini, C.; Gardiman, M.P.; et al. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget 2015, 6, 20829–20839. [Google Scholar] [CrossRef]
- Jurkovicova, D.; Lukackova, R.; Magyerkova, M.; Kulcsar, L.; Krivjanska, M.; Krivjansky, V.; Chovanec, M. microRNA expression profiling as supportive diagnostic and therapy prediction tool in chronic myeloid leukemia. Neoplasma 2015, 62, 949–958. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, Y.; Han, X.; Roy, M.; Liu, W.; Zhao, X.; Liu, J. Differential expression profiles and functional analysis of plasma miRNAs associated with chronic myeloid leukemia phases. Future Oncol. 2019, 15, 763–776. [Google Scholar] [CrossRef]
- Fallah, P.; Amirizadeh, N.; Poopak, B.; Toogeh, G.; Arefian, E.; Kohram, F.; Hosseini Rad, S.M.; Kohram, M.; Teimori Naghadeh, H.; Soleimani, M. Expression pattern of key microRNAs in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Int. J. Lab. Hematol. 2015, 37, 560–568. [Google Scholar] [CrossRef]
- Rashed, W.M.; Hammad, A.M.; Saad, A.M.; Shohdy, K.S. MicroRNA as a diagnostic biomarker in childhood acute lymphoblastic leukemia; systematic review, meta-analysis and recommendations. Crit. Rev. Oncol. Hematol. 2019, 136, 70–78. [Google Scholar] [CrossRef]
- Zhou, P.; Li, X. Serum miR-338-5p has potential for use as a tumor marker for retinoblastoma. Oncol. Lett. 2019, 18, 307–313. [Google Scholar] [CrossRef]
- Chen, Y.Z.; Liu, Z.P.; Zhou, K.Y.; Li, B. Value of serum miR-17-92 cluster in diagnosis of retinoblastoma. Zhongguo Dang Dai Er Ke Za Zhi 2017, 19, 776–780. [Google Scholar]
- Beta, M.; Venkatesan, N.; Vasudevan, M.; Vetrivel, U.; Khetan, V.; Krishnakumar, S. Identification and Insilico Analysis of Retinoblastoma Serum microRNA Profile and Gene Targets Towards Prediction of Novel Serum Biomarkers. Bioinform. Biol. Insights 2013, 7, 21–34. [Google Scholar] [CrossRef]
- Liu, S.S.; Wang, Y.S.; Sun, Y.F.; Miao, L.X.; Wang, J.; Li, Y.S.; Liu, H.Y.; Liu, Q.L. Plasma microRNA-320, microRNA-let-7e and microRNA-21 as novel potential biomarkers for the detection of retinoblastoma. Biomed. Rep. 2014, 2, 424–428. [Google Scholar] [CrossRef]
- Li, M.; Song, Q.; Li, H.; Lou, Y.; Wang, L. Circulating miR-25-3p and miR-451a May Be Potential Biomarkers for the Diagnosis of Papillary Thyroid Carcinoma. PLoS ONE 2015, 10, e0132403. [Google Scholar] [CrossRef]
- Wang, Z.; Lv, J.; Zou, X.; Huang, Z.; Zhang, H.; Liu, Q.; Jiang, L.; Zhou, X.; Zhu, W. A three plasma microRNA signature for papillary thyroid carcinoma diagnosis in Chinese patients. Gene 2019, 693, 37–45. [Google Scholar] [CrossRef]
- Yu, S.; Liu, X.; Zhang, Y.; Li, J.; Chen, S.; Zheng, H.; Reng, R.; Zhang, C.; Chen, J.; Chen, L. Circulating microRNA124-3p, microRNA9-3p and microRNA196b-5p may be potential signatures for differential diagnosis of thyroid nodules. Oncotarget 2016, 7, 84165–84177. [Google Scholar] [CrossRef][Green Version]
- Lee, Y.S.; Lim, Y.S.; Lee, J.C.; Wang, S.G.; Park, H.Y.; Kim, S.Y.; Lee, B.J. Differential expression levels of plasma-derived miR-146b and miR-155 in papillary thyroid cancer. Oral Oncol. 2015, 51, 77–83. [Google Scholar] [CrossRef]
- Lee, J.C.; Zhao, J.T.; Clifton-Bligh, R.J.; Gill, A.; Gundara, J.S.; Ip, J.C.; Glover, A.; Sywak, M.S.; Delbridge, L.W.; Robinson, B.G.; et al. MicroRNA-222 and microRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer 2013, 119, 4358–4365. [Google Scholar] [CrossRef]
- Shabani, N.; Sheikholeslami, S.; Paryan, M.; Zarif Yeganeh, M.; Tavangar, S.M.; Azizi, F.; Mohammadi-Yeganeh, S.; Hedayati, M. An investigation on the expression of miRNAs including miR-144 and miR-34a in plasma samples of RET-positive and RET-negative medullar thyroid carcinoma patients. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, J.; Xu, D.; Yang, Z.; Sun, J.; Sun, L.; Wu, Y.; Qiao, H. Combination of serum microRNAs and ultrasound profile as predictive biomarkers of diagnosis and prognosis for papillary thyroid microcarcinoma. Oncol. Rep. 2018, 40, 3611–3624. [Google Scholar] [CrossRef]
- Pilli, T.; Cantara, S.; Marzocchi, C.; Cardinale, S.; Santini, C.; Cevenini, G.; Pacini, F. Diagnostic Value of Circulating microRNA-95 and -190 in the Differential Diagnosis of Thyroid Nodules: A Validation Study in 1000 Consecutive Patients. Thyroid 2017, 27, 1053–1057. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, D.; Pan, J.; Yang, Z.; Chen, M.; Han, J.; Zhang, S.; Sun, L.; Qiao, H. Dynamic monitoring of circulating microRNAs as a predictive biomarker for the diagnosis and recurrence of papillary thyroid carcinoma. Oncol. Lett. 2017, 13, 4252–4266. [Google Scholar] [CrossRef]
- Igci, Y.Z.; Ozkaya, M.; Korkmaz, H.; Bozgeyik, E.; Bayraktar, R.; Ulasli, M.; Erkilic, S.; Eraydin, A.; Oztuzcu, S. Expression Levels of miR-30a-5p in Papillary Thyroid Carcinoma: A Comparison Between Serum and Fine Needle Aspiration Biopsy Samples. Genet. Test. Mol. Biomark. 2015, 19, 418–423. [Google Scholar] [CrossRef]
- Graham, M.E.; Hart, R.D.; Douglas, S.; Makki, F.M.; Pinto, D.; Butler, A.L.; Bullock, M.; Rigby, M.H.; Trites, J.R.; Taylor, S.M.; et al. Serum microRNA profiling to distinguish papillary thyroid cancer from benign thyroid masses. J. Otolaryngol. Head Neck Surg. 2015, 44, 33. [Google Scholar] [CrossRef]
- Cantara, S.; Pilli, T.; Sebastiani, G.; Cevenini, G.; Busonero, G.; Cardinale, S.; Dotta, F.; Pacini, F. Circulating miRNA95 and miRNA190 are sensitive markers for the differential diagnosis of thyroid nodules in a Caucasian population. J. Clin. Endocrinol. Metab. 2014, 99, 4190–4198. [Google Scholar] [CrossRef]
- Yoruker, E.E.; Terzioglu, D.; Teksoz, S.; Uslu, F.E.; Gezer, U.; Dalay, N. MicroRNA Expression Profiles in Papillary Thyroid Carcinoma, Benign Thyroid Nodules and Healthy Controls. J. Cancer 2016, 7, 803–809. [Google Scholar] [CrossRef]
- Yu, S.; Liu, Y.; Wang, J.; Guo, Z.; Zhang, Q.; Yu, F.; Zhang, Y.; Huang, K.; Li, Y.; Song, E.; et al. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2012, 97, 2084–2092. [Google Scholar] [CrossRef]
- Mahmoudian-Sani, M.R.; Mehri-Ghahfarrokhi, A.; Asadi-Samani, M.; Mobini, G.R. Serum miRNAs as Biomarkers for the Diagnosis and Prognosis of Thyroid Cancer: A Comprehensive Review of the Literature. Eur. Thyroid J. 2017, 6, 171–177. [Google Scholar] [CrossRef][Green Version]
- Samsonov, R.; Burdakov, V.; Shtam, T.; Radzhabova, Z.; Vasilyev, D.; Tsyrlina, E.; Titov, S.; Ivanov, M.; Berstein, L.; Filatov, M.; et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumour Biol. 2016, 37, 12011–12021. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Q.; Ma, S.; Sun, Y.; Vadamootoo, A.S.; Jin, C. A four serum-miRNA panel serves as a potential diagnostic biomarker of osteosarcoma. Int. J. Clin. Oncol. 2019, 24, 976–982. [Google Scholar] [CrossRef]
- Fujiwara, T.; Uotani, K.; Yoshida, A.; Morita, T.; Nezu, Y.; Kobayashi, E.; Yoshida, A.; Uehara, T.; Omori, T.; Sugiu, K.; et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget 2017, 8, 33375–33392. [Google Scholar] [CrossRef]
- Monterde-Cruz, L.; Ramirez-Salazar, E.G.; Rico-Martinez, G.; Linares-Gonzalez, L.M.; Guzman-Gonzalez, R.; Delgado-Cedillo, E.; Estrada-Villasenor, E.; Valdes-Flores, M.; Velazquez-Cruz, R.; Hidalgo-Bravo, A. Circulating miR-215-5p and miR-642a-5p as potential biomarker for diagnosis of osteosarcoma in Mexican population. Hum. Cell 2018, 31, 292–299. [Google Scholar] [CrossRef]
- Yao, Z.S.; Li, C.; Liang, D.; Jiang, X.B.; Tang, J.J.; Ye, L.Q.; Yuan, K.; Ren, H.; Yang, Z.D.; Jin, D.X.; et al. Diagnostic and prognostic implications of serum miR-101 in osteosarcoma. Cancer Biomark. Sect. A Dis. Markers 2018, 22, 127–133. [Google Scholar] [CrossRef]
- Cong, C.; Wang, W.; Tian, J.; Gao, T.; Zheng, W.; Zhou, C. Identification of serum miR-124 as a biomarker for diagnosis and prognosis in osteosarcoma. Cancer Biomark. 2018, 21, 449–454. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, X.; Zhang, Y.J.; Fang, G.W.; Xue, Y. MicroRNA-375 as a potential serum biomarker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J. Int. Med. Res. 2018, 46, 975–983. [Google Scholar] [CrossRef]
- Liu, K.; Sun, X.; Zhang, Y.; Liu, L.; Yuan, Q. MiR-598: A tumor suppressor with biomarker significance in osteosarcoma. Life Sci. 2017, 188, 141–148. [Google Scholar] [CrossRef]
- Niu, J.; Sun, Y.; Guo, Q.; Niu, D.; Liu, B. Serum miR-95-3p is a diagnostic and prognostic marker for osteosarcoma. Springerplus 2016, 5, 1947. [Google Scholar] [CrossRef]
- Liu, J.D.; Xin, Q.; Tao, C.S.; Sun, P.F.; Xu, P.; Wu, B.; Qu, L.; Li, S.Z. Serum miR-300 as a diagnostic and prognostic biomarker in osteosarcoma. Oncol. Lett. 2016, 12, 3912–3918. [Google Scholar] [CrossRef][Green Version]
- Pang, P.C.; Shi, X.Y.; Huang, W.L.; Sun, K. miR-497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3765–3769. [Google Scholar]
- Cao, L.; Wang, J.; Wang, P.Q. MiR-326 is a diagnostic biomarker and regulates cell survival and apoptosis by targeting Bcl-2 in osteosarcoma. Biomed. Pharmacother. 2016, 84, 828–835. [Google Scholar] [CrossRef]
- Dong, J.; Liu, Y.; Liao, W.; Liu, R.; Shi, P.; Wang, L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J. Bone Oncol. 2016, 5, 74–79. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, B.; Hu, J.; Zhou, Y.; Jiang, M.; Wu, M.; Qin, L.; Yang, X. miR-421 is a diagnostic and prognostic marker in patients with osteosarcoma. Tumour Biol. 2016, 37, 9001–9007. [Google Scholar] [CrossRef]
- Li, H.; Zhang, K.; Liu, L.H.; Ouyang, Y.; Guo, H.B.; Zhang, H.; Bu, J.; Xiao, T. MicroRNA screening identifies circulating microRNAs as potential biomarkers for osteosarcoma. Oncol. Lett. 2015, 10, 1662–1668. [Google Scholar] [CrossRef][Green Version]
- Wang, N.G.; Wang, D.C.; Tan, B.Y.; Wang, F.; Yuan, Z.N. Down-regulation of microRNA152 is associated with the diagnosis and prognosis of patients with osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 9314–9319. [Google Scholar]
