Collagen 1A1 (COL1A1) Is a Reliable Biomarker and Putative Therapeutic Target for Hepatocellular Carcinogenesis and Metastasis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Consent to Participate
2.2. Analyses of Cancer Microarray and RNAseq Dataset
2.3. Cell Lines and Cell Culture
2.4. siRNA-Mediated Knockdown of COL1A1
2.5. Western Blotting Analysis
2.6. Colony Formation Assay
2.7. Wound Healing Migration Assay
2.8. Matrigel Invasion Assay
2.9. Immunohistochemistry
2.10. Sphere Formation Assay
2.11. Statistical Analysis
3. Results
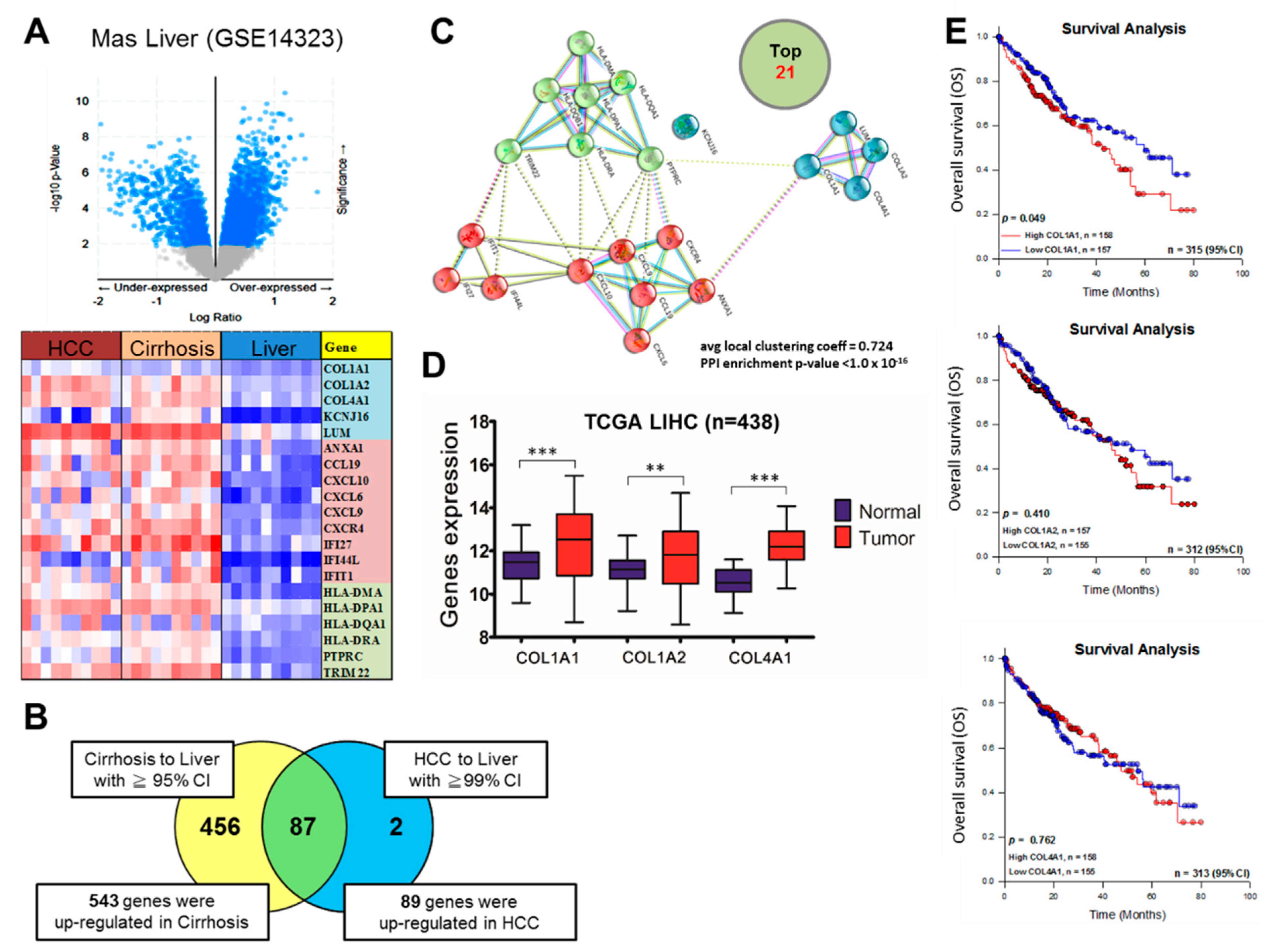
3.1. COL1A1 Is Highly Expressed in HCC and Confers Significant Survival Disadvantage
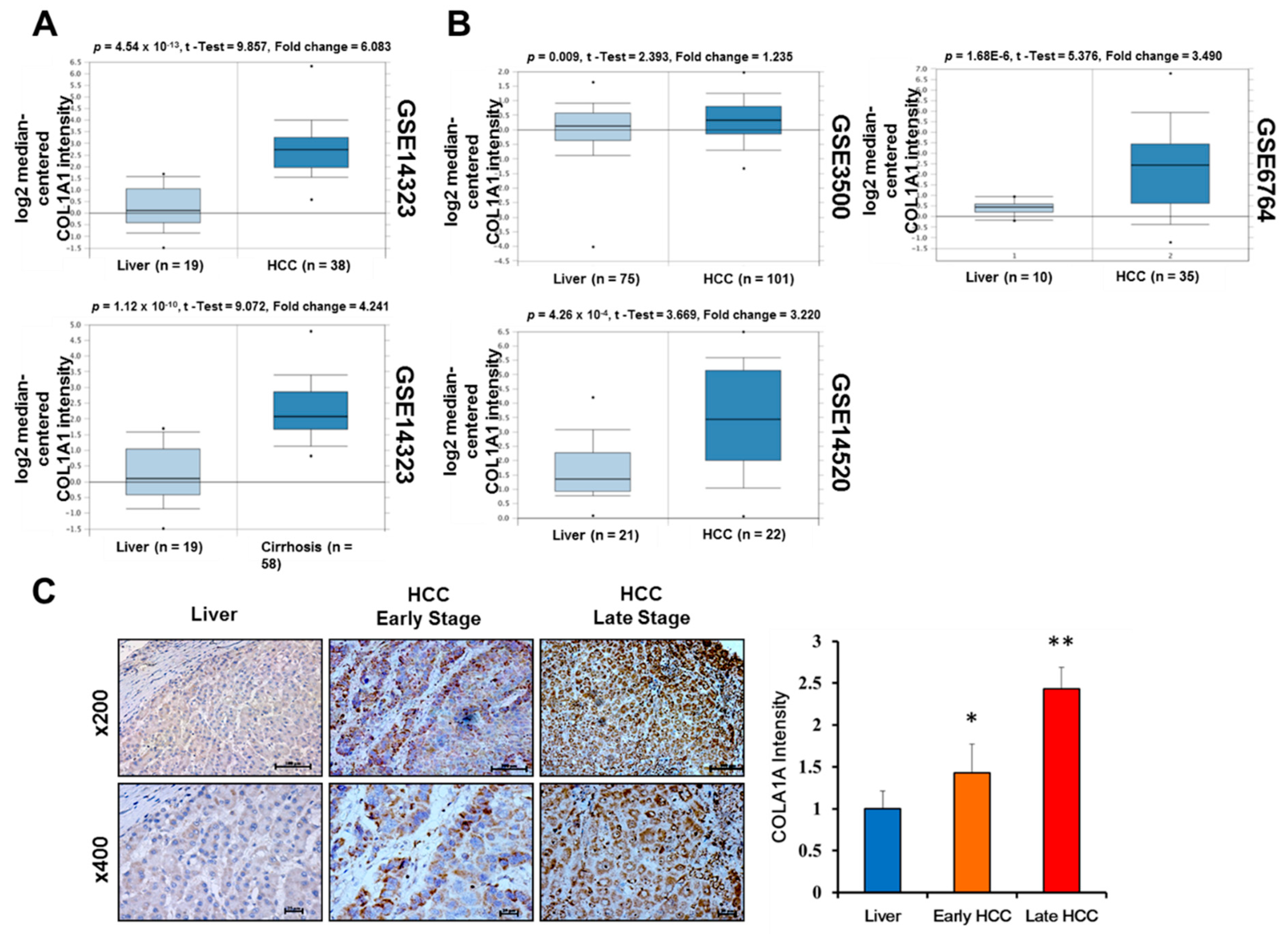
3.2. Upregulated COL1A1 Expression at Both mRNA and Protein Levels Strongly Correlates with Disease Progression
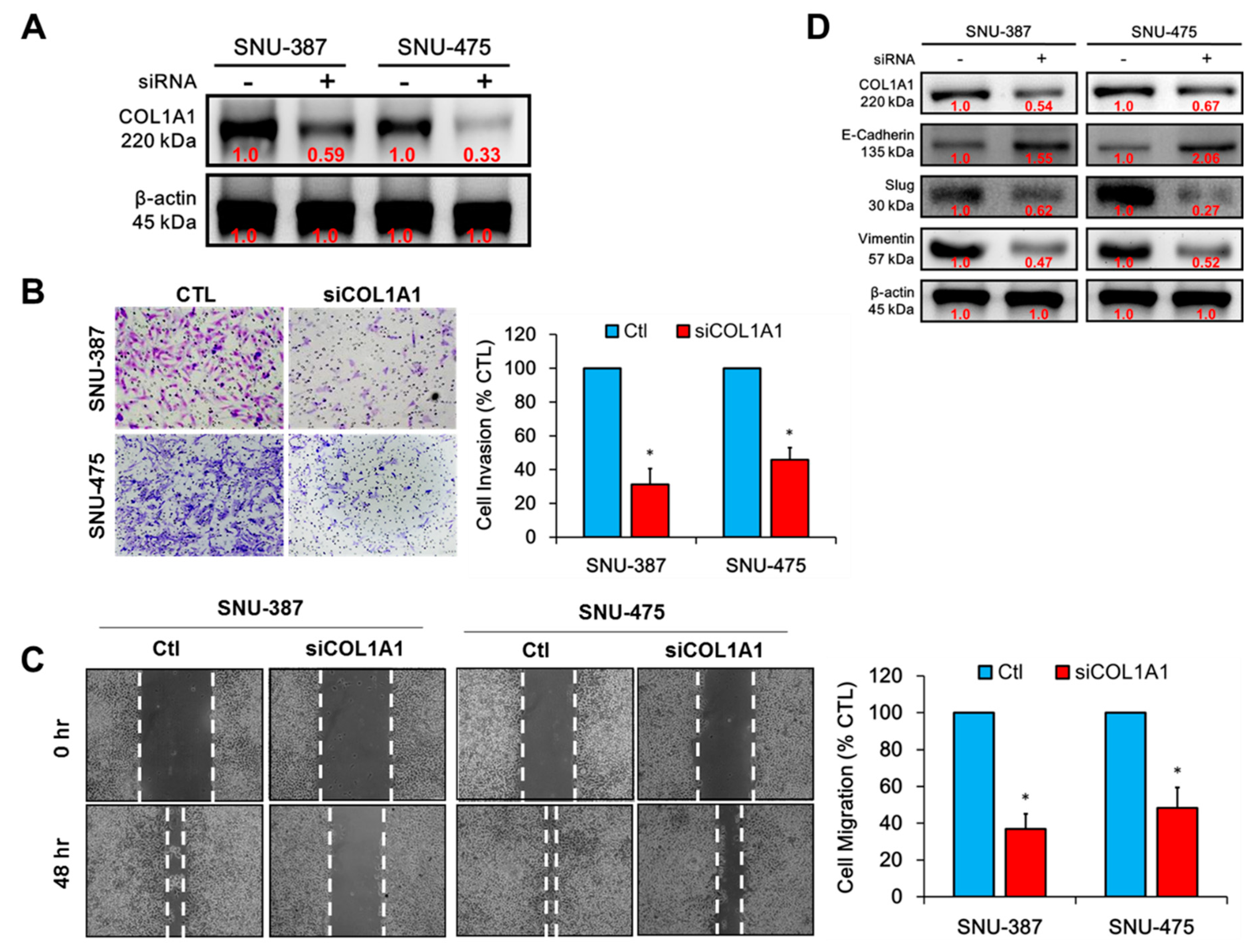
3.3. Knockdown of COL1A1 Suppressed HCC Cell Migration and Invasion through Deregulated Epithelial-to-Mesenchymal Transition (EMT), In Vitro
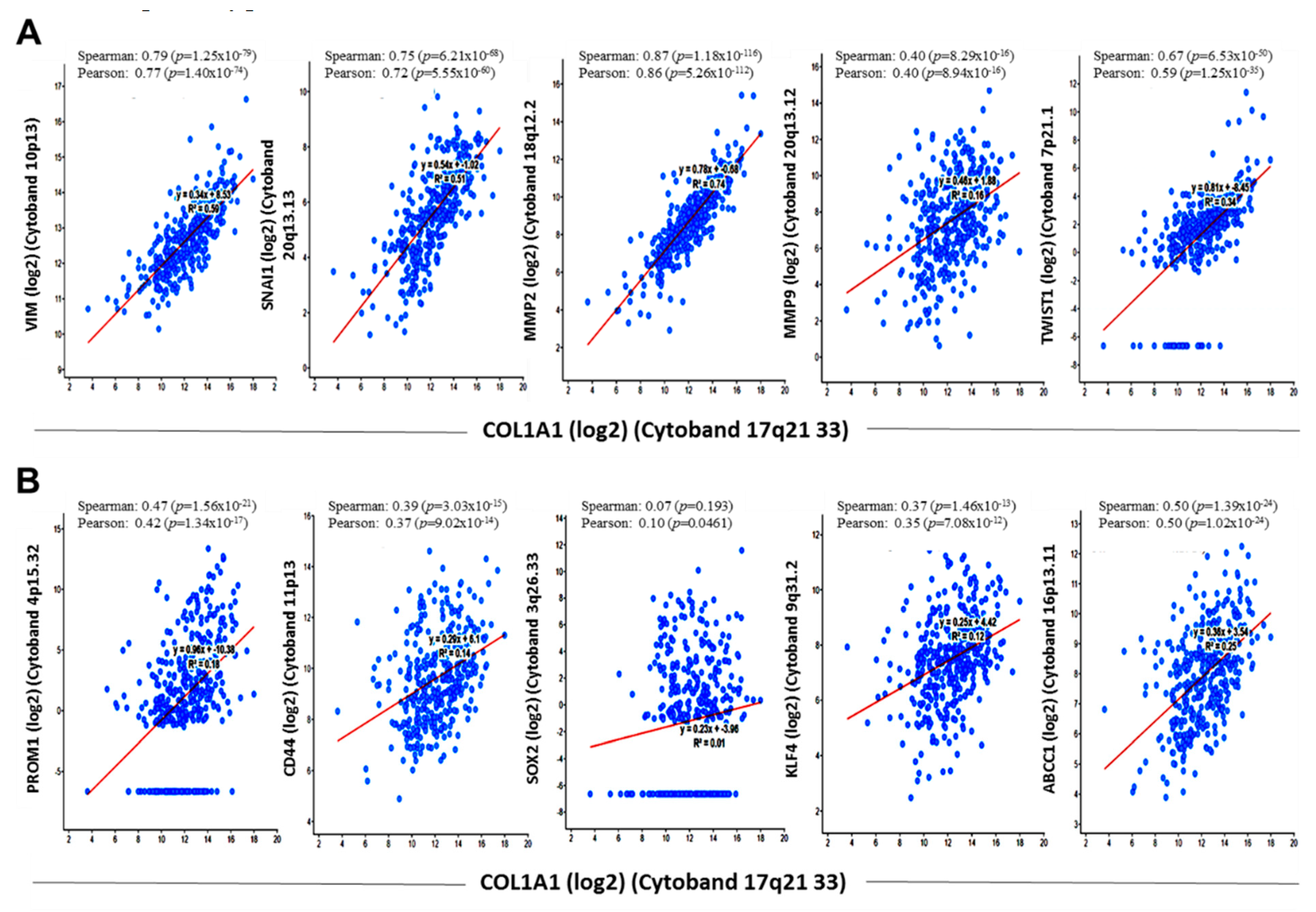
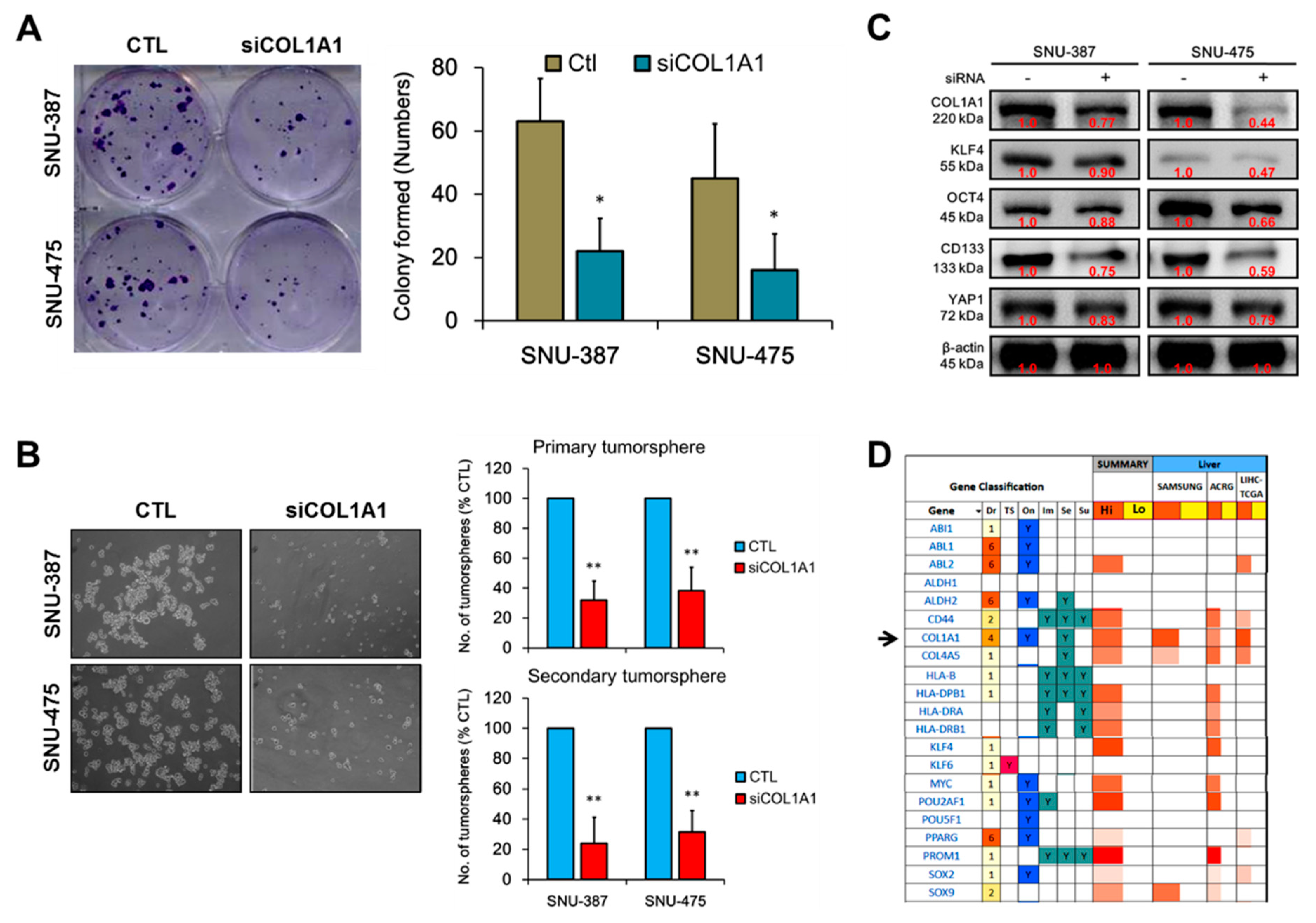
3.4. COL1A1 Is Strongly Associated with and Is a Probable Bridge between the Metastatic and Cancer Stem Cell-Like Phenotypes of HCC
3.5. Loss of COL1A1 Function Significantly Impair Colony and Tumorsphere Formation of HCC Cells In Vitro
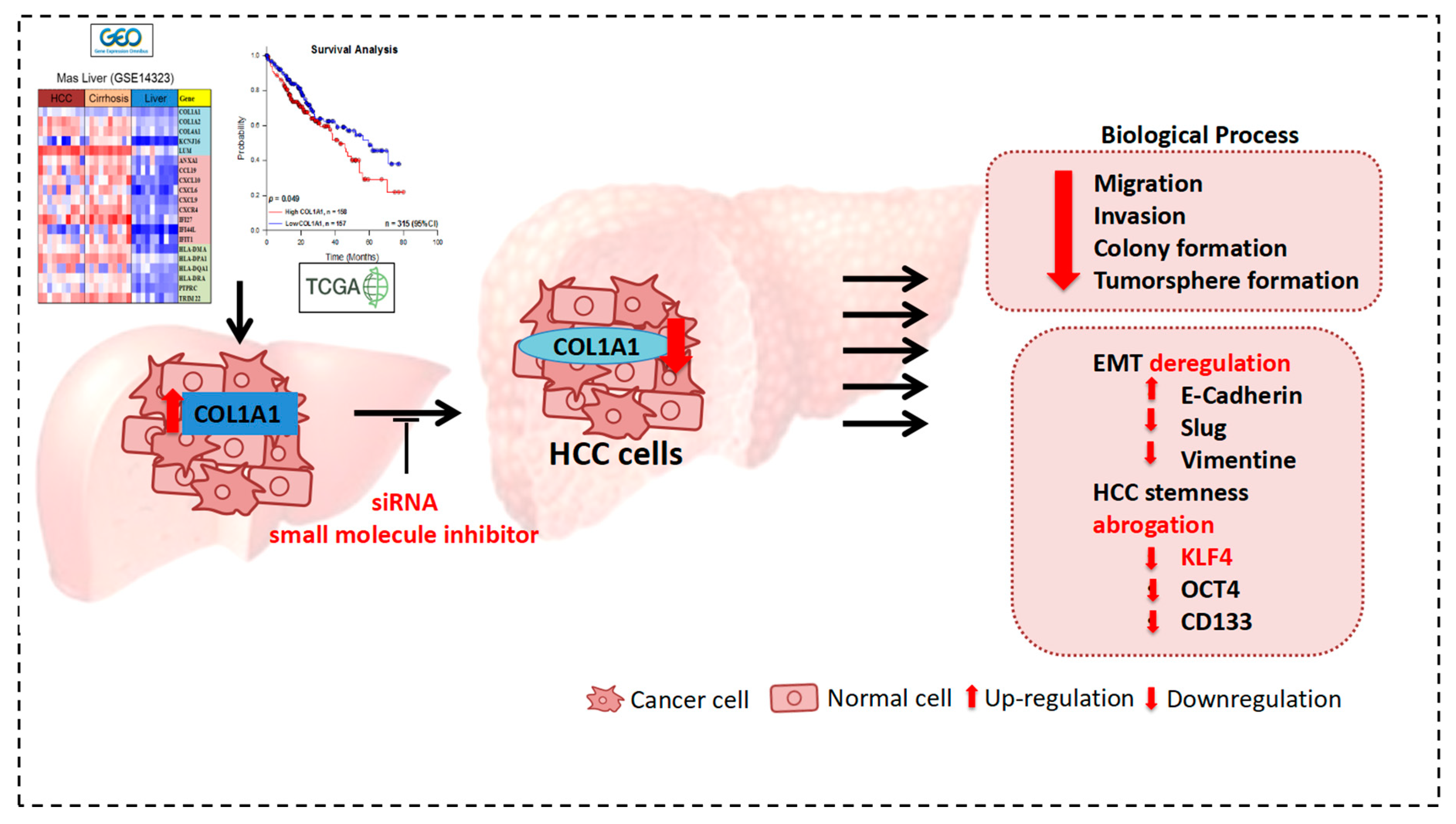
