Immune Resistance and EGFR Antagonists in Colorectal Cancer
Abstract
1. Introduction
2. The Roles of EGFR/ERBB Therapy in Colorectal Cancer
3. Acquired Resistance to Anti-EGFR Treatment in CRC Patients
4. Contribution of Tumor Microenvironment to Acquired Resistance to EGFR Blockade
5. Alternative Non-Genetic Mechanisms That Evade EGFR-Targeted Agents
6. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACT | adoptive cell transfer |
| AKT | protein kinase B |
| AREG | amphiregulin |
| CAR | chimeric antigen receptor |
| CMS | consensus molecular subtypes |
| CSC | colon cancer stem cell |
| CRC | colorectal cancer |
| ctDNA | circulation tumor DNA |
| EGFR | epidermal growth factor receptor |
| EREG | epiregulin |
| ERKs | extracellular signal–regulated kinases |
| FOXO3a | Forkhead box class O 3a |
| HDACs | histone deacetylases |
| IGF1R | insulin like growth factor 1 receptor |
| JAK/STAT | Janus kinase/signal transducer and activator of transcription |
| JNKs | c-Jun N-terminal kinase |
| mAbs | monoclonal antibodies |
| mCRC | metastatic colorectal cancer |
| MEK | mitogen-activated protein kinase |
| MMR | mismatch repair |
| MSI | microsatellite instability |
| MSS | microsatellite stable |
| mTORC1 | mammalian target of rapamycin complex 1 |
| NK | natural killer |
| PD-1 | programmed cell death protein 1 |
| PI3K | phosphoinositide 3-kinase |
| PKC | protein kinase C |
| PKM2 | pyruvate kinase muscle isozyme M2 |
| PLCγ | phospholipase C gamma |
| p38 MAPK | p38 mitogen-activated protein kinases |
| RAS | rat sarcoma viral oncogene homolog |
| RAF | rapidly accelerated fibrosarcoma |
| SRC | Rous sarcoma virus proto-oncogene homolog |
| TKIs | tyrosine kinase inhibitors |
| Treg | regulatory T cells |
| VEGFR | vascular endothelial growth factor receptor |
| TME | tumor microenvironment |
References
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 638–691. [Google Scholar] [CrossRef] [PubMed]
- Murphy, N.; Moreno, V.; Hughes, D.J.; Vodicka, L.; Vodicka, P.; Aglago, E.K.; Gunter, M.J.; Jenab, M. Lifestyle and dietary environmental factors in colorectal cancer susceptibility. Mol. Asp. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Nordlinger, B.; Arnold, D. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up dagger. Ann. Oncol. 2014, 25. [Google Scholar] [CrossRef] [PubMed]
- Sartore-Bianchi, A.; Siena, S.; Tonini, G.; Bardelli, A.; Santini, D. Overcoming dynamic molecular heterogeneity in metastatic colorectal cancer: Multikinase inhibition with regorafenib and the case of rechallenge with anti-EGFR. Cancer Treat Rev. 2016, 51, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, J.; Lavingia, V.; Fakih, M. Systemic treatment for metastatic colorectal cancer in the era of precision medicine. J. Surg. Oncol. 2019, 119, 564–582. [Google Scholar] [CrossRef] [PubMed]
- Rachiglio, A.M.; Lambiase, M.; Fenizia, F.; Roma, C.; Cardone, C.; Iannaccone, A.; De Luca, A.; Carotenuto, M.; Frezzetti, D.; Martinelli, E.; et al. Genomic Profiling of KRAS/NRAS/BRAF/PIK3CA Wild-Type Metastatic Colorectal Cancer Patients Reveals Novel Mutations in Genes Potentially Associated with Resistance to Anti-EGFR Agents. Cancers (Basel) 2019, 11, 859. [Google Scholar] [CrossRef] [PubMed]