- Wang, T.; Ji, F.; Dai, Z.; Xie, Y.; Yuan, D. Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer Biomark. 2015, 15, 543–550. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Zhang, X.; Zhang, M.; Liu, H.; Zhang, S.; Qi, B.; Sun, X. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed. Pharmacother. 2015, 75, 153–158. [Google Scholar] [CrossRef]
- Zhou, G.; Lu, M.; Chen, J.; Li, C.; Zhang, J.; Chen, J.; Shi, X.; Wu, S. Identification of miR-199a-5p in serum as noninvasive biomarkers for detecting and monitoring osteosarcoma. Tumour Biol. 2015, 36, 8845–8852. [Google Scholar] [CrossRef]
- Tang, J.; Zhao, H.; Cai, H.; Wu, H. Diagnostic and prognostic potentials of microRNA-27a in osteosarcoma. Biomed. Pharmacother. 2015, 71, 222–226. [Google Scholar] [CrossRef]
- Cai, H.; Zhao, H.; Tang, J.; Wu, H. Serum miR-195 is a diagnostic and prognostic marker for osteosarcoma. J. Surg. Res. 2015, 194, 505–510. [Google Scholar] [CrossRef]
- Zhang, C.; Yao, C.; Li, H.; Wang, G.; He, X. Serum levels of microRNA-133b and microRNA-206 expression predict prognosis in patients with osteosarcoma. Int. J. Clin. Exp. Pathol. 2014, 7, 4194–4203. [Google Scholar]
- Hong, Q.; Fang, J.; Pang, Y.; Zheng, J. Prognostic value of the microRNA-29 family in patients with primary osteosarcomas. Med. Oncol. 2014, 31, 37. [Google Scholar] [CrossRef]
- Huang, C.; Sun, Y.; Ma, S.; Vadamootoo, A.S.; Wang, L.; Jin, C. Identification of circulating miR-663a as a potential biomarker for diagnosing osteosarcoma. Pathol. Res. Pract. 2019, 215, 152411. [Google Scholar] [CrossRef]
- Lian, F.; Cui, Y.; Zhou, C.; Gao, K.; Wu, L. Identification of a plasma four-microRNA panel as potential noninvasive biomarker for osteosarcoma. PLoS ONE 2015, 10, e0121499. [Google Scholar] [CrossRef]
- Zhou, L.; Ma, X.; Yue, J.; Chen, T.; Wang, X.Y.; Wang, Z.W.; Pan, J.; Lin, Y. The diagnostic effect of serum miR-139-5p as an indicator in osteosarcoma. Cancer Biomark. 2018, 23, 561–567. [Google Scholar] [CrossRef]
- Hua, J.; Liu, D.; Cao, L.; Wang, D.; Wu, T.; Lin, F.; Su, P.; Niu, Y.; Sun, Y. Diagnostic and prognostic values of blood microRNA-Let7A for osteosarcoma. J. Bone Oncol. 2018, 12, 65–68. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, X.; Chai, J.; Chen, P.; Ren, P.; Gong, M. Circulating miR-148a is a significant diagnostic and prognostic biomarker for patients with osteosarcoma. Tumour Biol. 2014, 35, 12467–12472. [Google Scholar] [CrossRef]
- Polini, B.; Carpi, S.; Romanini, A.; Breschi, M.C.; Nieri, P.; Podesta, A. Circulating cell-free microRNAs in cutaneous melanoma staging and recurrence or survival prognosis. Pigment Cell Melanoma Res. 2019, 32, 486–499. [Google Scholar] [CrossRef]
- Mumford, S.L.; Towler, B.P.; Pashler, A.L.; Gilleard, O.; Martin, Y.; Newbury, S.F. Circulating MicroRNA Biomarkers in Melanoma: Tools and Challenges in Personalised Medicine. Biomolecules 2018, 8, 21. [Google Scholar] [CrossRef]
- Margue, C.; Reinsbach, S.; Philippidou, D.; Beaume, N.; Walters, C.; Schneider, J.G.; Nashan, D.; Behrmann, I.; Kreis, S. Comparison of a healthy miRNome with melanoma patient miRNomes: Are microRNAs suitable serum biomarkers for cancer? Oncotarget 2015, 6, 12110–12127. [Google Scholar] [CrossRef]
- Van Laar, R.; Lincoln, M.; Van Laar, B. Development and validation of a plasma-based melanoma biomarker suitable for clinical use. Br. J. Cancer 2018, 118, 857–866. [Google Scholar] [CrossRef]
- Sole, C.; Tramonti, D.; Schramm, M.; Goicoechea, I.; Armesto, M.; Hernandez, L.I.; Manterola, L.; Fernandez-Mercado, M.; Mujika, K.; Tuneu, A.; et al. The Circulating Transcriptome as a Source of Biomarkers for Melanoma. Cancers (Basel) 2019, 11, 70. [Google Scholar] [CrossRef]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef]
- Tosar, J.P.; Rovira, C.; Cayota, A. Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun. Biol. 2018, 1, 2. [Google Scholar] [CrossRef]
- Halic, M.; Moazed, D. Transposon silencing by piRNAs. Cell 2009, 138, 1058–1060. [Google Scholar] [CrossRef]
- Weick, E.M.; Miska, E.A. piRNAs: From biogenesis to function. Development 2014, 141, 3458–3471. [Google Scholar] [CrossRef]
- Weng, W.; Li, H.; Goel, A. Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 160–169. [Google Scholar] [CrossRef]
- Ha, H.; Song, J.; Wang, S.; Kapusta, A.; Feschotte, C.; Chen, K.C.; Xing, J. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genom. 2014, 15, 545. [Google Scholar] [CrossRef]
- Bahn, J.H.; Zhang, Q.; Li, F.; Chan, T.M.; Lin, X.; Kim, Y.; Wong, D.T.; Xiao, X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015, 61, 221–230. [Google Scholar] [CrossRef]
- Slaby, O. Non-coding RNAs as Biomarkers for Colorectal Cancer Screening and Early Detection. Adv. Exp. Med. Biol. 2016, 937, 153–170. [Google Scholar] [CrossRef]
- Kiss, T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 2002, 109, 145–148. [Google Scholar] [CrossRef]
- Klinge, S.; Woolford, J.L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Sloan, K.E. Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biol. Chem. 2018, 399, 1265–1276. [Google Scholar] [CrossRef]
- Dieci, G.; Preti, M.; Montanini, B. Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics 2009, 94, 83–88. [Google Scholar] [CrossRef]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.P.; Zhang, B.X.; Chu, L. Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef]
- Yamamura, S.; Imai-Sumida, M.; Tanaka, Y.; Dahiya, R. Interaction and cross-talk between non-coding RNAs. Cell. Mol. Life Sci. 2018, 75, 467–484. [Google Scholar] [CrossRef]
- Ender, C.; Krek, A.; Friedlander, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Chow, R.D.; Chen, S. Sno-derived RNAs are prevalent molecular markers of cancer immunity. Oncogene 2018, 37, 6442–6462. [Google Scholar] [CrossRef]
- Stepanov, G.A.; Filippova, J.A.; Komissarov, A.B.; Kuligina, E.V.; Richter, V.A.; Semenov, D.V. Regulatory role of small nucleolar RNAs in human diseases. BioMed Res. Int. 2015, 2015, 206849. [Google Scholar] [CrossRef]
- Su, Y.; Guarnera, M.A.; Fang, H.; Jiang, F. Small non-coding RNA biomarkers in sputum for lung cancer diagnosis. Mol. Cancer 2016, 15, 36. [Google Scholar] [CrossRef]
- Schmitz, U.; Naderi-Meshkin, H.; Gupta, S.K.; Wolkenhauer, O.; Vera, J. The RNA world in the 21st century-a systems approach to finding non-coding keys to clinical questions. Brief. Bioinform. 2016, 17, 380–392. [Google Scholar] [CrossRef]
- Han, D.; Wang, M.; Ma, N.; Xu, Y.; Jiang, Y.; Gao, X. Long noncoding RNAs: Novel players in colorectal cancer. Cancer Lett. 2015, 361, 13–21. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, L.; Yu, F.; Zhang, Y.; Li, P.; Wang, K. The functional roles of exosomal long non-coding RNAs in cancer. Cell. Mol. Life Sci. 2019, 76, 2059–2076. [Google Scholar] [CrossRef]
- Lin, T.; Hou, P.F.; Meng, S.; Chen, F.; Jiang, T.; Li, M.L.; Shi, M.L.; Liu, J.J.; Zheng, J.N.; Bai, J. Emerging Roles of p53 Related lncRNAs in Cancer Progression: A Systematic Review. Int. J. Biol. Sci. 2019, 15, 1287–1298. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Li, Q.; Shao, Y.; Zhang, X.; Zheng, T.; Miao, M.; Qin, L.; Wang, B.; Ye, G.; Xiao, B.; Guo, J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 2007–2012. [Google Scholar] [CrossRef]
- Naderi-Meshkin, H.; Lai, X.; Amirkhah, R.; Vera, J.; Rasko, J.E.J.; Schmitz, U. Exosomal lncRNAs and cancer: Connecting the missing links. Bioinformatics 2019, 35, 352–360. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, F.; Chen, H.; Tan, Q.; Qiu, S.; Chen, S.; Jing, W.; Yu, M.; Liang, C.; Ye, S.; et al. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging (Albany N. Y.) 2016, 8, 2023–2038. [Google Scholar] [CrossRef]
- Xie, H.; Ma, H.; Zhou, D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed Res. Int. 2013, 2013, 136106. [Google Scholar] [CrossRef]
- Tong, Y.S.; Wang, X.W.; Zhou, X.L.; Liu, Z.H.; Yang, T.X.; Shi, W.H.; Xie, H.W.; Lv, J.; Wu, Q.Q.; Cao, X.F. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol. Cancer 2015, 14, 3. [Google Scholar] [CrossRef]
- Tang, Q.; Ni, Z.; Cheng, Z.; Xu, J.; Yu, H.; Yin, P. Three circulating long non-coding RNAs act as biomarkers for predicting NSCLC. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2015, 37, 1002–1009. [Google Scholar] [CrossRef]
- Tang, J.; Jiang, R.; Deng, L.; Zhang, X.; Wang, K.; Sun, B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget 2015, 6, 4505–4515. [Google Scholar] [CrossRef]
- Qu, L.; Ding, J.; Chen, C.; Wu, Z.J.; Liu, B.; Gao, Y.; Chen, W.; Liu, F.; Sun, W.; Li, X.F.; et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016, 29, 653–668. [Google Scholar] [CrossRef]
- Liu, X.F.; Thin, K.Z.; Ming, X.L.; Shuo, L.; Ping, L.; Man, Z.; Li, N.D.; Tu, J.C. Small Nucleolar RNA Host Gene 18 Acts as a Tumor Suppressor and a Diagnostic Indicator in Hepatocellular Carcinoma. Technol. Cancer Res. Treat. 2018, 17. [Google Scholar] [CrossRef]
- Liu, M.; Xing, L.Q.; Liu, Y.J. A three-long noncoding RNA signature as a diagnostic biomarker for differentiating between triple-negative and non-triple-negative breast cancers. Medicine 2017, 96, e6222. [Google Scholar] [CrossRef]
- Kamel, L.M.; Atef, D.M.; Mackawy, A.M.; Shalaby, S.M.; Abdelraheim, N. Circulating long non-coding RNA GAS5 and SOX2OT as potential biomarkers for diagnosis and prognosis of non-small cell lung cancer. Biotechnol. Appl. Biochem. 2019. [Google Scholar] [CrossRef]
- Isin, M.; Ozgur, E.; Cetin, G.; Erten, N.; Aktan, M.; Gezer, U.; Dalay, N. Investigation of circulating lncRNAs in B-cell neoplasms. Clin. Chim. Acta Int. J. Clin. Chem. 2014, 431, 255–259. [Google Scholar] [CrossRef]
- Gao, S.; Xu, X.; Wang, Y.; Zhang, W.; Wang, X. Diagnostic utility of plasma lncRNA small nucleolar RNA host gene 1 in patients with hepatocellular carcinoma. Mol. Med. Rep. 2018, 18, 3305–3313. [Google Scholar] [CrossRef]
- Dong, L.; Qi, P.; Xu, M.D.; Ni, S.J.; Huang, D.; Xu, Q.H.; Weng, W.W.; Tan, C.; Sheng, W.Q.; Zhou, X.Y.; et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int. J. Cancer J. Int. Cancer 2015, 137, 1128–1135. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, Y.; Wang, Z.; Zheng, J.; Chen, Y.; Li, X.; Wang, Y.; Ming, H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017, 490, 406–414. [Google Scholar] [CrossRef]