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Golabi, P.; Fazel, S.; Otgonsuren, M.; Sayiner, M.; Locklear, C.T.; Younossi, Z.M. Mortality assessment of patients with hepatocellular carcinoma according to underlying disease and treatment modalities. Medicine 2017, 96, e5904. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.A.; Priolli, D.G.; Tralhao, J.G.; Botelho, M.F. Hepatocellular carcinoma: Epidemiology, biology, diagnosis, and therapies. Rev. Assoc. Med. Bras. 2013, 59, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-J.; Hu, M.-H.; Xu, F.-G.; Xu, H.-J.; She, J.-J.; Xia, H.-P. Understanding the inflammation-cancer transformation in the development of primary liver cancer. Hepatoma Res. 2018, 4, 29. [Google Scholar] [CrossRef]
- Kudo, M. Targeted and immune therapies for hepatocellular carcinoma: Predictions for 2019 and beyond. World J. Gastroenterol. 2019, 25, 789–807. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef]
- Mantovani, A.; Targher, G. Type 2 diabetes mellitus and risk of hepatocellular carcinoma: Spotlight on nonalcoholic fatty liver disease. Ann. Transl. Med. 2017, 5, 270. [Google Scholar] [CrossRef]
- Malhotra, R.; Soin, D.; Grover, P.; Galhotra, S.; Khutan, H.; Kaur, N. Hepatitis B virus and hepatitis C virus co-infection in hemodialysis patients: A retrospective study from a tertiary care hospital of North India. J. Nat. Sci. Biol. Med. 2016, 7, 72–74. [Google Scholar] [CrossRef]
- Birke, B. Hepatitis B and C viruses and hepatocellular carcinoma. Viruses 2010, 2, 1504–1509. [Google Scholar]