- Cremolini, C.; Benelli, M.; Fontana, E.; Pagani, F.; Rossini, D.; Fucà, G.; Busico, A.; Conca, E.; Di Donato, S.; Loupakis, F.; et al. Benefit from anti-EGFRs in RAS and BRAF wild-type metastatic transverse colon cancer: a clinical and molecular proof of concept study. ESMO Open 2019, 4, e000489. [Google Scholar] [CrossRef] [PubMed]
- Gbenedio, O.M.; Bonnans, C.; Grun, D.; Wang, C.Y.; Hatch, A.J.; Mahoney, M.R.; Barras, D.; Matli, M.; Miao, Y.; Garcia, K.C.; et al. RasGRP1 is a potential biomarker to stratify anti-EGFR therapy response in colorectal cancer. JCI Insight 2019, 5. [Google Scholar] [CrossRef]
- Gao, Y.; Maria, A.; Na, N.; Da Cruz Paula, A.; Gorelick, A.N.; Hechtman, J.J.; Carson, J.; Lefkowitz, R.R.; Weigelt, B.; Taylor, B.B.; et al. V211D mutation in MEK1 causes resistance to MEK inhibitors in colon cancer. Cancer Discov. 2019. [Google Scholar] [CrossRef]
- García-Albéniz, X.; Alonso, V.; Escudero, P.; Méndez, M.; Gallego, J.; Rodríguez, J.J.; Salud, A.; Fernández-Plana, J.; Manzano, H.; Zanui, M.; et al. Prospective Biomarker Study in Advanced RAS Wild-Type Colorectal Cancer: POSIBA Trial (GEMCAD 10-02). Oncologist 2019. [Google Scholar] [CrossRef]
- Montagut, C.; Tsui, D.D.; Diaz, L.A., Jr. Detection of somatic RAS mutations in circulating tumor DNA from metastatic colorectal cancer patients: Are we ready for clinical use? Ann. Oncol. 2018, 29, 1083–1084. [Google Scholar] [CrossRef] [PubMed]
- Mauri, G.; Pizzutilo, E.E.; Amatu, A.; Bencardino, K.; Palmeri, L.; Bonazzina, E.E.; Tosi, F.; Carlo Stella, G.; Burrafato, G.; Scaglione, F.; et al. Retreatment with anti-EGFR monoclonal antibodies in metastatic colorectal cancer: Systematic review of different strategies. Cancer Treat Rev. 2019, 73, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, L.; Ma, K.; Zhao, Y.; Liu, X.; Wang, Y.; Liu, M.; Liang, S.; Zhu, H.; Xu, N. Polarization of macrophages in the tumor microenvironment is influenced by EGFR signaling within colon cancer cells. Oncotarget 2016, 7, 75366–75378. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Yu, X.; Yang, B.; Zhang, Y.; Zhang, L.; Li, X.; Sun, H. Colorectal cancer heterogeneity and targeted therapy: Clinical implications, challenges and solutions for treatment resistance. Semin. Cell Dev. Biol. 2017, 64, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Klesmith, J.J.; Su, L.; Wu, L.; Schrack, I.I.; Dufort, F.F.; Birt, A.; Ambrose, C.; Hackel, B.B.; Lobb, R.R.; Rennert, P.D. Retargeting CD19 CAR T cells via engineered CD19-fusion proteins. Mol. Pharm. 2019. [Google Scholar] [CrossRef] [PubMed]
- Srinivas Patnaik, A. Drugs Targeting Epigenetic Modifications and Plausible Therapeutic Strategies Against Colorectal Cancer. Front Pharm. 2019, 10, 588. [Google Scholar] [CrossRef]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signaling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Hynes, N.N.; Lane, H.A. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat. Rev. Cancer 2005, 5, 341–354. [Google Scholar] [CrossRef]
- Lipsick, J.A. History of Cancer Research: Tyrosine Kinases. Cold Spring Harb Perspect Biol. 2019, 11. [Google Scholar] [CrossRef]
- Ciardiello, F.; Tortora, G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008, 358, 1160–1174. [Google Scholar] [CrossRef]