- Wang, L.; Duan, W.; Yan, S.; Xie, Y.; Wang, C. Circulating long non-coding RNA colon cancer-associated transcript 2 protected by exosome as a potential biomarker for colorectal cancer. Biomed. Pharmacother. 2019, 113, 108758. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, X.; Gao, S.; Jing, F.; Yang, Y.; Du, L.; Zheng, G.; Li, P.; Li, C.; Wang, C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 2016, 7, 85551–85563. [Google Scholar] [CrossRef]
- Berrondo, C.; Flax, J.; Kucherov, V.; Siebert, A.; Osinski, T.; Rosenberg, A.; Fucile, C.; Richheimer, S.; Beckham, C.J. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS ONE 2016, 11, e0147236. [Google Scholar] [CrossRef]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef]
- Isin, M.; Uysaler, E.; Ozgur, E.; Koseoglu, H.; Sanli, O.; Yucel, O.B.; Gezer, U.; Dalay, N. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front. Genet. 2015, 6, 168. [Google Scholar] [CrossRef]
- Donovan, M.J.; Noerholm, M.; Bentink, S.; Belzer, S.; Skog, J.; O’Neill, V.; Cochran, J.S.; Brown, G.A. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015, 18, 370–375. [Google Scholar] [CrossRef]
- Tang, H.; Wu, Z.; Zhang, J.; Su, B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol. Med. Rep. 2013, 7, 761–766. [Google Scholar] [CrossRef]
- Xie, Z.; Chen, X.; Li, J.; Guo, Y.; Li, H.; Pan, X.; Jiang, J.; Liu, H.; Wu, B. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget 2016, 7, 25408–25419. [Google Scholar] [CrossRef]
- Ge, X.; Wang, Y.; Nie, J.; Li, Q.; Tang, L.; Deng, X.; Wang, F.; Xu, B.; Wu, X.; Zhang, X.; et al. The diagnostic/prognostic potential and molecular functions of long non-coding RNAs in the exosomes derived from the bile of human cholangiocarcinoma. Oncotarget 2017, 8, 69995–70005. [Google Scholar] [CrossRef]
- Shao, Y.; Ye, M.; Jiang, X.; Sun, W.; Ding, X.; Liu, Z.; Ye, G.; Zhang, X.; Xiao, B.; Guo, J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer 2014, 120, 3320–3328. [Google Scholar] [CrossRef]
- Shah, M.Y.; Ferracin, M.; Pileczki, V.; Chen, B.; Redis, R.; Fabris, L.; Zhang, X.; Ivan, C.; Shimizu, M.; Rodriguez-Aguayo, C.; et al. Cancer-associated rs6983267 SNP and its accompanying long noncoding RNA CCAT2 induce myeloid malignancies via unique SNP-specific RNA mutations. Genome Res. 2018, 28, 432–447. [Google Scholar] [CrossRef]
- Redis, R.S.; Vela, L.E.; Lu, W.; Ferreira de Oliveira, J.; Ivan, C.; Rodriguez-Aguayo, C.; Adamoski, D.; Pasculli, B.; Taguchi, A.; Chen, Y.; et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol. Cell 2016, 61, 520–534. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Taheri, M. Colon Cancer-Associated Transcripts 1 and 2: Roles and functions in human cancers. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef]
- Shao, T.; Huang, J.; Zheng, Z.; Wu, Q.; Liu, T.; Lv, X. SCCA, TSGF, and the Long Non-Coding RNA AC007271.3 are Effective Biomarkers for Diagnosing Oral Squamous Cell Carcinoma. Cell. Physiol. Biochem. 2018, 47, 26–38. [Google Scholar] [CrossRef]
- Zhao, S.Y.; Wang, J.; Ouyang, S.B.; Huang, Z.K.; Liao, L. Salivary Circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as Novel Biomarkers for the Diagnosis of Oral Squamous Cell Carcinoma. Cell. Physiol. Biochem. 2018, 47, 2511–2521. [Google Scholar] [CrossRef]
- Fan, C.M.; Wang, J.P.; Tang, Y.Y.; Zhao, J.; He, S.Y.; Xiong, F.; Guo, C.; Xiang, B.; Zhou, M.; Li, X.L.; et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019, 110, 2180–2188. [Google Scholar] [CrossRef]
- He, B.; Zeng, J.; Chao, W.; Chen, X.; Huang, Y.; Deng, K.; Huang, Z.; Li, J.; Dai, M.; Chen, S.; et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget 2017, 8, 41166–41177. [Google Scholar] [CrossRef]
- Shuai, M.; Hong, J.; Huang, D.; Zhang, X.; Tian, Y. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol. Lett. 2018, 16, 6495–6501. [Google Scholar] [CrossRef]
- Sun, K.; Zhao, X.; Wan, J.; Yang, L.; Chu, J.; Dong, S.; Yin, H.; Ming, L.; He, F. The diagnostic value of long non-coding RNA MIR31HG and its role in esophageal squamous cell carcinoma. Life Sci. 2018, 202, 124–130. [Google Scholar] [CrossRef]
- Wang, W.; He, X.; Zheng, Z.; Ma, X.; Hu, X.; Wu, D.; Wang, M. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol. Cancer 2017, 16, 75. [Google Scholar] [CrossRef]
- Wang, Y.; Jing, W.; Ma, W.; Liang, C.; Chai, H.; Tu, J. Down-regulation of long non-coding RNA GAS5-AS1 and its prognostic and diagnostic significance in hepatocellular carcinoma. Cancer Biomark. 2018, 22, 227–236. [Google Scholar] [CrossRef]
- Luo, P.; Liang, C.; Zhang, X.; Liu, X.; Wang, Y.; Wu, M.; Feng, X.; Tu, J. Identification of long non-coding RNA ZFAS1 as a novel biomarker for diagnosis of HCC. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Ma, X.; Wang, X.; Yang, C.; Wang, Z.; Han, B.; Wu, L.; Zhuang, L. DANCR Acts as a Diagnostic Biomarker and Promotes Tumor Growth and Metastasis in Hepatocellular Carcinoma. Anticancer Res. 2016, 36, 6389–6398. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Tang, J.; Jiang, R.; Zhang, W.; Ji, J.; Sun, B. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2015, 37, 687–696. [Google Scholar] [CrossRef]
- Jing, W.; Gao, S.; Zhu, M.; Luo, P.; Jing, X.; Chai, H.; Tu, J. Potential diagnostic value of lncRNA SPRY4-IT1 in hepatocellular carcinoma. Oncol. Rep. 2016, 36, 1085–1092. [Google Scholar] [CrossRef]
- Shaker, O.G.; Ayoub, S.E.; Abdelwahed, M.Y.; Ahmed, N.A.; Hassan, E.A.; Ahmed, T.I.; Abousarie, M.A. Evaluation of serum long noncoding RNA NEAT and MiR-129-5p in hepatocellular carcinoma. IUBMB Life 2019. [Google Scholar] [CrossRef]
- Wang, Z.F.; Hu, R.; Pang, J.M.; Zhang, G.Z.; Yan, W.; Li, Z.N. Serum long noncoding RNA LRB1 as a potential biomarker for predicting the diagnosis and prognosis of human hepatocellular carcinoma. Oncol. Lett. 2018, 16, 1593–1601. [Google Scholar] [CrossRef]
- Zheng, Z.K.; Pang, C.; Yang, Y.; Duan, Q.; Zhang, J.; Liu, W.C. Serum long noncoding RNA urothelial carcinoma-associated 1: A novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. J. Int. Med. Res. 2018, 46, 348–356. [Google Scholar] [CrossRef]
- Zeng, Z.; Dong, J.; Li, Y.; Dong, Z.; Liu, Z.; Huang, J.; Wang, Y.; Zhen, Y.; Lu, Y. The expression level and clinical significance of lncRNA X91348 in hepatocellular carcinoma. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3067–3071. [Google Scholar] [CrossRef]
- Yu, J.; Han, J.; Zhang, J.; Li, G.; Liu, H.; Cui, X.; Xu, Y.; Li, T.; Liu, J.; Wang, C. The long noncoding RNAs PVT1 and uc002mbe.2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Medicine (Baltim.) 2016, 95, e4436. [Google Scholar] [CrossRef]
- Wang, K.; Guo, W.X.; Li, N.; Gao, C.F.; Shi, J.; Tang, Y.F.; Shen, F.; Wu, M.C.; Liu, S.R.; Cheng, S.Q. Serum LncRNAs Profiles Serve as Novel Potential Biomarkers for the Diagnosis of HBV-Positive Hepatocellular Carcinoma. PLoS ONE 2015, 10, e0144934. [Google Scholar] [CrossRef]
- Kamel, M.M.; Matboli, M.; Sallam, M.; Montasser, I.F.; Saad, A.S.; El-Tawdi, A.H.F. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl. Res. 2016, 168, 134–145. [Google Scholar] [CrossRef]
- Sun, L.; Su, Y.; Liu, X.; Xu, M.; Chen, X.; Zhu, Y.; Guo, Z.; Bai, T.; Dong, L.; Wei, C.; et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J. Cancer 2018, 9, 2631–2639. [Google Scholar] [CrossRef]
- Xu, H.; Chen, Y.; Dong, X.; Wang, X. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol. Prev. Biomark. 2018, 27, 710–716. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, H.; Jing, W.; Luo, P.; Qiu, S.; Liu, X.; Zhu, M.; Liang, C.; Yu, M.; Tu, J. The Circular RNA hsa_circ_0001445 Regulates the Proliferation and Migration of Hepatocellular Carcinoma and May Serve as a Diagnostic Biomarker. Dis. Markers 2018, 2018, 3073467. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, Y.; Yang, G.; He, S.; Qiu, X.; Zhang, L.; Deng, Q.; Zheng, F. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin. Chim. Acta 2019, 492, 37–44. [Google Scholar] [CrossRef]
- Shi, J.; Li, X.; Zhang, F.; Kong, L.; Zhang, X.; Cheng, Y.; Guan, Q.; Cao, X.; Zhu, W.; Ou, K.; et al. The Plasma LncRNA Acting as Fingerprint in Hilar Cholangiocarcinoma. Cell. Physiol. Biochem. 2018, 49, 1694–1702. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, W.; Liu, W.; Kong, X.; Li, L.; He, J.; Wang, D.; Zhang, M.; Zhou, G.; Xu, W.; et al. Circulating lncRNA ABHD11-AS1 serves as a biomarker for early pancreatic cancer diagnosis. J. Cancer 2019, 10, 3746–3756. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Zheng, S.; Zhou, Y.; Zhao, L.; Ye, H.; Zhao, X.; Gao, W.; Fu, Z.; Zhou, Q.; et al. Expression profile of long non-coding RNAs in pancreatic cancer and their clinical significance as biomarkers. Oncotarget 2015, 6, 35684–35698. [Google Scholar] [CrossRef]
- Yang, F.; Liu, D.Y.; Guo, J.T.; Ge, N.; Zhu, P.; Liu, X.; Wang, S.; Wang, G.X.; Sun, S.Y. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J. Gastroenterol. 2017, 23, 8345–8354. [Google Scholar] [CrossRef]
- Yang, Z.; Sun, Y.; Liu, R.; Shi, Y.; Ding, S. Plasma long noncoding RNAs PANDAR, FOXD2-AS1, and SMARCC2 as potential novel diagnostic biomarkers for gastric cancer. Cancer Manag. Res. 2019, 11, 6175–6184. [Google Scholar] [CrossRef]
- Zheng, R.; Liang, J.; Lu, J.; Li, S.; Zhang, G.; Wang, X.; Liu, M.; Wang, W.; Chu, H.; Tao, G.; et al. Genome-wide long non-coding RNAs identified a panel of novel plasma biomarkers for gastric cancer diagnosis. Gastric Cancer 2019, 22, 731–741. [Google Scholar] [CrossRef]
- Ghaedi, H.; Mozaffari, M.A.N.; Salehi, Z.; Ghasemi, H.; Zadian, S.S.; Alipoor, S.; Hadianpour, S.; Alipoor, B. Co-expression profiling of plasma miRNAs and long noncoding RNAs in gastric cancer patients. Gene 2019, 687, 135–142. [Google Scholar] [CrossRef]
- Tan, L.; Yang, Y.; Shao, Y.; Zhang, H.; Guo, J. Plasma lncRNA-GACAT2 is a valuable marker for the screening of gastric cancer. Oncol. Lett. 2016, 12, 4845–4849. [Google Scholar] [CrossRef][Green Version]