- Zamor, P.J.; de Lemos, A.S.; Russo, M.W. Russo Viral hepatitis and hepatocellular carcinoma: Etiology and management. J. Gastrointest. Oncol. 2017, 8, 229–242. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- Nissen, N.I.; Karsdal, M.; Willumsen, N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J. Exp. Clin. Cancer Res. 2019, 38, 115. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, K.; Karsdal, M.A. Chapter 1-Type I Collagen. Biochemistry of Collagens, Laminins and Elastin; Karsdal, M.A., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 1–11. ISBN 9780128098479. [Google Scholar] [CrossRef]
- Gelse, K.; Poschl, E.; Aigner, T. Collagens--structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef] [PubMed]
- Exposito, J.-Y.; Valcourt, U.; Cluzel, C.; Lethias, C. The fibrillar collagen family. Int. J. Mol. Sci. 2010, 11, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, S.; Ross, K.N.; Lander, E.S.; Golub, T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003, 33, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Ibanez de Caceres, I.; Dulaimi, E.; Hoffman, A.M.; Al-Saleem, T.; Uzzo, R.G.; Cairns, P. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res. 2006, 66, 5021–5028. [Google Scholar] [CrossRef] [PubMed]
- Bonazzi, V.F.; Nancarrow, D.J.; Stark, M.S.; Moser, R.J.; Boyle, G.M.; Aoude, L.G.; Schmidt, C.; Hayward, N.K. Cross-platform array screening identifies COL1A2, THBS1, TNFRSF10D and UCHL1 as genes frequently silenced by methylation in melanoma. PLoS ONE 2011, 6, e26121. [Google Scholar] [CrossRef]