- Pancione, M.; Giordano, G.; Parcesepe, P.; Cerulo, L.; Coppola, L.; Curatolo, A.A.; Conciatori, F.; Milella, M.; Porras, A. Emerging Insight into MAPK Inhibitors and Immunotherapy in Colorectal Cancer. Curr. Med. Chem. 2017, 24, 1383–1402. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Punt, C.C.; Koopman, M.; Vermeulen, L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol. 2017, 14, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Raghav, K.; Loree, J.M.; Morris, J.S.; Overman, M.J.; Yu, R.; Meric-Bernstam, F.; Menter, D.; Korphaisarn, K.; Kee, B.; Singh, S.; et al. Validation of HER2 Amplification as a Predictive Biomarker for Anti–Epidermal Growth Factor Receptor Antibody Therapy in Metastatic Colorectal Cancer. JCO Precis. Oncol. 2019, 3, 1–13. [Google Scholar] [CrossRef]
- Saito, R.; Suzuki, H.; Yamada, T.; Endo, S.; Moriwaki, T.; Ueno, T.; Hirose, M.; Hirai, S.; Yamato, K.; Mizokami, Y.; et al. Predicting skin toxicity according to EGFR polymorphisms in patients with colorectal cancer receiving antibody against EGFR. Anticancer Res. 2013, 33, 4995–4998. [Google Scholar] [PubMed]
- Borrero-Palacios, A.; Cebrián, A.; del Pulgar, M.G.; García-Carbonero, R.; García, P.; Aranda, E.; Elez, E.; López-López, R.; Cervantes, A.; Nadal, C. Combination of KIR2DS4 and FcγRIIa polymorphisms predicts the response to cetuximab in KRAS mutant metastatic colorectal cancer. Sci. Rep. 2019, 9, 7706. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.H.; Wang, F.; Chen, X.X.; He, B.B.; Pan, Y.Y.; Jie, C.; Liu, X.; Cao, W.W.; Peng, H.H.; Lin, K.; et al. FCGR2A, FCGR3A polymorphisms and therapeutic efficacy of anti-EGFR monoclonal antibody in metastatic colorectal cancer. Oncotarget 2015, 6, 28071–28083. [Google Scholar] [CrossRef] [PubMed]
- Jaka, A.; Gutiérrez-Rivera, A.; Ormaechea, N.; Blanco, J.; La Casta, A.; Sarasqueta, C.; Izeta, A.; Tuneu, A. Association between EGFR gene polymorphisms, skin rash and response to anti-EGFR therapy in metastatic colorectal cancer patients. Exp. Dermatol. 2014, 23, 751–753. [Google Scholar] [CrossRef] [PubMed]
- Bonin, S.; Donada, M.; Bussolati, G.; Nardon, E.; Annaratone, L.; Pichler, M.; Chiaravalli, A.A.; Capella, C.; Hoefler, G.; Stanta, G. A synonymous EGFR polymorphism predicting responsiveness to anti-EGFR therapy in metastatic colorectal cancer patients. Tumour. Biol. 2016, 7295–7303. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Stintzing, S.; Sunakawa, Y.; Cao, S.; Zhang, W.; Yang, D.; Ning, Y.; Matsusaka, S.; Berger, M.M.; Miyamoto, Y.; et al. Predictive value of TLR7 polymorphism for cetuximab-based chemotherapy in patients with metastatic colorectal cancer. Int. J. Cancer 2017, 141, 1222–1230. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Kaltenbrun, E.; Anderson, G.G.; Stephens, S.S.; Arena, S.; Bardelli, A.; Counter, C.C.; Wood, K.C. Codon bias imposes a targetable limitation on KRAS-driven therapeutic resistance. Nat. Commun. 2017, 8, 15617. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.G.; Winter, P.P.; Lin, K.K.; Nussbaum, D.D.; Cakir, M.; Stein, E.E.; Soderquist, R.R.; Crawford, L.; Leeds, J.J.; Newcomb, R.; et al. A Landscape of Therapeutic Cooperativity in KRAS Mutant Cancers Reveals Principles for Controlling Tumor Evolution. Cell Rep. 2017, 20, 999–1015. [Google Scholar] [CrossRef] [PubMed]