- Elsayed, E.T.; Salem, P.E.; Darwish, A.M.; Fayed, H.M. Plasma long non-coding RNA HOTAIR as a potential biomarker for gastric cancer. Int. J. Biol. Markers 2018, 33, 528–533. [Google Scholar] [CrossRef]
- Liu, J.; Wang, J.; Song, Y.; Ma, B.; Luo, J.; Ni, Z.; Gao, P.; Sun, J.; Zhao, J.; Chen, X.; et al. A panel consisting of three novel circulating lncRNAs, is it a predictive tool for gastric cancer? J. Cell. Mol. Med. 2018, 22, 3605–3613. [Google Scholar] [CrossRef]
- Gao, J.; Cao, R.; Mu, H. Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 12936–12942. [Google Scholar]
- Zhang, K.; Shi, H.; Xi, H.; Wu, X.; Cui, J.; Gao, Y.; Liang, W.; Hu, C.; Liu, Y.; Li, J.; et al. Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer. Theranostics 2017, 7, 213–227. [Google Scholar] [CrossRef]
- Jin, C.; Shi, W.; Wang, F.; Shen, X.; Qi, J.; Cong, H.; Yuan, J.; Shi, L.; Zhu, B.; Luo, X.; et al. Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget 2016, 7, 51763–51772. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.M.; Ma, F.B.; Pan, S.R.; Liu, B.Z. Long noncoding RNA HOXA11-AS promotes gastric cancer cell proliferation and invasion via SRSF1 and functions as a biomarker in gastric cancer. World J. Gastroenterol. 2019, 25, 2763–2775. [Google Scholar] [CrossRef]
- Mohamed, W.A.; Schaalan, M.F.; Ramadan, B. The expression profiling of circulating miR-204, miR-182, and lncRNA H19 as novel potential biomarkers for the progression of peptic ulcer to gastric cancer. J. Cell. Biochem. 2019, 120, 13464–13477. [Google Scholar] [CrossRef]
- Zhou, X.; Yin, C.; Dang, Y.; Ye, F.; Zhang, G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep. 2015, 5, 11516. [Google Scholar] [CrossRef]
- Zong, W.; Feng, W.; Jiang, Y.; Ju, S.; Cui, M.; Jing, R. Evaluating the diagnostic and prognostic value of serum long non-coding RNA CTC-497E21.4 in gastric cancer. Clin. Chem. Lab. Med. 2019, 57, 1063–1072. [Google Scholar] [CrossRef]
- Fu, M.; Huang, Z.; Zang, X.; Pan, L.; Liang, W.; Chen, J.; Qian, H.; Xu, W.; Jiang, P.; Zhang, X. Long noncoding RNA LINC00978 promotes cancer growth and acts as a diagnostic biomarker in gastric cancer. Cell Prolif. 2018, 51, e12425. [Google Scholar] [CrossRef]
- Lu, Q.; Yu, T.; Ou, X.; Cao, D.; Xie, T.; Chen, X. Potential lncRNA diagnostic biomarkers for early gastric cancer. Mol. Med. Rep. 2017, 16, 9545–9552. [Google Scholar] [CrossRef]
- Arita, T.; Ichikawa, D.; Konishi, H.; Komatsu, S.; Shiozaki, A.; Shoda, K.; Kawaguchi, T.; Hirajima, S.; Nagata, H.; Kubota, T.; et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013, 33, 3185–3193. [Google Scholar]
- Lin, L.Y.; Yang, L.; Zeng, Q.; Wang, L.; Chen, M.L.; Zhao, Z.H.; Ye, G.D.; Luo, Q.C.; Lv, P.Y.; Guo, Q.W.; et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol. Cancer 2018, 17, 84. [Google Scholar] [CrossRef]
- Virgilio, E.; Giarnieri, E.; Giovagnoli, M.R.; Montagnini, M.; Proietti, A.; D’Urso, R.; Mercantini, P.; Balducci, G.; Cavallini, M. Long non-coding RNAs in the gastric juice of gastric cancer patients. Pathol. Res. Pract. 2018, 214, 1239–1246. [Google Scholar] [CrossRef]
- Li, W.H.; Song, Y.C.; Zhang, H.; Zhou, Z.J.; Xie, X.; Zeng, Q.N.; Guo, K.; Wang, T.; Xia, P.; Chang, D.M. Decreased Expression of Hsa_circ_00001649 in Gastric Cancer and Its Clinical Significance. Dis. Markers 2017, 2017, 4587698. [Google Scholar] [CrossRef]
- Li, T.; Shao, Y.; Fu, L.; Xie, Y.; Zhu, L.; Sun, W.; Yu, R.; Xiao, B.; Guo, J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J. Mol. Med. (Berl.) 2018, 96, 85–96. [Google Scholar] [CrossRef]
- Huang, M.; He, Y.R.; Liang, L.C.; Huang, Q.; Zhu, Z.Q. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J. Gastroenterol. 2017, 23, 6330–6338. [Google Scholar] [CrossRef]
- Sun, H.; Tang, W.; Rong, D.; Jin, H.; Fu, K.; Zhang, W.; Liu, Z.; Cao, H.; Cao, X. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018, 21, 299–306. [Google Scholar] [CrossRef]
- Tang, W.; Fu, K.; Sun, H.; Rong, D.; Wang, H.; Cao, H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol. Cancer 2018, 17, 137. [Google Scholar] [CrossRef]
- Chen, S.; Li, T.; Zhao, Q.; Xiao, B.; Guo, J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta 2017, 466, 167–171. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, P.Y.; Xie, J.W.; Wang, J.B.; Lin, J.X.; Chen, Q.Y.; Cao, L.L.; Huang, C.M.; Li, P.; Zheng, C.H. Hsa_circ_0000467 promotes cancer progression and serves as a diagnostic and prognostic biomarker for gastric cancer. J. Clin. Lab. Anal. 2019, 33, e22726. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, S.; Li, T.; Xiao, B.; Zhang, X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J. Clin. Lab. Anal. 2018, 32, e22333. [Google Scholar] [CrossRef]
- Abedini, P.; Fattahi, A.; Agah, S.; Talebi, A.; Beygi, A.H.; Amini, S.M.; Mirzaei, A.; Akbari, A. Expression analysis of circulating plasma long noncoding RNAs in colorectal cancer: The relevance of lncRNAs ATB and CCAT1 as potential clinical hallmarks. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef]
- Liu, H.; Ye, D.; Chen, A.; Tan, D.; Zhang, W.; Jiang, W.; Wang, M.; Zhang, X. A pilot study of new promising non-coding RNA diagnostic biomarkers for early-stage colorectal cancers. Clin. Chem. Lab. Med. 2019, 57, 1073–1083. [Google Scholar] [CrossRef]
- Shaker, O.G.; Senousy, M.A.; Elbaz, E.M. Association of rs6983267 at 8q24, HULC rs7763881 polymorphisms and serum lncRNAs CCAT2 and HULC with colorectal cancer in Egyptian patients. Sci. Rep. 2017, 7, 16246. [Google Scholar] [CrossRef]
- Dai, M.; Chen, X.; Mo, S.; Li, J.; Huang, Z.; Huang, S.; Xu, J.; He, B.; Zou, Y.; Chen, J.; et al. Meta-signature LncRNAs serve as novel biomarkers for colorectal cancer: Integrated bioinformatics analysis, experimental validation and diagnostic evaluation. Sci. Rep. 2017, 7, 46572. [Google Scholar] [CrossRef]
- Feng, W.; Zong, W.; Li, Y.; Shen, X.; Cui, X.; Ju, S. Abnormally expressed long noncoding RNA B3GALT5-AS1 may serve as a biomarker for the diagnostic and prognostic of gastric cancer. J. Cell. Biochem. 2019. [Google Scholar] [CrossRef]
- Wang, C.; Yu, J.; Han, Y.; Li, L.; Li, J.; Li, T.; Qi, P. Long non-coding RNAs LOC285194, RP11-462C24.1 and Nbla12061 in serum provide a new approach for distinguishing patients with colorectal cancer from healthy controls. Oncotarget 2016, 7, 70769–70778. [Google Scholar] [CrossRef]
- Wang, R.; Du, L.; Yang, X.; Jiang, X.; Duan, W.; Yan, S.; Xie, Y.; Zhu, Y.; Wang, Q.; Wang, L.; et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J. Cancer Res. Clin. Oncol. 2016, 142, 2291–2301. [Google Scholar] [CrossRef]
- Zhao, W.; Song, M.; Zhang, J.; Kuerban, M.; Wang, H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 14131–14140. [Google Scholar]
- Hu, D.; Zhan, Y.; Zhu, K.; Bai, M.; Han, J.; Si, Y.; Zhang, H.; Kong, D. Plasma Exosomal Long Non-Coding RNAs Serve as Biomarkers for Early Detection of Colorectal Cancer. Cell. Physiol. Biochem. 2018, 51, 2704–2715. [Google Scholar] [CrossRef]
- Dong, L.; Lin, W.; Qi, P.; Xu, M.D.; Wu, X.; Ni, S.; Huang, D.; Weng, W.W.; Tan, C.; Sheng, W.; et al. Circulating Long RNAs in Serum Extracellular Vesicles: Their Characterization and Potential Application as Biomarkers for Diagnosis of Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1158–1166. [Google Scholar] [CrossRef]
- Ji, W.; Qiu, C.; Wang, M.; Mao, N.; Wu, S.; Dai, Y. Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem. Biophy. Res. Commun. 2018, 497, 122–126. [Google Scholar] [CrossRef]
- Li, X.N.; Wang, Z.J.; Ye, C.X.; Zhao, B.C.; Huang, X.X.; Yang, L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed. Pharmacother. 2019, 112, 108611. [Google Scholar] [CrossRef]
- Luo, J.; Li, Q.; Pan, J.; Li, L.; Fang, L.; Zhang, Y. Expression level of long noncoding RNA H19 in plasma of patients with nonsmall cell lung cancer and its clinical significance. J. Cancer Res. Ther. 2018, 14, 860–863. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; Liu, X.; Luo, P.; Jing, W.; Zhu, M.; Tu, J. Identification of Circulating Long Noncoding RNA HOTAIR as a Novel Biomarker for Diagnosis and Monitoring of Non-Small Cell Lung Cancer. Technol. Cancer Res. Treat. 2017, 16, 1060–1066. [Google Scholar] [CrossRef]
- Li, N.; Feng, X.B.; Tan, Q.; Luo, P.; Jing, W.; Zhu, M.; Liang, C.; Tu, J.; Ning, Y. Identification of Circulating Long Noncoding RNA Linc00152 as a Novel Biomarker for Diagnosis and Monitoring of Non-Small-Cell Lung Cancer. Dis. Markers 2017, 2017, 7439698. [Google Scholar] [CrossRef]
- Lin, Y.; Leng, Q.; Zhan, M.; Jiang, F. A Plasma Long Noncoding RNA Signature for Early Detection of Lung Cancer. Transl. Oncol. 2018, 11, 1225–1231. [Google Scholar] [CrossRef]
- Tan, Q.; Zuo, J.; Qiu, S.; Yu, Y.; Zhou, H.; Li, N.; Wang, H.; Liang, C.; Yu, M.; Tu, J. Identification of circulating long non-coding RNA GAS5 as a potential biomarker for non-small cell lung cancer diagnosisnon-small cell lung cancer, long non-coding RNA, plasma, GAS5, biomarker. Int. J. Oncol. 2017, 50, 1729–1738. [Google Scholar] [CrossRef]
- Hu, X.; Bao, J.; Wang, Z.; Zhang, Z.; Gu, P.; Tao, F.; Cui, D.; Jiang, W. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumour Biol. 2016, 37, 3497–3504. [Google Scholar] [CrossRef]
- Wang, H.M.; Lu, J.H.; Chen, W.Y.; Gu, A.Q. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int. J. Clin. Exp. Med. 2015, 8, 11824–11830. [Google Scholar]
- Liu, H.; Zhou, G.; Fu, X.; Cui, H.; Pu, G.; Xiao, Y.; Sun, W.; Dong, X.; Zhang, L.; Cao, S.; et al. Long noncoding RNA TUG1 is a diagnostic factor in lung adenocarcinoma and suppresses apoptosis via epigenetic silencing of BAX. Oncotarget 2017, 8, 101899–101910. [Google Scholar] [CrossRef]
- Wan, L.; Zhang, L.; Fan, K.; Wang, J.J. Diagnostic significance of circulating long noncoding RNA PCAT6 in patients with non-small cell lung cancer. Onco Targets Ther. 2017, 10, 5695–5702. [Google Scholar] [CrossRef]
- Tantai, J.; Hu, D.; Yang, Y.; Geng, J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 7887–7895. [Google Scholar]