- Hayashi, M.; Nomoto, S.; Hishida, M.; Inokawa, Y.; Kanda, M.; Okamura, Y.; Nishikawa, Y.; Tanaka, C.; Kobayashi, D.; Yamada, S.; et al. Identification of the collagen type 1 alpha 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma. BMC Cancer 2014, 14, 108. [Google Scholar] [CrossRef] [PubMed]
- Zang, S.; Guo, R.; Xing, R.; Zhang, L.; Li, W.; Zhao, M.; Fang, J.; Hu, F.; Kang, B.; Ren, Y.; et al. Identification of differentially-expressed genes in intestinal gastric cancer by microarray analysis. Genom. Proteom. Bioinform. 2014, 12, 276–283. [Google Scholar] [CrossRef]
- Koilan, S.; Hamilton, D.; Baburyan, N.; Padala, M.K.; Weber, K.T.; Guntaka, R.V. Prevention of liver fibrosis by triple helix-forming oligodeoxyribonucleotides targeted to the promoter region of type I collagen gene. Oligonucleotides 2010, 20, 231–237. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef]
- Jayachandran, A.; Dhungel, B.; Steel, J.C. Epithelial-to-mesenchymal plasticity of cancer stem cells: Therapeutic targets in hepatocellular carcinoma. J. Hematol. Oncol. 2016, 9, 74. [Google Scholar] [CrossRef] [PubMed]
- Flores-Téllez, T.N.; Villa-Treviño, S.; Piña-Vázquez, C. Road to stemness in hepatocellular carcinoma. World J. Gastroenterol. 2017, 23, 6750–6776. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, W.; Jia, Y.; Yu, Q.; Grau, G.E.; Peng, L.; Ran, Y.; Yang, Z.; Deng, H.; Lou, J. Single-cell clones of liver cancer stem cells have the potential of differentiating into different types of tumor cells. Cell Death Dis. 2013, 4, e857. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Gao, Y.; Li, Z.; Jia, J.; Fang, M.; Wang, M.; Feng, H.; Chen, Q.; Guan, W.; Wang, Z.; et al. A Nomogram Predicting Extrahepatic Metastases for Patients with Adjuvant Transarterial Chemoembolization after Hepatectomy. J. Cancer 2018, 9, 4223–4233. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; He, X.; Andayani, D.; Yang, L.; Ye, J.; Li, Y.; Chen, Y.; Li, L. Pattern of distant extrahepatic metastases in primary liver cancer: A SEER based study. J. Cancer 2017, 8, 2312–2318. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pineda, I.; Sandoval, J.A.; Davidoff, A.M. Hepatic metastatic disease in pediatric and adolescent solid tumors. World J. Hepatol. 2015, 7, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, T.; Ikoma, H.; Okamoto, K.; Kokuba, Y.; Sonoyama, T.; Otsuji, E. Clinicopathologic features and risk factors for extrahepatic recurrences of hepatocellular carcinoma after curative resection. World J. Surg. 2012, 36, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; He, Y.; Wei, Z.; Lu, Y.; Du, F.; Ou, G.; Wang, N.; Luo, X.G.; Ma, W.; Zhang, T.C.; et al. MRTF-A mediates the activation of COL1A1 expression stimulated by multiple signaling pathways in human breast cancer cells. BioMed. Pharmacother. 2018, 104, 718–728. [Google Scholar] [CrossRef]