- Montagut, C.; Dalmases, A.; Bellosillo, B.; Crespo, M.; Pairet, S.; Iglesias, M.; Salido, M.; Gallen, M.; Marsters, S.; Tsai, S.S.; et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat. Med. 2012, 18, 221–223. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Siravegna, G.; Blaszkowsky, L.L.; Corti, G.; Crisafulli, G.; Ahronian, L.L.; Mussolin, B.; Kwak, E.L.; Buscarino, M.; Lazzari, L.; et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov. 2016, 6, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Pietrantonio, F.; Vernieri, C.; Siravegna, G.; Mennitto, A.; Berenato, R.; Perrone, F.; Gloghini, A.; Tamborini, E.; Lonardi, S.; Morano, F.; et al. Heterogeneity of acquired resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer. Clin. Cancer Res. 2017, 23, 2414–2422. [Google Scholar] [CrossRef] [PubMed]
- Takegawa, N.; Tsurutani, J.; Kawakami, H.; Yonesaka, K.; Kato, R.; Haratani, K.; Hayashi, H.; Takeda, M.; Nonagase, Y.; Maenishi, O.; et al. [fam-] trastuzumab deruxtecan, antitumor activity is dependent on HER2 expression level rather than on HER2 amplification. Int. J. Cancer 2019. [Google Scholar] [CrossRef]
- Kanat, O.; Ertas, H.; Caner, B. Dual HER2 inhibition strategies in the management of treatment-refractory metastatic colorectal cancer: History and status. World J. Clin. Cases. 2018, 6, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Saenz, A.; Dreyer, C.; Campbell, M.M.; Steri, V.; Gulizia, N.; Moasser, M.M. HER2 Amplification in Tumors Activates PI3K/Akt Signaling Independent of HER3. Cancer Res. 2018, 78, 3645–3658. [Google Scholar] [CrossRef]
- Jia, J.; Morse, M.M.; Nagy, R.R.; Lanman, R.R.; Strickler, J.H. Cell-Free DNA Profiling to Discover Mechanisms of Exceptional Response to Cabozantinib Plus Panitumumab in a Patient with Treatment Refractory Metastatic Colorectal Cancer. Front. Oncol. 2018, 8, 305. [Google Scholar] [CrossRef]
- Lanaya, H.; Natarajan, A.; Komposch, K.; Li, L.; Amberg, N.; Chen, L.; Wculek, S.K.; Hammer, M.; Zenz, R.; Peck-Radosavljevic, M.; et al. EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat. Cell Biol. 2014, 16, 972–981. [Google Scholar] [CrossRef]
- Giordano, G.; Febbraro, A.; Tomaselli, E.; Sarnicola, M.M.; Parcesepe, P.; Parente, D.; Forte, N.; Fabozzi, A.; Remo, A.; Bonetti, A.; et al. Cancer-related CD15/FUT4 overexpression decreases benefit to agents targeting EGFR or VEGF acting as a novel RAF-MEK-ERK kinase downstream regulator in metastatic colorectal cancer. J. Exp. Clin. Cancer Res. 2015, 34, 108. [Google Scholar] [CrossRef] [PubMed]
- Giordano, G.; Parcesepe, P.; D’Andrea, M.M.; Coppola, L.; Di Raimo, T.; Remo, A.; Manfrin, E.; Fiorini, C.; Scarpa, A.; Amoreo, C.C.; et al. JAK/STAT5-mediated subtype-specific lymphocyte antigen 6 complex, locus G6D (LY6G6D) expression drives mismatch repair proficient colorectal cancer. J. Exp. Clin. Cancer Res. 2019, 38, 28. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Guo, M.; Wei, Y.; Yu, S.; Li, H.; Wang, Y.; Xu, X.; Cui, Y.; Tian, J.; Liang, L.; et al. FoxO3a confers cetuximab resistance in RAS wild-type metastatic colorectal cancer through c-Myc. Oncotarget 2016, 7, 80888–80900. [Google Scholar] [CrossRef] [PubMed]