- Li, W.; Li, N.; Kang, X.; Shi, K. Circulating long non-coding RNA AFAP1-AS1 is a potential diagnostic biomarker for non-small cell lung cancer. Clin. Chim. Acta 2017, 475, 152–156. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, Y.; Du, L.; Jiang, X.; Yan, S.; Duan, W.; Li, J.; Zhan, Y.; Wang, L.; Zhang, S.; et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol. Oncol. 2018, 12, 648–658. [Google Scholar] [CrossRef]
- Jiang, N.; Meng, X.; Mi, H.; Chi, Y.; Li, S.; Jin, Z.; Tian, H.; He, J.; Shen, W.; Tian, H.; et al. Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin. Chim. Acta 2018, 486, 26–33. [Google Scholar] [CrossRef]
- Li, C.; Lv, Y.; Shao, C.; Chen, C.; Zhang, T.; Wei, Y.; Fan, H.; Lv, T.; Liu, H.; Song, Y. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J. Cell. Physiol. 2019, 234, 20721–20727. [Google Scholar] [CrossRef]
- Teng, Y.; Kang, H.; Chu, Y. Identification of an Exosomal Long Noncoding RNA SOX2-OT in Plasma as a Promising Biomarker for Lung Squamous Cell Carcinoma. Genet. Test. Mol. Biomark. 2019, 23, 235–240. [Google Scholar] [CrossRef]
- Bai, Y.; Qu, Y.; Wu, Z.; Ren, Y.; Cheng, Z.; Lu, Y.; Hu, J.; Lou, J.; Zhao, J.; Chen, C.; et al. Absolute quantification and analysis of extracellular vesicle lncRNAs from the peripheral blood of patients with lung cancer based on multi-colour fluorescence chip-based digital PCR. Biosens. Bioelectron. 2019, 142, 111523. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Y.; Lu, J.; Sun, Y.; Xiao, H.; Liu, M.; Tian, L. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med. Oncol. 2014, 31, 148. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.Q.; Weng, W.W.; Zhang, Q.Y.; Yang, X.Q.; Gan, H.L.; Yang, Y.S.; Zhang, P.P.; Sun, M.H.; Xu, M.D.; et al. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis 2016, 5, e192. [Google Scholar] [CrossRef]
- Li, G.; Zhang, Y.; Mao, J.; Hu, P.; Chen, Q.; Ding, W.; Pu, R. lncRNA TUC338 is a potential diagnostic biomarker for bladder cancer. J. Cell. Biochem. 2019. [Google Scholar] [CrossRef]
- Wang, J.; Yang, K.; Yuan, W.; Gao, Z. Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Bladder Cancer Diagnosis and Prognosis. Med. Sci. Monit. 2018, 24, 9307–9316. [Google Scholar] [CrossRef]
- Zhang, S.; Du, L.; Wang, L.; Jiang, X.; Zhan, Y.; Li, J.; Yan, K.; Duan, W.; Zhao, Y.; Wang, L.; et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J. Cell. Mol. Med. 2019, 23, 1396–1405. [Google Scholar] [CrossRef]
- Wen, J.J.; Ma, Y.D.; Yang, G.S.; Wang, G.M. Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 498–503. [Google Scholar]
- Du, L.; Duan, W.; Jiang, X.; Zhao, L.; Li, J.; Wang, R.; Yan, S.; Xie, Y.; Yan, K.; Wang, Q.; et al. Cell-free lncRNA expression signatures in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. J. Cell. Mol. Med. 2018, 22, 2838–2845. [Google Scholar] [CrossRef]
- Yazarlou, F.; Modarressi, M.H.; Mowla, S.J.; Oskooei, V.K.; Motevaseli, E.; Tooli, L.F.; Nekoohesh, L.; Eghbali, M.; Ghafouri-Fard, S.; Afsharpad, M. Urinary exosomal expression of long non-coding RNAs as diagnostic marker in bladder cancer. Cancer Manag. Res. 2018, 10, 6357–6365. [Google Scholar] [CrossRef]
- Zhan, Y.; Du, L.; Wang, L.; Jiang, X.; Zhang, S.; Li, J.; Yan, K.; Duan, W.; Zhao, Y.; Wang, L.; et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol. Cancer 2018, 17, 142. [Google Scholar] [CrossRef]
- Duan, W.; Du, L.; Jiang, X.; Wang, R.; Yan, S.; Xie, Y.; Yan, K.; Wang, Q.; Wang, L.; Zhang, X.; et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget 2016, 7, 78850–78858. [Google Scholar] [CrossRef]
- Chen, X.; Chen, R.X.; Wei, W.S.; Li, Y.H.; Feng, Z.H.; Tan, L.; Chen, J.W.; Yuan, G.J.; Chen, S.L.; Guo, S.J.; et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin. Cancer Res. 2018, 24, 6319–6330. [Google Scholar] [CrossRef]
- Li, G.; Zhang, Y.; Mao, J.; Hu, P.; Chen, Q.; Ding, W.; Pu, R. LncRNA TUC338 is overexpressed in prostate carcinoma and downregulates miR-466. Gene 2019, 707, 224–230. [Google Scholar] [CrossRef]
- Chen, Z.; Zhen, M.; Zhou, J. LncRNA BRE-AS1 interacts with miR-145-5p to regulate cancer cell proliferation and apoptosis in prostate carcinoma and has early diagnostic values. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Ren, S.; Wang, F.; Shen, J.; Sun, Y.; Xu, W.; Lu, J.; Wei, M.; Xu, C.; Wu, C.; Zhang, Z.; et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 2013, 49, 2949–2959. [Google Scholar] [CrossRef]
- Wang, T.; Qu, X.; Jiang, J.; Gao, P.; Zhao, D.; Lian, X.; Li, X. Diagnostic significance of urinary long non-coding PCA3 RNA in prostate cancer. Oncotarget 2017, 8, 58577–58586. [Google Scholar] [CrossRef]
- Bayat, H.; Narouie, B.; Ziaee, S.M.; Mowla, S.J. Two long non-coding RNAs, Prcat17.3 and Prcat38, could efficiently discriminate benign prostate hyperplasia from prostate cancer. Prostate 2018, 78, 812–818. [Google Scholar] [CrossRef]
- Zhang, W.; Ren, S.C.; Shi, X.L.; Liu, Y.W.; Zhu, Y.S.; Jing, T.L.; Wang, F.B.; Chen, R.; Xu, C.L.; Wang, H.Q.; et al. A novel urinary long non-coding RNA transcript improves diagnostic accuracy in patients undergoing prostate biopsy. Prostate 2015, 75, 653–661. [Google Scholar] [CrossRef]
- Zhang, K.; Luo, Z.; Zhang, Y.; Zhang, L.; Wu, L.; Liu, L.; Yang, J.; Song, X.; Liu, J. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016, 17, 187–194. [Google Scholar] [CrossRef]
- Miao, Y.; Fan, R.; Chen, L.; Qian, H. Clinical Significance of Long Non-coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Ann. Clin. Lab. Sci. 2016, 46, 418–424. [Google Scholar]
- Xu, N.; Chen, F.; Wang, F.; Lu, X.; Wang, X.; Lv, M.; Lu, C. Clinical significance of high expression of circulating serum lncRNA RP11-445H22.4 in breast cancer patients: A Chinese population-based study. Tumour Biol. 2015, 36, 7659–7665. [Google Scholar] [CrossRef]
- Yin, W.B.; Yan, M.G.; Fang, X.; Guo, J.J.; Xiong, W.; Zhang, R.P. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin. Chim. Acta 2018, 487, 363–368. [Google Scholar] [CrossRef]
- Xu, H.; Gong, Z.; Shen, Y.; Fang, Y.; Zhong, S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics 2018, 10, 187–197. [Google Scholar] [CrossRef]
- Naizhaer, G.; Kuerban, A.; Kuerban, R.; Zhou, P. Up-regulation of lncRNA FALEC indicates prognosis and diagnosis values in cervical cancer. Pathol. Res. Pract. 2019, 215, 152495. [Google Scholar] [CrossRef]
- Sun, W.; Wang, L.; Zhao, D.; Wang, P.; Li, Y.; Wang, S. Four Circulating Long Non-Coding RNAs Act as Biomarkers for Predicting Cervical Cancer. Gynecol. Obstet. Investig. 2018, 83, 533–539. [Google Scholar] [CrossRef]
- Zhang, X.; Mao, L.; Li, L.; He, Z.; Wang, N.; Song, Y. Long noncoding RNA GIHCG functions as an oncogene and serves as a serum diagnostic biomarker for cervical cancer. J. Cancer 2019, 10, 672–681. [Google Scholar] [CrossRef]
- Yang, J.P.; Yang, X.J.; Xiao, L.; Wang, Y. Long noncoding RNA PVT1 as a novel serum biomarker for detection of cervical cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3980–3986. [Google Scholar]
- Shen, X.; Zhang, Y.; Wu, X.; Guo, Y.; Shi, W.; Qi, J.; Cong, H.; Wang, X.; Wu, X.; Ju, S. Upregulated lncRNA-PCAT1 is closely related to clinical diagnosis of multiple myeloma as a predictive biomarker in serum. Cancer Biomark. 2017, 18, 257–263. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, H.; Shen, X.; Wang, X.; Ju, S.; Lu, M.; Cong, H. Serum level of long noncoding RNA H19 as a diagnostic biomarker of multiple myeloma. Clin. Chim. Acta 2018, 480, 199–205. [Google Scholar] [CrossRef]
- Min, W.; Dai, D.; Wang, J.; Zhang, D.; Zhang, Y.; Han, G.; Zhang, L.; Chen, C.; Li, X.; Li, Y.; et al. Long Noncoding RNA miR210HG as a Potential Biomarker for the Diagnosis of Glioma. PLoS ONE 2016, 11, e0160451. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, X.; Wei, B.; Qiao, G.; Jiang, T.; Chen, Z. Plasma lncRNA GAS8-AS1 as a Potential Biomarker of Papillary Thyroid Carcinoma in Chinese Patients. Int. J. Endocrinol. 2017, 2017, 2645904. [Google Scholar] [CrossRef]
- Yang, C.; Wei, Y.; Yu, L.; Xiao, Y. Identification of Altered Circular RNA Expression in Serum Exosomes from Patients with Papillary Thyroid Carcinoma by High-Throughput Sequencing. Med. Sci. Monit. 2019, 25, 2785–2791. [Google Scholar] [CrossRef]
- Zhu, K.; Niu, L.; Wang, J.; Wang, Y.; Zhou, J.; Wang, F.; Cheng, Y.; Zhang, Q.; Li, H. Circular RNA hsa_circ_0000885 Levels are Increased in Tissue and Serum Samples from Patients with Osteosarcoma. Med. Sci. Monit. 2019, 25, 1499–1505. [Google Scholar] [CrossRef]
- Chen, X.; Gao, G.; Liu, S.; Yu, L.; Yan, D.; Yao, X.; Sun, W.; Han, D.; Dong, H. Long Noncoding RNA PVT1 as a Novel Diagnostic Biomarker and Therapeutic Target for Melanoma. BioMed Res. Int. 2017, 2017, 7038579. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, Y.; Zhang, C.; Duan, C. Multiple Roles of Exosomal Long Noncoding RNAs in Cancers. BioMed Res. Int. 2019, 2019, 1460572. [Google Scholar] [CrossRef]
- Jiang, N.; Pan, J.; Fang, S.; Zhou, C.; Han, Y.; Chen, J.; Meng, X.; Jin, X.; Gong, Z. Liquid biopsy: Circulating exosomal long noncoding RNAs in cancer. Clin. Chim. Acta 2019, 495, 331–337. [Google Scholar] [CrossRef]
- Sarfi, M.; Abbastabar, M.; Khalili, E. Long noncoding RNAs biomarker-based cancer assessment. J. Cell. Physiol. 2019, 234, 16971–16986. [Google Scholar] [CrossRef]
- Zhou, Y.; Xia, L.; Lin, J.; Wang, H.; Oyang, L.; Tan, S.; Tian, Y.; Su, M.; Wang, H.; Cao, D.; et al. Exosomes in Nasopharyngeal Carcinoma. J. Cancer 2018, 9, 767–777. [Google Scholar] [CrossRef]
- Peng, W.; Wang, J.; Shan, B.; Peng, Z.; Dong, Y.; Shi, W.; He, D.; Cheng, Y.; Zhao, W.; Zhang, C.; et al. Diagnostic and Prognostic Potential of Circulating Long Non-Coding RNAs in Non Small Cell Lung Cancer. Cell. Physiol. Biochem. 2018, 49, 816–827. [Google Scholar] [CrossRef]
- Martignano, F.; Rossi, L.; Maugeri, A.; Galla, V.; Conteduca, V.; De Giorgi, U.; Casadio, V.; Schepisi, G. Urinary RNA-based biomarkers for prostate cancer detection. Clin. Chim. Acta 2017, 473, 96–105. [Google Scholar] [CrossRef]
- Wu, P.G.; Zhang, Y.X. Long non-coding RNAs in prostate cancer: An update. Zhonghua Nan Ke Xue 2018, 24, 735–739. [Google Scholar]