- Li, J.; Ding, Y.; Li, A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J. Surg. 2016, 14, 297. [Google Scholar] [CrossRef]
- Klutstein, M.; Nejman, D.; Greenfield, R.; Cedar, H. DNA Methylation in Cancer and Aging. Cancer Res. 2016, 76, 3446–3450. [Google Scholar] [CrossRef]
- Warton, K.; Mahon, K.L.; Samimi, G. Methylated circulating tumor DNA in blood: Power of cancer prognosis and response. Endocr. Relat. Cancer 2016, 23, R157–R171. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shen, J.X.; Wu, H.T.; Li, X.L.; Wen, X.F.; Du, C.W.; Zhang, G.J. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov. Med. 2018, 25, 211–223. [Google Scholar] [PubMed]
- Wang, Q.; Yu, J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem. Cell Biol. 2018, 96, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Yasuda, H.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Goel, A.; Kusunoki, M. Increased expression of Slug and Vimentin as novel predictive biomarkers for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis 2013, 34, 2548–2557. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C. Cancer stem cells: Role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine 2016, 95, S20–S25. [Google Scholar] [CrossRef]
| Clinicopathological Variable | No. | High COL1A1 | Low COL1A1 | X2 | p-Value |
|---|---|---|---|---|---|
| Age, years | |||||
| ≤60 | 33 | 19 | 14 | 0.014 | 0.905 |