- Weitsman, G.; Mitchell, N.N.; Evans, R.; Cheung, A.; Kalber, T.T.; Bofinger, R.; Fruhwirth, G.G.; Keppler, M.; Wright, Z.V.F.; Barber, P.P.; et al. Detecting intratumoral heterogeneity of EGFR activity by liposome-based in vivo transfection of a fluorescent biosensor. Oncogene 2017, 36, 3618–3628. [Google Scholar] [CrossRef] [PubMed]
- Marzi, L.; Combes, E.; Vié, N.; Ayrolles-Torro, A.; Tosi, D.; Desigaud, D.; Perez-Gracia, E.; Larbouret, C.; Montagut, C.; Iglesias, M.; et al. FOXO3a and the MAPK p38 are activated by cetuximab to induce cell death and inhibit cell proliferation and their expression predicts cetuximab efficacy in colorectal cancer. Br. J. Cancer 2016, 115, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Xu, Y.; Chang, L.; Gong, Y.; Li, L.; Mo, X.; Zhang, X.; Lin, G.; Zhou, J.; Liu, D.; et al. Genotyping of Circulating Tumor DNA Reveals the Clinically Actionable Mutation Landscape of Advanced Colorectal Cancer. Mol. Cancer 2019, 18, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, P.P.; Cardone, C.; Martini, G.; Ciardiello, D.; Belli, V.; Matrone, N.; Barra, G.; Napolitano, S.; Della Corte, C.; Turano, M.; et al. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J. Exp. Clin. Cancer Res. 2019, 38, 41. [Google Scholar] [CrossRef]
- Sartore-Bianchi, A.; Amatu, A.; Porcu, L.; Ghezzi, S.; Lonardi, S.; Leone, F.; Bergamo, F.; Fenocchio, E.; Martinelli, E.; Borelli, B.; et al. HER2 Positivity Predicts Unresponsiveness to EGFR-Targeted Treatment in Metastatic Colorectal Cancer. Oncologist 2019. [Google Scholar] [CrossRef]
- Belli, V.; Matrone, N.; Napolitano, S.; Migliardi, G.; Cottino, F.; Bertotti, A.; Trusolino, L.; Martinelli, E.; Morgillo, F.; Ciardiello, D.; et al. Combined blockade of MEK and PI3KCA as an effective antitumor strategy in HER2 gene amplified human colorectal cancer models. J. Exp. Clin. Cancer Res. 2019, 38, 236. [Google Scholar] [CrossRef]
- Rimassa, L.; Bozzarelli, S.; Pietrantonio, F.; Cordio, S.; Lonardi, S.; Toppo, L.; Zaniboni, A.; Bordonaro, R.; Di Bartolomeo, M.; Tomasello, G.; et al. Phase II Study of Tivantinib and Cetuximab in Patients with KRAS Wild-type Metastatic Colorectal Cancer with Acquired Resistance to EGFR Inhibitors and Emergence of MET Overexpression: Lesson Learned for Future Trials with EGFR/MET Dual Inhibition. Clin. Colorectal. Cancer 2019, 18, 125–132. [Google Scholar] [CrossRef]
- Della Corte, C.C.; Fasano, M.; Papaccio, F.; Ciardiello, F.; Morgillo, F. Role of HGF-MET Signaling in Primary and Acquired Resistance to Targeted Therapies in Cancer. Biomedicines 2014, 2, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Papaccio, F.; Della Corte, C.C.; Viscardi, G.; Di Liello, R.; Esposito, G.; Sparano, F.; Ciardiello, F.; Morgillo, F. HGF/MET and the Immune System: Relevance for Cancer Immunotherapy. Int. J. Mol. Sci. 2018, 19, 3595. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Lee, J.; Park, S.S.; Park, J.J.; Lim, H.H.; Kang, W.W.; Park, Y.Y.; Kim, S.T. c-MET Overexpression in Colorectal Cancer: A Poor Prognostic Factor for Survival. Clin. Colorectal Cancer 2018, 17, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Wang, B. Caught in the “Akt”: Cross-talk between EphA2 and EGFR through the Akt-PIKfyve axis maintains cellular sensitivity to EGF. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Gelfo, V.; Rodia, M.M.; Pucci, M.; Dall’Ora, M.; Santi, S.; Solmi, R.; Roth, L.; Lindzen, M.; Bonafè, M.; Bertotti, A.; et al. A module of inflammatory cytokines defines resistance of colorectal cancer to EGFR inhibitors. Oncotarget 2016, 7, 72167–72183. [Google Scholar] [CrossRef]