- Vlaeminck-Guillem, V. Extracellular Vesicles in Prostate Cancer Carcinogenesis, Diagnosis, and Management. Front. Oncol. 2018, 8, 222. [Google Scholar] [CrossRef]
- Kahroba, H.; Hejazi, M.S.; Samadi, N. Exosomes: From carcinogenesis and metastasis to diagnosis and treatment of gastric cancer. Cell. Mol. Life Sci. 2019, 76, 1747–1758. [Google Scholar] [CrossRef]
- Yang, C.; Kim, H.S.; Song, G.; Lim, W. The potential role of exosomes derived from ovarian cancer cells for diagnostic and therapeutic approaches. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef]
- Wieczorek, E.; Reszka, E. mRNA, microRNA and lncRNA as novel bladder tumor markers. Clin. Chim. Acta 2018, 477, 141–153. [Google Scholar] [CrossRef]
- Nobili, L.; Ronchetti, D.; Agnelli, L.; Taiana, E.; Vinci, C.; Neri, A. Long Non-Coding RNAs in Multiple Myeloma. Genes (Basel) 2018, 9, 69. [Google Scholar] [CrossRef]
- Dai, S.P.; Jin, J.; Li, W.M. Diagnostic efficacy of long non-coding RNA in lung cancer: A systematic review and meta-analysis. Postgrad. Med. J. 2018, 94, 578–587. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, Y.; Wu, X.; Zhang, C.; Li, G. Diagnostic Value of lncRNAs as Biomarker in Hepatocellular Carcinoma: An Updated Meta-Analysis. Can. J. Gastroenterol. Hepatol. 2018, 2018, 8410195. [Google Scholar] [CrossRef]
- Ning, L.; Hu, Y.C.; Wang, S.; Lang, J.H. Altered long noncoding RNAs and survival outcomes in ovarian cancer: A systematic review and meta-analysis (PRISMA Compliant). Medicine (Baltim.) 2018, 97, e11481. [Google Scholar] [CrossRef]
- Gong, Y.B.; Zou, Y.F. Clinical significance of lncRNA FAM83H-AS1 in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4656–4662. [Google Scholar] [CrossRef]
- Marks, L.S.; Fradet, Y.; Lim Deras, I.; Blase, A.; Mathis, J.; Aubin, S.M.J.; Cancio, A.T.; Desaulniers, M.; Ellis, W.J.; Rittenhouse, H.; et al. PCA3 Molecular Urine Assay for Prostate Cancer in Men Undergoing Repeat Biopsy. Urology 2007, 69, 532–535. [Google Scholar] [CrossRef]
- Newcomb, L.F.; Zheng, Y.; Faino, A.V.; Bianchi-Frias, D.; Cooperberg, M.R.; Brown, M.D.; Brooks, J.D.; Dash, A.; Fabrizio, M.D.; Gleave, M.E.; et al. Performance of PCA3 and TMPRSS2:ERG urinary biomarkers in prediction of biopsy outcome in the Canary Prostate Active Surveillance Study (PASS). Prostate Cancer Prostatic Dis. 2019. [Google Scholar] [CrossRef]
- Rittenhouse, H.; Blase, A.; Shamel, B.; Schalken, J.; Groskopf, J. The Long and Winding Road to FDA Approval of a Novel Prostate Cancer Test: Our Story. Clin. Chem. 2013, 59, 32–34. [Google Scholar] [CrossRef]
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.N. Circular RNA Splicing. Adv. Exp. Med. Biol. 2018, 1087, 41–52. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Arnaiz, E.; Sole, C.; Manterola, L.; Iparraguirre, L.; Otaegui, D.; Lawrie, C.H. CircRNAs and cancer: Biomarkers and master regulators. Semin. Cancer Biol. 2018. [Google Scholar] [CrossRef]
- Dragomir, M.; Calin, G.A. Circular RNAs in Cancer—Lessons Learned From microRNAs. Front. Oncol. 2018, 8. [Google Scholar] [CrossRef]
- Hsu, M.-T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- Ragan, C.; Goodall, G.J.; Shirokikh, N.E.; Preiss, T. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. 2019, 9, 2048. [Google Scholar] [CrossRef]
- Schneider, T.; Hung, L.H.; Schreiner, S.; Starke, S.; Eckhof, H.; Rossbach, O.; Reich, S.; Medenbach, J.; Bindereif, A. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci. Rep. 2016, 6, 31313. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Ma, J.; Sun, T.; Zhou, Q.; Wang, W.; Wang, G.; Wu, P.; Wang, H.; Jiang, L.; et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer 2019, 18, 116. [Google Scholar] [CrossRef]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Circular RNAs: Biogenesis, Function, and a Role as Possible Cancer Biomarkers. Int. J. Genom. 2017, 2017, 6218353. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, X.; Wei, S.; Chen, Y.; Chen, Y.; Fan, X.; Han, S.; Wu, G. hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017, 284, 2170–2182. [Google Scholar] [CrossRef]
- Hang, D.; Zhou, J.; Qin, N.; Zhou, W.; Ma, H.; Jin, G.; Hu, Z.; Dai, J.; Shen, H. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018, 7, 2783–2791. [Google Scholar] [CrossRef]
- Liu, X.X.; Yang, Y.E.; Liu, X.; Zhang, M.Y.; Li, R.; Yin, Y.H.; Qu, Y.Q. A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J. Transl. Med. 2019, 17, 50. [Google Scholar] [CrossRef]
- Li, J.; Li, H.; Lv, X.; Yang, Z.; Gao, M.; Bi, Y.; Zhang, Z.; Wang, S.; Cui, Z.; Zhou, B.; et al. Diagnostic performance of circular RNAs in human cancers: A systematic review and meta-analysis. Mol. Genet. Genom. Med. 2019, 7, e00749. [Google Scholar] [CrossRef]
- Jiang, F.; Hong, F.; Shah, M.W.; Shen, X. Circular RNAs as diagnostic biomarkers in gastric cancer: A meta-analysis review. Pathol. Res. Pract. 2019, 215, 152419. [Google Scholar] [CrossRef]
- Song, F.; Luo, H.; Xie, M.; Zhu, H.; Hou, Y. Microarray expression profile of circular RNAs in human body fluids. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e55–e56. [Google Scholar] [CrossRef]
- Jafari Ghods, F. Circular RNA in Saliva. Adv. Exp. Med. Biol. 2018, 1087, 131–139. [Google Scholar] [CrossRef]
- Shao, Y.; Li, J.; Lu, R.; Li, T.; Yang, Y.; Xiao, B.; Guo, J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017, 6, 1173–1180. [Google Scholar] [CrossRef]
- Krishnan, P.; Damaraju, S. The Challenges and Opportunities in the Clinical Application of Noncoding RNAs: The Road Map for miRNAs and piRNAs in Cancer Diagnostics and Prognostics. Int. J. Genom. 2018, 2018, 5848046. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Boland, C.R. Erratum to: Non-coding RNA: It’s Not Junk. Dig. Dis. Sci. 2017, 62, 3260. [Google Scholar] [CrossRef]
- Beyret, E.; Lin, H. Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev. Biol. 2011, 355, 215–226. [Google Scholar] [CrossRef]
- Liu, S.-L.; Sun, P.; Li, Y.; Liu, S.-S.; Lu, Y. Exosomes as critical mediators of cell-to-cell communication in cancer pathogenesis and their potential clinical application. Transl. Cancer Res. 2019, 8, 298–311. [Google Scholar] [CrossRef]
- Wu, J.; Li, H.; Xie, H.; Wu, X.; Lan, P. The malignant role of exosomes in the communication among colorectal cancer cell, macrophage and microbiome. Carcinogenesis 2019, 40, 601–610. [Google Scholar] [CrossRef]
- Pichler, M.; Calin, G.A. MicroRNAs in cancer: From developmental genes in worms to their clinical application in patients. Br. J. Cancer 2015, 113, 569–573. [Google Scholar] [CrossRef]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. microRNA Therapeutics in Cancer—An Emerging Concept. EBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef]
- Wang, W.T.; Han, C.; Sun, Y.M.; Chen, T.Q.; Chen, Y.Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 55. [Google Scholar] [CrossRef]
- Van Roosbroeck, K.; Fanini, F.; Setoyama, T.; Ivan, C.; Rodriguez-Aguayo, C.; Fuentes-Mattei, E.; Xiao, L.; Vannini, I.; Redis, R.S.; D’Abundo, L.; et al. Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clin. Cancer Res. 2017, 23, 2891–2904. [Google Scholar] [CrossRef]
- Nishimura, M.; Jung, E.J.; Shah, M.Y.; Lu, C.; Spizzo, R.; Shimizu, M.; Han, H.D.; Ivan, C.; Rossi, S.; Zhang, X.; et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013, 3, 1302–1315. [Google Scholar] [CrossRef]
- Cortez, M.A.; Nicoloso, M.S.; Shimizu, M.; Rossi, S.; Gopisetty, G.; Molina, J.R.; Carlotti, C., Jr.; Tirapelli, D.; Neder, L.; Brassesco, M.S.; et al. miR-29b and miR-125a regulate podoplanin and suppress invasion in glioblastoma. Genes Chromosomes Cancer 2010, 49, 981–990. [Google Scholar] [CrossRef]
- Bayraktar, R.; Bertilaccio, M.T.S.; Calin, G.A. The Interaction Between Two Worlds: MicroRNAs and Toll-Like Receptors. Front. Immunol. 2019, 10, 1053. [Google Scholar] [CrossRef]
- Cortez, M.A.; Anfossi, S.; Ramapriyan, R.; Menon, H.; Atalar, S.C.; Aliru, M.; Welsh, J.; Calin, G.A. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer 2019, 58, 244–253. [Google Scholar] [CrossRef]
- Smolle, M.A.; Calin, H.N.; Pichler, M.; Calin, G.A. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. FEBS J. 2017, 284, 1952–1966. [Google Scholar] [CrossRef]
- Slaby, O.; Laga, R.; Sedlacek, O. Therapeutic targeting of non-coding RNAs in cancer. Biochem. J. 2017, 474, 4219–4251. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Srivastava, S.K.; Khan, M.A.; Prajapati, V.K.; Singh, S.; Carter, J.E.; Singh, A.P. Racial disparities in prostate cancer: A molecular perspective. Front. Biosci. (Landmark Ed.) 2017, 22, 772–782. [Google Scholar] [CrossRef]
- Krishnan, A.R.; Zheng, H.; Kwok, J.G.; Qu, Y.; Zou, A.E.; Korrapati, A.; Li, P.X.; Califano, J.A.; Hovell, M.F.; Wang-Rodriguez, J.; et al. A comprehensive study of smoking-specific microRNA alterations in head and neck squamous cell carcinoma. Oral Oncol. 2017, 72, 56–64. [Google Scholar] [CrossRef]
- Hasakova, K.; Bezakova, J.; Vician, M.; Reis, R.; Zeman, M.; Herichova, I. Gender-dependent expression of leading and passenger strand of miR-21 and miR-16 in human colorectal cancer and adjacent colonic tissues. Physiol. Res. 2017, 66, S575–S582. [Google Scholar]
- Sazanov, A.A.; Kiselyova, E.V.; Zakharenko, A.A.; Romanov, M.N.; Zaraysky, M.I. Plasma and saliva miR-21 expression in colorectal cancer patients. J. Appl. Genet. 2017, 58, 231–237. [Google Scholar] [CrossRef]
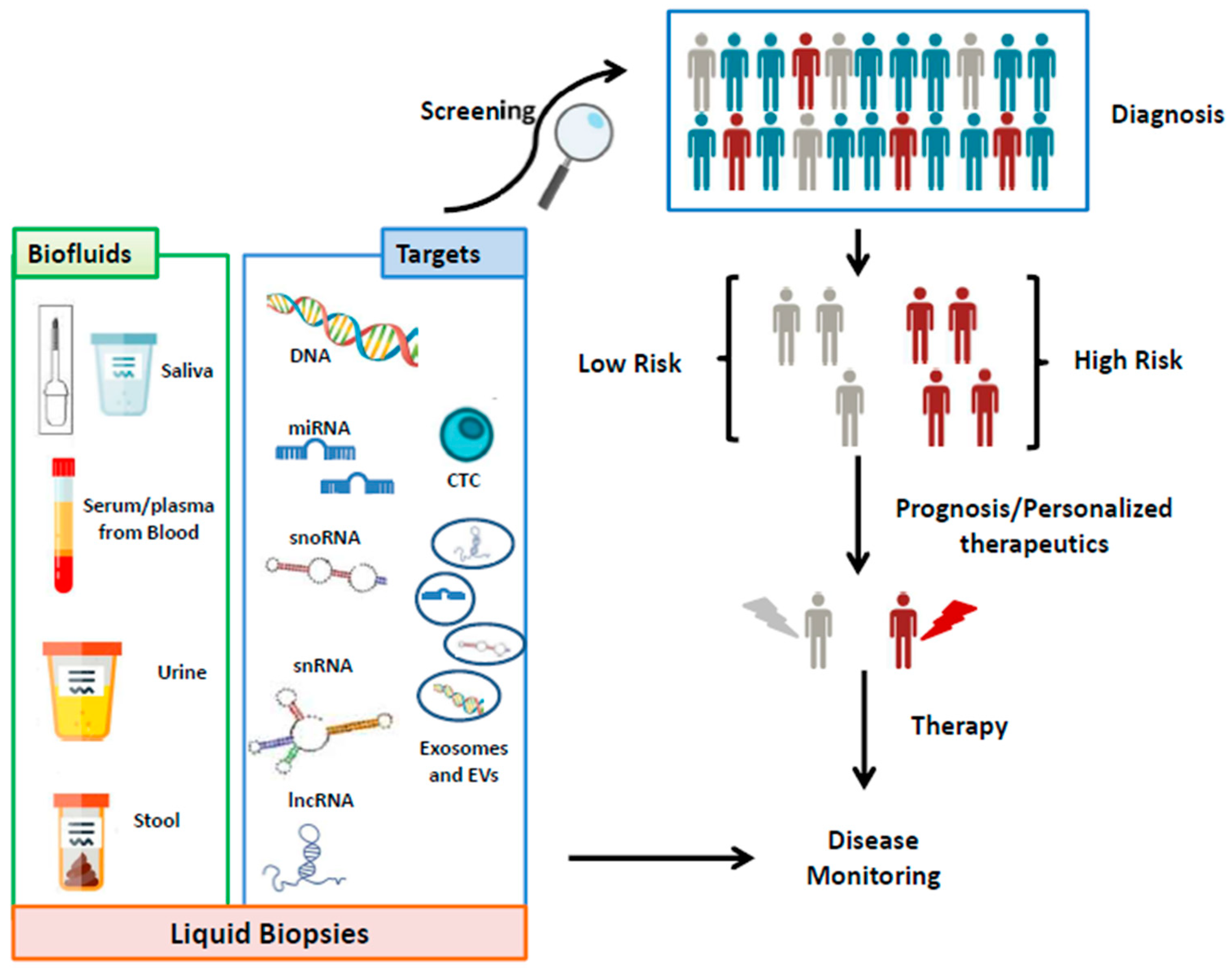
| Cat. | Cancer Type | miRNA | piRNA | sn/snoRNA |
|---|---|---|---|---|
| Digestive | Oral cancer | miR-125a, miR-200a (SA) [97] miR-16, miR-29a (S) [66] miR-125b (P) [66] | ||
| Nasopharyngeal cancer | miR-548q, miR-630, miR940 (P) [134] miR-16, miR-21, miR-24, miR-155, miR-378 (P) [135], miR-548q, miR-483-5p (P) [136], miR-24 (P) [137] miR-17, miR-20a, miR-29c, miR-223 (S) [138], miR-486-5p, miR-45, miR-100 (S) [139] miR-188-5p, miR-1908, miR-3196, miR-3935, miR-4284, miR-4433-5p, miR-4665-3p, miR-513b (B) [140] miR-31-5p (B) [141] miR-34c-3p, miR-18a-5p (TEP) [142] | |||
| Esophageal cancer | miR-16, miR-17, miR-20a, miR-30a, miR-155, miR-375 (P) [66] miR-10, miR-18a (S) [66] miR-21, miR-1246 (SE) [67] miR-30a (SE) [67] | |||