| >60 | 39 | 23 | 16 | ||
| Gender | |||||
| Male | 45 | 21 | 24 | 0.533 | 0.465 |
| Female | 27 | 15 | 12 | ||
| HBsAg | |||||
| negative (−) | 11 | 4 | 7 | 2.578 | 0.108 |
| positive (+) | 61 | 38 | 23 | ||
| α-fetoprotein (AFP) | |||||
| <400 ng/dL | 25 | 12 | 8 | 6.254 | 0.017 * |
| ≥400 ng/dL | 47 | 30 | 22 | ||
| Cirrhosis | |||||
| Absent | 23 | 9 | 14 | 4.374 | 0.036 * |
| Present | 49 | 32 | 17 | ||
| Tumor size (mm) | |||||
| <20 | 19 | 8 | 11 | 1.891 | 0.169 |
| ≥20 | 53 | 32 | 21 | ||
| Lymph node metastasis | |||||
| negative (−) | 31 | 13 | 18 | 5.002 | 0.025 * |
| positive (+) | 41 | 28 | 13 | ||
| TNM tumor stage | |||||
| I+II | 29 | 10 | 19 | 4.677 | 0.031 * |
| III+IV | 43 | 26 | 17 |
| Clinicopathological Variables | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Age (≤60 vs >60) | 0.401 | 0.215–1.003 | 0.037 * | 0.649 | 0.776–1.019 | 0.041 * |
| Gender (male vs female) | 1.049 | 0.630–1.315 | 0.764 | |||
| HBsAg (positive vs negative) | 0.414 | 0.206–0.983 | 0.071 | 0.705 | 0.374–1.028 | 0.673 |
| Cirrhosis # | 1.013 | 0.881–1.045 | <0.001 * | 1.009 | 0.796–1.042 | <0.001 * |
| Tumor size (mm) # | 1.430 | 1.030–1.719 | <0.001 * | 1.602 | 1.343–2.152 | 0.021 * |
| α-fetoprotein (AFP) # | 2.316 | 1.683–3.473 | <0.001 * | 2.016 | 1.476–2.861 | <0.001 * |
| Lymph node metastasis # | 0.875 | 0.652–0.967 | 0.017 * | 0.784 | 0.569–1.029 | 0.849 |
| TNM tumor stage # | 1.543 | 1.241–2.068 | <0.001 * | 1.314 | 1.050–1.826 | 0.010 * |