- Zaiss, D.M.W.; van Loosdregt, J.; Gorlani, A.; Bekker, C.P.J.; Gröne, A.; Sibilia, M.; van Bergen en Henegouwen, P.M.; Roovers, R.C.; Coffer, P.J.; Sijts, A.J. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity 2013, 38, 275–284. [Google Scholar] [CrossRef]
- Srivatsa, S.; Paul, M.M.; Cardone, C.; Holcmann, M.; Amberg, N.; Pathria, P.; Diamanti, M.M.; Linder, M.; Timelthaler, G.; Dienes, H.H.; et al. EGFR in Tumor-Associated Myeloid Cells Promotes Development of Colorectal Cancer in Mice and Associates with Outcomes of Patients. Gastroenterology 2017, 178–190. [Google Scholar] [CrossRef]
- Ledys, F.; Klopfenstein, Q.; Truntzer, C.; Arnould, L.; Vincent, J.; Bengrine, L.; Remark, R.; Boidot, R.; Ladoire, S.; Ghiringhelli, F.; et al. RAS status and neoadjuvant chemotherapy impact CD8+ cells and tumor HLA class I expression in liver metastatic colorectal cancer. J. Immunother. Cancer 2018, 6, 123. [Google Scholar] [CrossRef]
- Veluchamy, J.J.; Spanholtz, J.; Tordoir, M.; Thijssen, V.V.; Heideman, D.D.; Verheul, H.H.; de Gruijl, T.T.; van der Vliet, H.J. Combination of NK Cells and Cetuximab to Enhance Anti-Tumor Responses in RAS Mutant Metastatic Colorectal Cancer. PLoS ONE 2016, 11, e0157830. [Google Scholar] [CrossRef]
- Bae, J.J.; Kho, D.D.; Sun, E.E.; Ko, Y.Y.; Yoon, S.; Lee, K.K.; Ahn, K.K.; Lee, J.J.; Joo, Y.Y.; Chung, I.I.; et al. Elevated Coexpression of KITENIN and the ErbB4 CYT-2 Isoform Promotes the Transition from Colon Adenoma to Carcinoma Following APC loss. Clin. Cancer Res. 2016, 22, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Kang, Y. Complex interplay between tumor microenvironment and cancer therapy. Front. Med. 2018, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Liang, C.; Ren, L.; Zhu, D.; Feng, Q.; Chang, W.; He, G.; Ye, L.; Chen, J.; Lin, Q.; et al. Additional Biomarkers beyond RAS That Impact the Efficacy of Cetuximab plus Chemotherapy in mCRC: A Retrospective Biomarker Analysis. J. Oncol. 2018, 2018, 5072987. [Google Scholar] [CrossRef] [PubMed]
- Kubach, J.; Hubo, M.; Amendt, C.; Stroh, C.; Jonuleit, H. IgG1 anti-epidermal growth factor receptor antibodies induce CD8-dependent antitumor activity. Int. J. Cancer 2014, 136, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.S.; Nicholls, A.A.; Wilding, J.J.; Ntouroupi, T.T.; Mortensen, N.N.; Bodmer, W.F. Direct and immune mediated antibody targeting of ERBB receptors in a colorectal cancer cell-line panel. Proc. Natl. Acad. Sci. USA 2012, 109, 21046–21051. [Google Scholar] [CrossRef] [PubMed]
- Vigano, S.; Alatzoglou, D.; Irving, M.; Ménétrier-Caux, C.; Caux, C.; Romero, P.; Coukos, G. Targeting Adenosine in Cancer Immunotherapy to Enhance T-Cell Function. Front. Immunol. 2019, 10, 925. [Google Scholar] [CrossRef] [PubMed]
- Kohrt, H.H.; Colevas, A.A.; Houot, R. Targeting CD137 enhances the efficacy of cetuximab. J. Clin. Investig. 2014, 124, 2668–2682. [Google Scholar] [CrossRef] [PubMed]
- Xynos, I.I.; Karadima, M.M.; Voutsas, I.F.; Amptoulach, S.; Skopelitis, E.; Kosmas, C.; Gritzapis, A.D.; Tsavaris, N. Chemotherapy ± cetuximab modulates peripheral immune responses in metastatic colorectal cancer. Oncology 2013, 84, 273–283. [Google Scholar] [CrossRef]