| Hepatocellular carcinoma | let-7b, let-7f, miR-16, miR-17, miR-30e, miR-125a, miR-221, miR-375 (S) [66] miR-18a, miR-221, miR-222, miR-224 (SE) [68] miR-106b (P, SE) [66] | |||
| Cholangiocarcinoma | miR-29, miR-122, miR-155, miR-192 (S) [143] miR-483-5p, miR-194 (S) [144] miR-150-5p (S) [145] miR-26a (S) [146] miR-106a (S) [147] miR-1281, miR-126, miR-26a, miR-30b, miR-122 (S) [148] miR-21, miR-221 (P) [149] miR-150 [150] (P) miR-412, miR-640, miR-1537, miR-3189 (BI) [148] miR-9, miR-145 * (BI) [151] Pancreato-biliary cancers vs. Healthy Controls: miR-6075, miR-4294^, miR-6880-5p, miR-6799-5p, miR-125a-3p, miR-4530^, miR-6836-3p, miR-4476 ^ (S) [152] ^ = reported, but no sufficient available data (www.miRbase.org) | RNU2-1f (BI) [153] | ||
| Gallbladder carcinoma | let-7a, miR-21, miR-187, miR-143, miR-202, and miR-335 (P) [154] | |||
| Pancreatic cancer | miR-486-5p, miR-938 (P) [69] miR-486-5p, miR-126-3p, miR-106b-3p (P) [70] miR-223 (P) [71] miR-182 (P) [72] miR-10b, miR-30c, miR-106b, miR-155, miR-212 (P) [73] miR-1290 (S) [74] miR-192 (S) [75] miR-21, miR-17-5p (S) [67] miR-210 (P, U) [66] | piR-016658, piR-001311 (PV) [26] | SNORA74A, SNORA25 (S/SE) [155] RNU2-1f (P/S) [156] | |
| Gastric cancer | miR-199-3p, miR-20a, miR-106b, miR-221, miR-486, miR-451, miR-17-5p, miR-106a, miR-106b, miR-21, let-7a, miR-195-5p (P) [76] miR-21, miR-221, miR-376c, miR-744, miR-378, miR-195-5p, let-7a, miR-196a, miR-451, miR-486 (S) [76] let-7c, let-7i, let-7f, miR-17, miR-17, miR-106b (S) [66] miR-200c (P, S), miR-16 (P, S) [66] let-7a, miR-18a, miR-19b, miR-106a, miR-17, miR-106a/b, miR-106b (P) [66] | piR-823, piR-651 (B) [157] | ||
| Colorectal cancer (CRC) | miR-25-3p [77] (SE) miR-27a, miR-130a (SE) [78] miR-17, miR-18a, miR-18b, miR-19a, miR-19b, miR-20a, miR-20b, miR-106a (P, SE) [79] miR-21, miR-23a, miR-224, miR-92a, miR-155 (SE) [80] miR-196b-5p (SE) [81] miR-217 [158] (SE) miR-182-5p, miR-149, miR-96-5p (SE) [82] miR-19a, miR-29b, miR-29c, miR-125b, miR-155 (S) [66] miR-92a, miR-17/-92, miR-200c, miR-141, miR-375 (P) [66] let-7f, miR-18a, miR-20a (F) [66] miR-29a (P, S) [66] miR-221 (P, F) [66] miR-106a (F, P) [66] let-7a (SE, P) [66] | piR-019825 (PV) [26] piR-5937; piR-28876 (S) [159] piR-001311, piR-004153, piR-017723, piR-017724, piR-020365 (S) [160] | RNU2-1f (P/S) [156] | |
| Respiratory | NSCLC | miR-193b, miR-301, miR-141, miR-200b (S) [83] miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, miR-214 (P) [67] miR-34a (B); let-7i, miR-10, miR-19a, miR-19b, miR-29c, miR-30c, miR-34c, miR-125a, miR-125b, miR-200c, miR-141, miR-429 (S); let-7a/b (P) [66] miR-20a (P) [66] miR-30a (P) [66] miR-375 (P) [66] let-7a/b (S/P) [66] let-7f (PV) [66] miR-125b (S, P) [66] miR-155 (S, S/P) [66] | snoRD33, snoRD66, snoRD76 (P) [161] snoRD33; snoRD66; snoRD42; snoRD78 (SP) [162] RNU2-1f (S) [163,164] | |
| Laryngeal cancer | miR-21, miR-155, miR-221 (P) [165] miR-155 (P) [166] miR-331-3p, miR-603, miR-1303, miR-660-5p, 212-3p (P) [167] miR-31, miR-141, miR-149a, miR-182, let-7a, miR-4853p, miR-122, miR-33, miR-145, miR-223, miR-133a (S) [168] let-7 (S) [169] miRNA-378 (S) [170] miR-221 (S) [171] | |||
| Urologic | Renal cell carcinoma | miR-210 (S) [66] miR-221 (P) [66] | piR-823 (S, U) [172] | |
| Bladder cancer | miR-375 (UE) [93] miR-21-5p (UE) [94] miR-30a-5p, let-7c-5p and miR-486-5p (U) [57] miR-19a (P); miR-106b (U); miR-210 (S, U) [66] miR-324-5p, miR-4738-3p (together with miR-497-HG and RCAN1 FOSB mRNA) (U) [173] | |||
| Prostatic cancer | let-7i, miR-16, miR-24, miR-26a, miR-26b, miR-34b, miR-92b, miR-93, miR-103, miR-106a, miR-141, miR-195, miR-197, miR-223, miR-298, miR-328, miR-346, miR-375, miR-1290 (S) [84,85] let-7e, let-7c, miR-20a, miR-21, miR-30c, miR-130b, miR-145, miR-1811a-2 *, miR-221, miR-301a, miR-326, miR-331-3p, miR-432, miR-574-3p, miR-622, miR-625 *, miR-1285, miR-2110, miR-141, miR-1290 (PE) [25,86,99] miR-107, miR-574-3p, miR-141-5p, miR-21-5p, miR-34a, miR-483-5p (PE) [87] miR-375, miR-21, let-7c (UE) [95] miR-196a-5p, miR-501-3p (UE) [96] miR-103a-3p, let-7a-5p (P) [89] let-7a (B) [66] miR-20a (P) [66] miR-30a (P) [66] miR-30c (P) [66] miR-375 (P) [66] miR-34b, miR-16 (S) [66] miR-141 (S, PV, SE) [66] | piR-016658, piR-020496 (PV) [26] | ||
| Gynaecologic | Breast cancer | miR-195, let-7, miR-155, miR-138 (B); miR-214, miR-15a, miR-18a, miR-107, miR-425, miR-133a, miR-139-5p, miR-143, miR-145, miR-365, miR-484, miR-1246, miR-1307-3p, miR-6861-5p, miR-4634, miR-6875-5p, miR-155, miR-19a, miR-181b, miR-24, miR-1, miR-92a, miR-133a, miR-133b, let-7c, miR-182, miR-155, miR-21, miR-126, miR-155, miR-199a, miR-335, miR-201, let-7b, miR-4270, miR-1225-5p, miR-188-5p, miR-1202, miR-148b-3p, miR-652-3p, miR-10b-5p, miR-18b, miR-103, miR-107, miR-652, miR-10b, miR-34a, miR-155, miR-29b-2, miR-155, miR-197, miR-205, miR-92a, miR-21, miR-21-5p, miR-375, miR-205-5p, miR-194-5p, miR-382-5p, miR-376c-3p, miR-411-5p, miR-34a, miR-93, miR-373, miR-17, miR-155, miR-125b, miR-122, miR-375, miR-155 (S); miR-127-3p, miR-376a, miR-148b, miR-409-3p, miR-652, miR-801, miR-148b, miR-133a, miR-409-3p, miR-505-5p, miR-125b-5p, miR-21-5p, miR-96-5p, miR-4281, miR-1207-5p, miR-642b-3p, miR-1290, miR-3141, miR-10b, miR-373 (P)[90] miR-101, miR-372, miR-373 (SE) [67] miR-10, miR-16, miR-18a, miR-19a, miR-29a, miR-34a, miR-34b/c, miR-125a, miR-375 (S) [66] miR-106b, miR-125b (P) [66] miR-34a (B) [66] miR-155 (S, S/P) [66] | RNU6 (S) [174] | |
| Ovarian cancer | miR-214, miR-140, miR-147, miR-135b, miR-205, miR-150, miR-149, miR-370, miR-206, miR-197, miR-634, miR-485-5p, miR-612, miR-608, miR-202, miR-373, miR-324-3p, miR-103, miR-593, miR-574, miR-483, miR-527, miR-603, miR-649, miR-18a, miR-595, miR-193b, miR-642, miR-557, miR-801, let-7e (SE) [100] miR-193a-5p, miR-21, miR-375, miR-210, miR-150-5p, miR-181-5p, miR-142-3p, miR-26a-5p, let-7d-5p, miR-374a-5p, miR-766-3p, miR-200a-3p, miR-130b-3p, miR-328-3p, miR-1246, miR-595, miR-2278, miR-376a, miR-125b, miR-199a, miR-7, miR-429, miR-25, miR-93, miR-200a, miR-200b, miR-200c, miR-145, miR-200c, miR-141, let-7i-5p, miR-122, miR-152-5p, miR-25-3p, miR-22, miR-93, miR-451, miR-106b, miR-21, miR-92, miR-221 (S) [92] miR-373, miR-200a, miR-200b, miR-200c, miR-21, miR-92, miR-155, miR-127, miR-93, miR-99b, miR-126, miR-29a, miR-21, miR-141, miR-203, miR-205, miR-214 (SE) [92] miR-148a, miR-200b, miR-26a, miR-625-3p, miR-720, miR-1274a, miR-19b, miR-223, miR-16, miR-150, miR-20a, miR-126, miR-1290, miR-205, let-7f, miR-16, miR-21, miR-191, miR-4284 (P) [92] let-7f (P) [66] let-7i (S/P) [66] let-7b [66] miR-92, miR-106b, miR-29a (S) [66] | RNU2-1f (S) [175] | ||
| Endometrial cancer | miR-9, miR-92a, miR-99a, miR-100, miR-199b, miR-1228 (P) [176] miR-9, miR-1228, miR-9, miR-92a (P) [177] miR-21, miR-222, miR-223, miR-186 and miR-204 (S) [178,179] miR-203 (S) [180] miR-21 (S) [181] miR-887-5p (S) [182] miR-106b (U) [176] miR-200c-3p (U) [183] | |||
| Cervical cancer | miR-101, miR-127, miR-142-3p, miR-150, miR-205, miR-21, miR-24, miR-425-5p, miR-486-5p, miR-494 (S) [184] miR-21, miR-146a, miR-155, miR-182, miR-200c, let-7b, miR-145 (S) [185] miR-9, miR-10a, miR-20a, miR-196a (S) [186] miR-21, miR-29a, miR-25, miR-200a, miR-486-5p (S) [187] miR-92a (S) [188] miR-646, miR-141-5p, miR-542-3p (S) [189] miR-205 (S) [190] | |||
| Haematologic | Hodgkin Lymphoma | miR-144, miR-143, miR-129-5p, miR-182, miR-411, miR-126-3p, miR-433, miR-23a, miR-24, miR-423-5p (B) [191] | piR-651 (S) [192] | |
| Non-Hodgkin Lymphoma | miR-92a, miR-638 (P) [128] DLBCL miR-155, miR-210 (S); miR-221 (P)[66] miR-15a, miR-16–1, miR-34a, miR-155, miR-29c (S) [128] miR-425, miR-141, miR-197, miR-145, miR-345, miR-200c, miR-324-5p, let-7i-5, miR-424, miR-222 (B), [191] miR-34a-5p, miR-323b-3p, miR-431-5p, miR-125b, miR-130a, miR-155, miR-200c, miR-29c, miR-145, miR-451, miR-17, miR-20b, miR-210, miR-296, miR-15a-3p, miR-21-5p, miR-210-5p, miR-181a-5p, miR-155-5p, miR-210-3p, miR-16-1, miR-29c, miR-155, miR-34a, miR-223, miR-155, miR-210 (S); miR-21 (S,P); miR-155 (EV from serum)[193] NTCL miR-221 (P) [128] PNCSL miR-21 (S) [128] miR-21, miR-19, and miR-92a (CSF) miR-451, miR-711, miR-935, miR-223, miR-125b [194] | |||
| Chronic leukemia | CLL let-7a, let-7c, let-7f, let-7g, let-7i, miR-10a, miR-15a, miR-19a, miR-20a, miR-21, miR-22, miR-24, miR-26b, miR-29a, miR-29b, miR-29c, miR-30b, miR-31-5p, miR-33, miR-34a, miR-34a-5p, miR-92, miR-95, miR-99a, miR-100, miR-101, miR-106b, miR-107, miR-123, miR-125a, miR-125a, miR-126, miR-126, miR-130a, miR-132, miR-134, miR-136, miR-138, miR-139, miR-140, miR-141, miR-142-5p, miR-143, miR-146a, miR-147a, miR-148a, miR-150, miR-150-5p, miR-151-3p, miR-154, miR-155, miR-155-5p, miR-181a, miR-181b, miR-182, miR-183, miR-184, miR-185, miR-190, miR-191, miR-192, miR-193b, miR-196-2, miR-197, miR-198, miR-199a, miR-206, miR-212, miR-217, miR-220, miR-223, miR-326, miR-323-3p, miR-632, miR-337-3p, miR-338, miR-342-3p, miR-363, miR-367, miR-370, miR-374b, miR-377, miR-424, miR-442, miR-451, miR-453, miR-484, miR-491, miR-494, miR-572, miR-582-5p, miR-636, miR-640, miR-660, miR-923, miR-1201, miR-1202, miR-1203, miR-3676 (B) [130] miR-195, miR-199b, miR-320, miR-432, (P) [130] CML miR-17, miR-18a, miR-20a, miR-21, miR-27a, miR-155 (B) [195] miR-122-5p, miR-16-5p, miR-451a and miR-92a-3p, miR-4644, miR-6075, miR-4656, miR-5739 (B) [196] miR-90, miR-150, miR-16, miR-17, miR-20, miR-21, miR-29, miR-92a, miR-101, miR-106, miR-126, miR-146, miR-150, miR-152, miR-155, miR-451, miR-568, miR-607 (B) [197] | |||
| Acute leukemia | AML miR-92a, miR-143, miR-342, miR-100, miR-196a, miR-146a, miR-29a, miR-142-3p, miR-29a, miR-142-3p, miR-150, miR-342, miR-150, miR-342 (P) [128] miR-21, miR-155, miR-210, miR-221, miR-10a-5p, miR-93-5p, miR-129-5p, miR-155-5p, miR-181b-5p, miR-320d (S) [128] ALL miR-155 (S, P) [66] miR-128b, miR-204, miR-218, miR-331, miR-135b, miR-132, miR-199, miR-139, miR-150, miR-519e; miR-487b; miR-361; miR-142-3p; miR-222; miR-339, miR-451; miR-373; miR-296; miR-485-3p; miR-483, miR-196a, miR-383, miR-542-5p, miR-133a (B), miR-708, miR-511, miR-708, let-7b, miR-708, miR-210, miR-181b, miR-345, miR-324-5p, and miR-125b, miR-23a, miR-27a, miR-23b, miR-24 (B) [198] | |||