| COL1A1 expression (high vs. low) | 2.008 | 0.644–5.316 | <0.001 * | 2.252 | 0.971–6.473 | <0.001 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, H.-P.; Chang, H.-L.; Bamodu, O.A.; Yadav, V.K.; Huang, T.-Y.; Wu, A.T.H.; Yeh, C.-T.; Tsai, S.-H.; Lee, W.-H. Collagen 1A1 (COL1A1) Is a Reliable Biomarker and Putative Therapeutic Target for Hepatocellular Carcinogenesis and Metastasis. Cancers 2019, 11, 786. https://doi.org/10.3390/cancers11060786
Ma H-P, Chang H-L, Bamodu OA, Yadav VK, Huang T-Y, Wu ATH, Yeh C-T, Tsai S-H, Lee W-H. Collagen 1A1 (COL1A1) Is a Reliable Biomarker and Putative Therapeutic Target for Hepatocellular Carcinogenesis and Metastasis. Cancers. 2019; 11(6):786. https://doi.org/10.3390/cancers11060786
Chicago/Turabian StyleMa, Hon-Ping, Hang-Lung Chang, Oluwaseun Adebayo Bamodu, Vijesh Kumar Yadav, Ting-Yi Huang, Alexander T. H. Wu, Chi-Tai Yeh, Shin-Han Tsai, and Wei-Hwa Lee. 2019. "Collagen 1A1 (COL1A1) Is a Reliable Biomarker and Putative Therapeutic Target for Hepatocellular Carcinogenesis and Metastasis" Cancers 11, no. 6: 786. https://doi.org/10.3390/cancers11060786
APA StyleMa, H.-P., Chang, H.-L., Bamodu, O. A., Yadav, V. K., Huang, T.-Y., Wu, A. T. H., Yeh, C.-T., Tsai, S.-H., & Lee, W.-H. (2019). Collagen 1A1 (COL1A1) Is a Reliable Biomarker and Putative Therapeutic Target for Hepatocellular Carcinogenesis and Metastasis. Cancers, 11(6), 786. https://doi.org/10.3390/cancers11060786