- Cardoso, A.A.; Pinto, M.M.; Pinto, A.A.; Oliveira, M.M.; Pinto, M.M.; Gonçalves, R.; Relvas, J.J.; Figueiredo, C.; Seruca, R.; Mantovani, A.; et al. Macrophages stimulate gastric and colorectal cancer invasion through EGFR Y(1086), c-Src, Erk1/2 and Akt phosphorylation and smallGTPase activity. Oncogene 2014, 33, 2123–2133. [Google Scholar] [CrossRef]
- Greening, D.D.; Lee, S.S.; Ji, H.; Simpson, R.R.; Rigopoulos, A.; Murone, C.; Fang, C.; Gong, S.; O’Keefe, G.; Scott, A.M. Molecular profiling of cetuximab and bevacizumab treatment of colorectal tumours reveals perturbations in metabolic and hypoxic response pathways. Oncotarget 2015, 6, 38166–38180. [Google Scholar] [CrossRef]
- Turin, I.; Delfanti, S.; Ferulli, F.; Brugnatelli, S.; Tanzi, M.; Maestri, M.; Cobianchi, L.; Lisini, D.; Luinetti, O.; Paulli, M.; et al. In Vitro Killing of Colorectal Carcinoma Cells by Autologous Activated NK Cells is Boosted by Anti-Epidermal Growth Factor Receptor-induced ADCC Regardless of RAS Mutation Status. J. Immunother. 2018, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Sharma, A.; Lin, Y.; Wu, Y.; He, Q.; Gu, Y.; Xu, Z.Z.; Monteiro, M.; Gu, W. Insluin and epithelial growth factor (EGF) promote programmed death ligand 1(PD-L1) production and transport in colon cancer stem cells. BMC Cancer 2019, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Mardiana, S.; John, L.L.; Henderson, M.M.; Slaney, C.C.; von Scheidt, B.; Giuffrida, L.; Davenport, A.A.; Trapani, J.J.; Neeson, P.P.; Loi, S.; et al. A Multifunctional Role for Adjuvant Anti-4-1BB Therapy in Augmenting Antitumor Response by Chimeric Antigen Receptor T Cells. Cancer Res. 2017, 77, 1296–1309. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, X.; Liu, Q.; Li, C.; Graves-Deal, R.; Cao, Z.; Singh, B.; Franklin, J.J.; Wang, J.; Hu, H.; et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat. Med. 2017, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Bormann, F.; Stinzing, S.; Tierling, S.; Morkel, M.; Markelova, M.M.; Walter, J.; Weichert, W.; Roßner, F.; Kuhn, N.; Perner, J.; et al. Epigenetic regulation of Amphiregulin and Epiregulin in colorectal cancer. Int. J. Cancer 2019, 144, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Kumar, B.; Pan, K.; Dhawan, P.; Singh, A.B. HDAC-4 regulates claudin-2 expression in EGFR-ERK1/2 dependent manner to regulate colonic epithelial cell differentiation. Oncotarget 2017, 8, 87718–87736. [Google Scholar] [CrossRef][Green Version]
- Lieu, C.C.; Corcoran, R.R.; Overman, M.J. Integrating Biomarkers and Targeted Therapy into Colorectal Cancer Management. Am. Soc. Clin. Oncol. Educ. Book 2019, 207–215. [Google Scholar] [CrossRef]
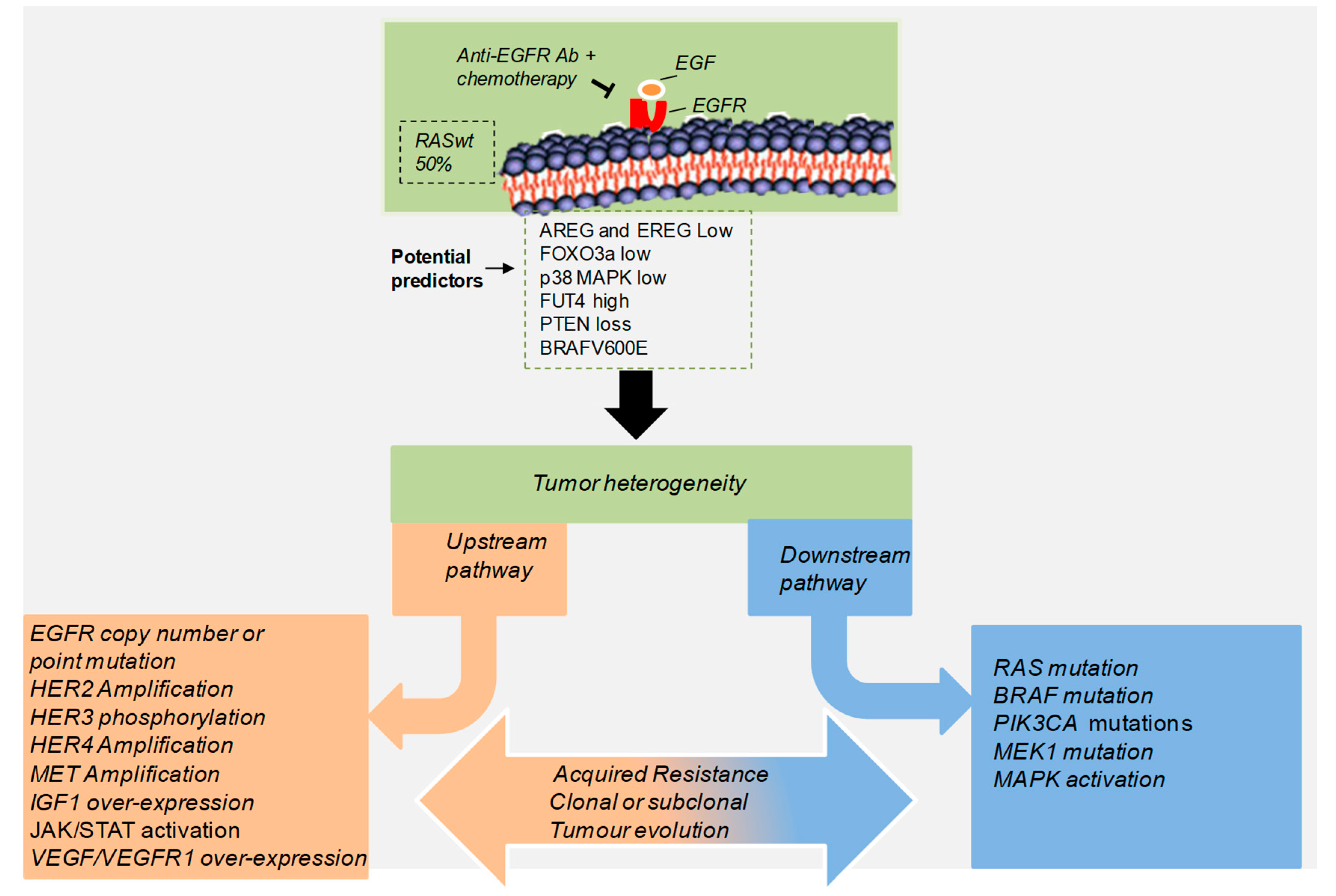
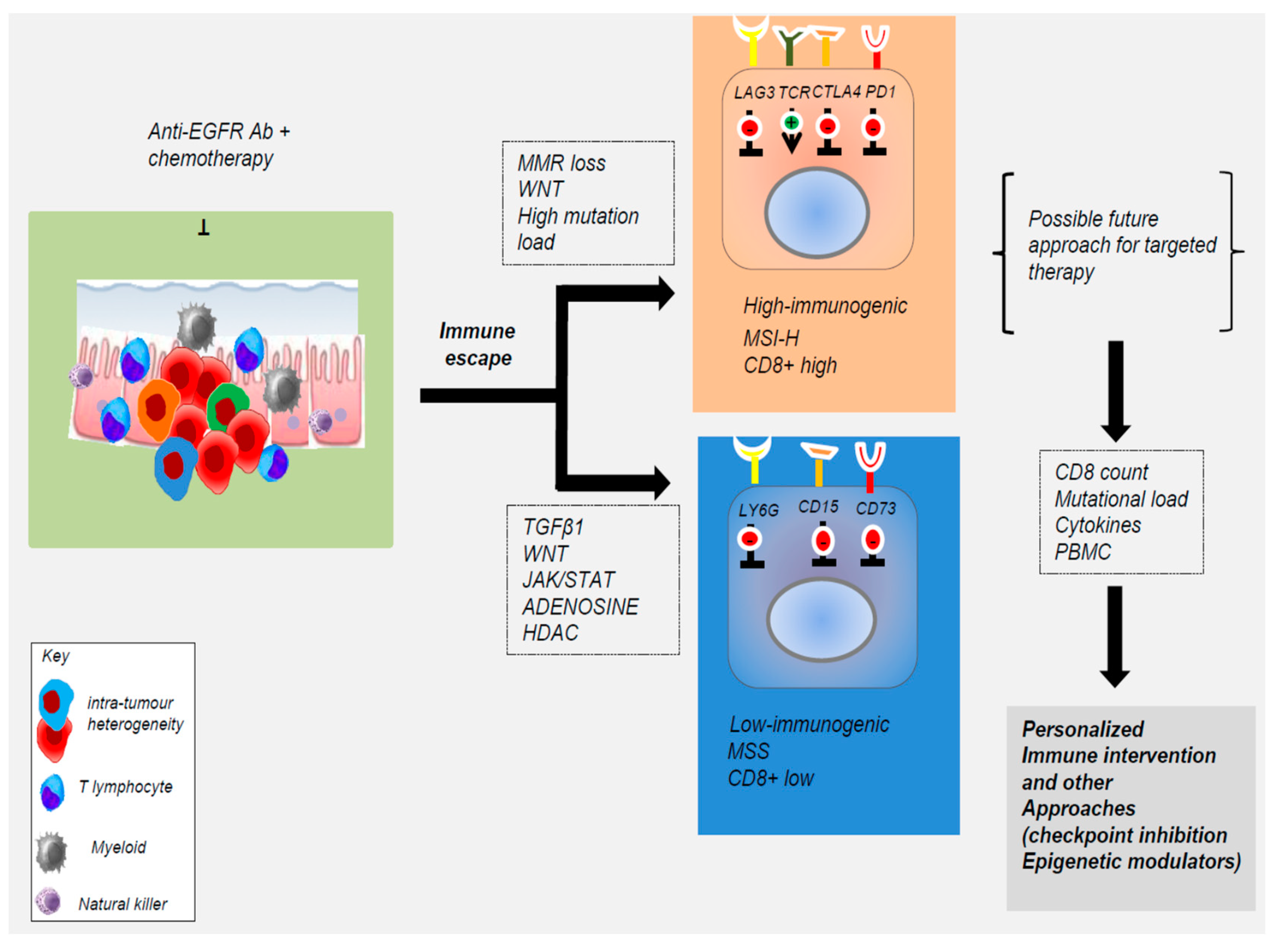
| Gene/Pathway | Genetic Evidence | Population | EGFR Abs | Reference |
|---|---|---|---|---|
| Ras/Raf/MEK | ||||
| KRAS and NRAS | Mutation | CRC | cetuximab andpanitumumb | [6,7] |
| BRAF | Mutation | RAS wild-type CRC | cetuximab and panitumumb | [31,32,33,34] |
| Receptors and ligands from EGFR family | ||||
| Epiregulin (EREG) | Low expression | RAS wild-type and mutant CRC | cetuximab | [35,36,37,38] |