| Multiple myeloma | miR-19a (S)[66] miR-92a, miR-483-5p, miR-483-5p, miR-20a (P) [128] miR-29a, miR-34a, let-7e, miR-19a, miR-92a, miR-202, miR-720, miR-1308 (S) [128] | |||
| Central Nervous System (CNS) | Glioma | miR-21 (B, P, S) miR-203, miR-137, miR-185, miR-210, miR-205, miR-221/222, miR-29, miR-397a/b/c, miR-125b, miR-497, miR-182, miR-128, miR-451a, miR-15b-5p, miR-23a, miR-133, miR-150 *, miR-197, miR-497, miR-548b-5p, miR-15b-5p, miR-16-5p, miR-19a-3p, miR-19b-3p, miR-20a-5p, miR-106a-5p, miR-130a-3p, miR-181b-5p, miR-208a-3p, miR-17, miR-130a, miR-10b, miR-93, miR-590-3p, miR-454 (S) miR-122, miR-128, miR-342-3p, miR-454-3p (P) miR-15b, miR-16, miR-128, miR-342-3p (B) miR-222, miR-124-3p, miR-301a, miR-320, miR-574-3p (SE) miR-21, miR-10b; miR-200, miR-21, miR-21, miR-15b, miR-451, miR-711, miR-935, miR-223, miR-125b, miR-21-5p, miR-218-5p, miR-193b-3p, miR-331-3p, miR-374a-5p, miR-548c-3p, miR-520f-3p, miR-27b-3p, miR-130b-3p (CSF) [91] | ||
| Medulloblastoma | Diagnostic flowchart to distinguish Medulloblastoma from other CNS cancers: miR-451, miR-711, miR-935, miR-125b, miR-223 (CSF) [194] | |||
| Retinoblastoma | miR-338-5p (S) [199] miR-18a (S) (S) [200] miR-17, miR-18a, miR-20a (S) [201] miR-320, miR-let-7e, miR-21(S) [202] | |||
| Endocrine | Thyroid cancer | miR-25-3p, miR-451a (P) [203] miR-346, miR-10a-5p, miR-34a-5p (P) [204] miR-124-3p and miR-9-3p (P) [205] miR-146b, miR-155 (P) [206] miR-222, miR146b (P) [207] miR-144, miR-34a (P) [208] miR-222, miR-221, miR-146b, miR-21 (S) [209] miR-95, miR-190 (S) [210] miR-222, miR-221 miR-146b (S) [211] miR-30a-5p (S) [212] miR-146a-5p, miR-199b-3p, let7b-5p, miR-10a-5p (S) [213] miR-95, miR-190 (S) [214] miR-222 (S) [215] let-7e, miR-151-5p, miR-222 (S) [216] miR-375, miR-34a, miR-145b, miR-221, miR-222, miR-155, Let-7, miR-181b (S) [217] miR-146b-5p, miR-222-3p, miR-155-5p, and miR-378a-3p (B) [218] miR-21, miR-31, miR-181a-5p (PE) [218] | ||
| Bone | Osteosarcoma | miR-487a, miR-493-5p, miR-501-3p and miR-502-5p (S) [219] miR-25-3p (S) [220] miR-215-5p, miR-642a-5p (S) [221] miR-101 (S) [222] miR-124 (S) [223] miR-375 (S) [224] miR-598 (S) [225] miR-95-3p (S) [226] miR-300 (S) [227] miR-497 (S) [228] miR-326 (S) [229] miRNA-223 (S) [230] miR-421 (S) [231] miR-106a-5p, miR16-5p, miR-20a-5p, miR-425-5p, miR451a, miR-25-3p, miR139-5p(S) [232] miR-152 (S) [233] miR-191 (S) [234] miR-221 (S) [235] miR-199a-5p (S) [236] miR-27a (S) [237] miR-195 (S) [238] miR-133b, miR-206 (S) [239] miR-29a, miR-29b, miR-29c (S) [240] miR-663a (P) [241] miR-195-5p, miR-199a-3p, miR-320a, miR-374a-5p (P) [242] miR-139-5p (P) [243] miR-Let7a (B) [244] miR-148a (B) [245] | ||
| Skin Cancers | Melanoma | miR-1260a, miR-126-5p, miR-1280, miR-145-5p, miR-150-5p, miR-155-5p, miR-16-5p, miR-182-5p, miR-193b-3p, miR-197-3p, miR-200c-3p, miR-204-5p, miR-205-5p, miR-206, miR-211-5p, miR-211-5p, miR-221-3p, miR-22-3p, miR-27a-3p, miR-28-5p, miR-301a-3p, miR-30b-5p, miR-373-5p, miR-374-5p, miR-432-3p, miR-4487, miR-451a, miR-4706, miR-4731, miR-509-3p, miR-509-5p, miR-550a-3p, miR-627-5p, miR-629-5p, miR629-5p, miR-720, miR-9-5p (S) [246] miR-126-5p, miR-211-5p, miR-206, miR-16 miR-211, miR-182-5p, miR-301a-3p, miR-193b-3p, miR-197-3p, miR-200c-3p, miR-204-5p, miR-28-5p, miR-27a-3p, miR-29c-5p, miR-324-3p, miR-374a-5p, miR-4487, miR-4706, miR-4731, miR-509-3p, miR-509-5p, miR-550a-3p, miR-627-5p, miR-629-5p, miR-720, miR-205-5p (S) [247] miR-182-5p, miR-193b-3p, miR-200c-3p, miR-204-5p, miR-205-5p, miR-211-5p, miR-301a-3p, miR-720 (S) [248] miR-150-5p, miR-146a, miR-149-3p, miR-155, miR-15b-5p, miR-181a, miR-193a-3p, miR-20a, miR-223, miR-524-5p, miR-125b (P) [247] miR-1258, miR-1264, miR-1269a, miR-1302, miR-1306-5p, miR-138-5p, miR-152-3p, miR-1537-3p, miR-154-5p, miR-1-5p, miR-181b-5p, miR-1910-5p, miR-1973, miR-205-5p, miR-219a-2-3p, miR-2682-5p, miR-27a-3p, miR-299-3p, miR-301a-3p, miR-3131, miR-337-5p, miR-34a-5p, miR-3928-3p, miR-424-5p, miR-431-5p, miR-450a-5p, miR-4532, miR-454-3p, miR-4787-3p, miR-497-5p, miR-520d-3p, miR-522-3p, miR-548a-5p, miR-548ad-3p, miR-548l, miR-553, miR-624-3p, miR-764 (P) [249] miR-134-5p, miR-320a-3p (P) [250] miR-21-5p (P) [246] miR-200c-3p (P, S) [246] miR-125b (SE) [247] |
| Cat. | Cancer Type | lncRNA | circRNA |
|---|---|---|---|
| Digestive | Oral cancer | AC007271.3 (S) [307] HOTAIR (SA) [300] | hsa_circ_0001874 (SA) [308] hsa_circ_0001971 (SA) [308] circMAN1A2 (S) [309] |
| Nasopharyngeal cancer | MALAT1, AFAP1-AS1, AL359062 (S) [310] | circRNA_0000285 (S) [311] circMAN1A2 (S) [309] | |
| Esophageal cancer | POU3F3; HNF1A-AS1; SPRY4-IT1 (P) [282] MIR31HG (P), [312] HOTAIR (S) [313] POU3F3, SCCA (P) [282] | - | |
| Hepatocellular carcinoma | HULC (P) [281] RP11-160H22.5, XLOC_014172, LOC149086 (P) [284] SNHG18 (P) [286] SNHG1 (P) [290] GAS5-AS1 (P) [314] ZFAS1 (P) [315] DANCR (P) [316] HULC, Linc00152 (P) [317] SPRY4-IT1 (P) [318] NEAT (S) [319] LRB1 (S) [320] UCA1 (S) [321] X91348 (S) [322] PVT1, uc002mbe.2 (S) [323] uc001ncr, AX800134 (S) [324] UCA1, WRAP53 (S) [325] LINC00161 (S, SE, U) [326] ENSG00000258332.1, LINC00635 (SE) [327] | hsa_circ_0001445 (P) [328] circSMARCA5 (P) [329] | |
| Cholangiocarcinoma | PCAT1, MALAT1, CPS1-IT1 (P) [330] ENST00000588480.1, ENST00000517758.1 (BI) [302] | - | |
| Gallbladder carcinoma | - | - | |
| Pancreatic cancer | ABHD11-AS1, LINC00176, SNHG11 (P) [331] HOTAIR, PVT1 (SA) [301] HOTTIP-005, RP11-567G11.1 (P/S) [332] | circLDLRAD3 (P) [333] | |
| Gastric cancer | PANDAR, FOXD2-AS1, SMARCC2 (P) [334] FAM49B-AS, GUSBP11, CTDHUT (P) [335] H19, MEG3 (P) [336] GACAT2 (P) [337] HOTAIR (P) [338] AK001058, INHBA-AS1, MIR4435-2HG, CEBPA-AS1, CTC-501O10.1, AC100830.4, RP11-210K20.5 (P) [339] ZFAS1 (P) [280] UCA1 (P) [340] TINCR, CCAT2, AOC4P, BANCR, LINC00857 (P) [341] HULC (S) [342] HOXA11-AS (S) [343] H19 (S) [344,345] CTC-497E21.4 (S) [346] LINC00978 (S) [347] XIST, BCYRN1, RRP1B, TDRG1 (S) [348] H19 (S) [349] CUDR; LSINCT-1; PTENP1 (S) [291] LINC00152 (P, E) [278] UEGC1 (PE) [350] LINC00152, AA174084, UCA1, RMRP, ABHD11-AS1, LINC00982, H19 [351] (GJ) AA174084 (GJ) [303] | hsa_circ_0001649 (P) [352] hsa_circ_0001017, hsa_circ_0061276 (P) [353] hsa_circ_0000745 (P) [354] hsa_circ_0000520 (P) [355] circ-KIAA1244 (P) [356] hsa_circ_0000190 (P) [357] hsa_circ_0000467 (P) [358] hsa_circ_0000181 (P) [359] | |
| Colorectal cancer (CRC) | ATB, CCAT1 (P) [360] 91H, PVT-1, MEG3 (P) [361] CCAT2, HULC (P) [362] BLACAT1 (P) [363] B3GALT5-AS1 (P) [364] CCAT2 (S) [293] LOC285194, RP11-462C24.1, Nbla12061 (S) [365] NR_029373, NR_034119 (S) [366] CCAT1, HOTAIR (S) [367] CRNDE-p (SE) [158] CRNDE-h (SE) [294] CCAT2 (SE) [293] LNCV6_116109, LNCV6_98390, LNCV6_38772, LNCV_108266, LNCV6_84003, LNCV6_98602 (PE) [368] CRNDE-h (SE) [294] BCAR4 (+mRNA KRTAP5-4 and MAGEA3) (SEV) [369] | hsa_circ_0001649 (S) [370] circVAPA (hsa_circ_0006990) (S) [371] | |
| Respiratory | NSCLC | GAS5, SOX2OT (P) [288] H19 (P) [372] HOTAIR (P) [373] Linc00152 (P) [374] SNHG1, RMRP (P) [375] GAS5 (P) [376] SPRY4-IT1, ANRIL, NEAT1 (P) [377] RP11-397D12.4, AC007403.1, ERICH1-AS1 (P) [283] UCA1 (P) [378] RP11-397D12.4, AC007403.1, ERICH1-AS1 (P) [283] TUG1 (S) [379] PCAT6 (S) [380] XIST, HIF1A-AS1 (S) [381] AFAP1-AS1 (S) [382] SOX2OT, ANRIL, CEA, CYFRA21-1, SCCA (S) [383] XLOC_009167 (B) [384] MALAT1 (SE) [292] GAS5 (SE) [385] SOX2-OT (PE) [386] SLC9A3-AS1, PCAT6 (EV from P) [387] | - |
| Laryngeal cancer | HOTAIR (S) [388] | - | |
| Urologic | Renal cell carcinoma | lncARSR (P) [285] LET, PVT1, PANDAR, PTENP1, linc00963 (S) [389] | - |
| Bladder cancer | TUC338 (P) [390] H19 (SE) [391] PCAT-1, UBC1, SNHG16 (SE) [392] H19 (U) [393] miR-497-HG (together with miR-324-5p, miR-4738-3p and RCAN1 FOSB mRNA) (U) [173] uc004cox.4, GAS5 (U) [394] HOTAIR, HOX-AS2, ANRIL, HYMA1, LINC00477, LOC100506688, OTX2-AS1 (UE) [295] UCA1-201, UCA1-203, MALAT1, LINC00355 (UE) [395] MALAT1, PCAT-1, SPRY4-IT1 (UE) [396] MEG3, SNHG16, MALAT1 (S) [397] | circPRMT5 (SE, UE) [398] | |
| Prostatic cancer | TUC338 (P) [399] BRE-AS1 (P) [400] MALAT1 (P/S) [401] PCA3/PSA (U) [402] PRCAT17.3, PRCAT38 (U) [403] PCA3, ERG (UE) [296,297,299] TP53COR1 (UE) [298] FR0348383 (U) [404] | - | |
| Gynaecologic | Breast cancer | ANRIL, HIF1A-AS2, UCA1 (P) [287] H19 (P) [405] MALAT1 (S) [406] RP11-445H22.4 (S) [407] | hsa_circ_0001785 (P) [408] |
| Ovarian cancer | - | circMAN1A2 (S) [309] | |
| Endometrial cancer | - | hsa_circ_0109046 (SEV) [409] hsa_circ_0002577 (SEV) [409] | |
| Cervical cancer | FALEC (P) [410] HOTAIR, PVT1, XLOC_000303, AL592284.1 (P) [411] GIHCG (S) [412] PVT1 (S) [413] | - | |
| Haematologic | Hodgkin Lymphoma | - | - |
| Non-Hodgkin Lymphoma | - | - | |
| Chronic leukemia | LincRNA-p21 (P) [289] | - | |
| Acute leukemia | - | - | |
| Multiple myeloma | TUG1; MALAT1; HOTAIR; GAS5 (P) [289] PCAT-1 (S) [414] H19 (S) [415] | - | |
| Central Nervous System (CNS) | Glioma | miR210HG (S) [416] | - |
| Medulloblastoma | - | - | |
| Retinoblastoma | - | - | |
| Endocrine | Thyroid cancer | GAS8-AS1 (P) [417] | circMAN1A2 (S) [309] hsacirc_007293, hsacirc_031752, hsacirc_020135 (SE) [418] |
| Bone | Osteosarcoma | UCA1 (S) [393] | hsa_circ_0000885 (S) [419] |
| Skin Cancers | Melanoma | PVT1 (S) [420] | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170. https://doi.org/10.3390/cancers11081170
Pardini B, Sabo AA, Birolo G, Calin GA. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers. 2019; 11(8):1170. https://doi.org/10.3390/cancers11081170
Chicago/Turabian StylePardini, Barbara, Alexandru Anton Sabo, Giovanni Birolo, and George Adrian Calin. 2019. "Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies" Cancers 11, no. 8: 1170. https://doi.org/10.3390/cancers11081170
APA StylePardini, B., Sabo, A. A., Birolo, G., & Calin, G. A. (2019). Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers, 11(8), 1170. https://doi.org/10.3390/cancers11081170