| Amphiregulin (AREG) | Low epression | RAS wild-type and mutant CRC | cetuximab | [35,36,37,38] |
| EGFR | low copy number | cetuximab | [5] | |
| HER2 | Amplification | RAS/RAF wild-type mCRC) | cetuximab and panitumumb | [24] |
| EGFR downstream | ||||
| PIK3CA | Mutation | RAS wild-type CRC | cetuximab | [17,35] |
| PTEN | loss | RAS wild-type CRC | cetuximab | [38,39] |
| JAK/STAT3 | Hyper-activated | CRC | cetuximab | [40] |
| CD15/LY6G6D | High expression | mCRC | cetuximab | [41,42] |
| EGFR independent | ||||
| p38 MAPK | Low expression | RAS wild-type CRC | cetuximab | [43,44] |
| FOXO3a | Low expression | RAS wild-type CRC | cetuximab | [43,45] |
| Gene | Polymorphism | Potential Effect | Patient Population |
|---|---|---|---|
| EGFR | C/C genotype (SNP-994) | less skin toxicity | RAS wild type |
| EGFR | T/T genotype (SNP-216) | Better response | RAS wild type |
| EGFR | G/G genotype rs1050171 * | predictive of response | RAS wild type |
| UBE2M (involved in EGFR Turnover) | C/C genotype rs895374 * | Predicts short PFS | RAS wild type |
| Fc gamma receptor 3a (FCGR3A) | F/F genotype (V158F) ** | longer PFS and OS | RAS wild type |
| Toll like receptor 7 (TLR7) | G/G genotype rs3853839 * | favorable PFS | RAS wild type |
| killer cell immunoglobulin-like receptor (KIR) | KIR2DS4 (full-lenght variant) | Predictive of response | RAS mutated |
| Gene/Pathway | Genetic Evidence | Study | Reference |
|---|---|---|---|
| Ras/Raf/MEK pathway | |||
| KRAS and NRAS | missense mutations | preclinical and clinical | [6,7] |
| BRAF | missense mutations | clinical and meta-analysis | [31,32] |
| MEK1 | missense mutations | preclinical and clinical | [34,35] |
| Receptors and ligands from EGFR family | |||
| EGFR | missense mutations | preclinical and clinical | [33] |
| HER2 | amplification | preclinical and clinical | [36,37] |
| HER3/4 ligand | overexpression | preclinical and clinical | [46] |
| Heregulin | overexpression | clinical | [46] |
| TGF-α | overexpression | preclinical | [17,18] |
| Other tyrosine kinase receptors | |||
| MET | amplification | preclinical and clinical | [5,6,7,8] |
| IGF1R | overexpression | preclinical | [47] |
| VEGF/VEGFR | overexpression | preclinical | [4,14,23] |
| EGFR downstream signaling | |||
| PI3K/Akt pathway | hyperactivation | preclinical and clinical | [43,45] |
| MEK/ERKs pathway | hyperactivation | preclinical and clinical | [34,35] |
| Foxo 3 | upregulation | preclinical and clinical | [43,45] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giordano, G.; Remo, A.; Porras, A.; Pancione, M. Immune Resistance and EGFR Antagonists in Colorectal Cancer. Cancers 2019, 11, 1089. https://doi.org/10.3390/cancers11081089
Giordano G, Remo A, Porras A, Pancione M. Immune Resistance and EGFR Antagonists in Colorectal Cancer. Cancers. 2019; 11(8):1089. https://doi.org/10.3390/cancers11081089
Chicago/Turabian StyleGiordano, Guido, Andrea Remo, Almudena Porras, and Massimo Pancione. 2019. "Immune Resistance and EGFR Antagonists in Colorectal Cancer" Cancers 11, no. 8: 1089. https://doi.org/10.3390/cancers11081089
APA StyleGiordano, G., Remo, A., Porras, A., & Pancione, M. (2019). Immune Resistance and EGFR Antagonists in Colorectal Cancer. Cancers, 11(8), 1089. https://doi.org/10.3390/cancers11081089







