Amplification of the EGFR and CCND1 Are Coordinated and Play Important Roles in the Progression of Oral Squamous Cell Carcinomas
Abstract
1. Introduction
2. Results
2.1. Identification of Common, Distinct CNAs in 72 OSCCs
2.2. Confirmation of EGFR and CCND1, Including in the CNA Region of 7p11.2 and 11q13.3
2.3. Clinical Implications of CNA’s in EGFR and CCND1
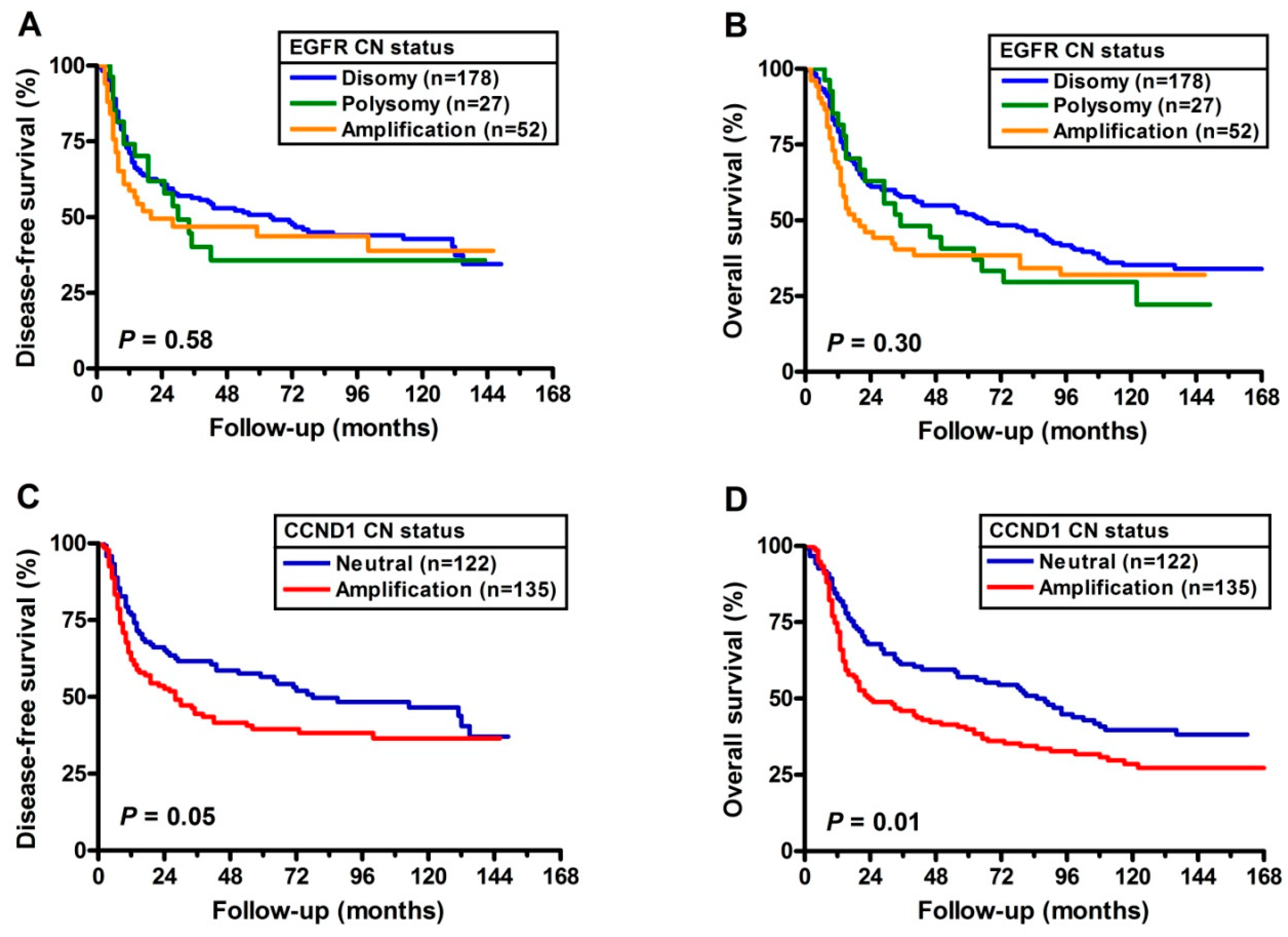
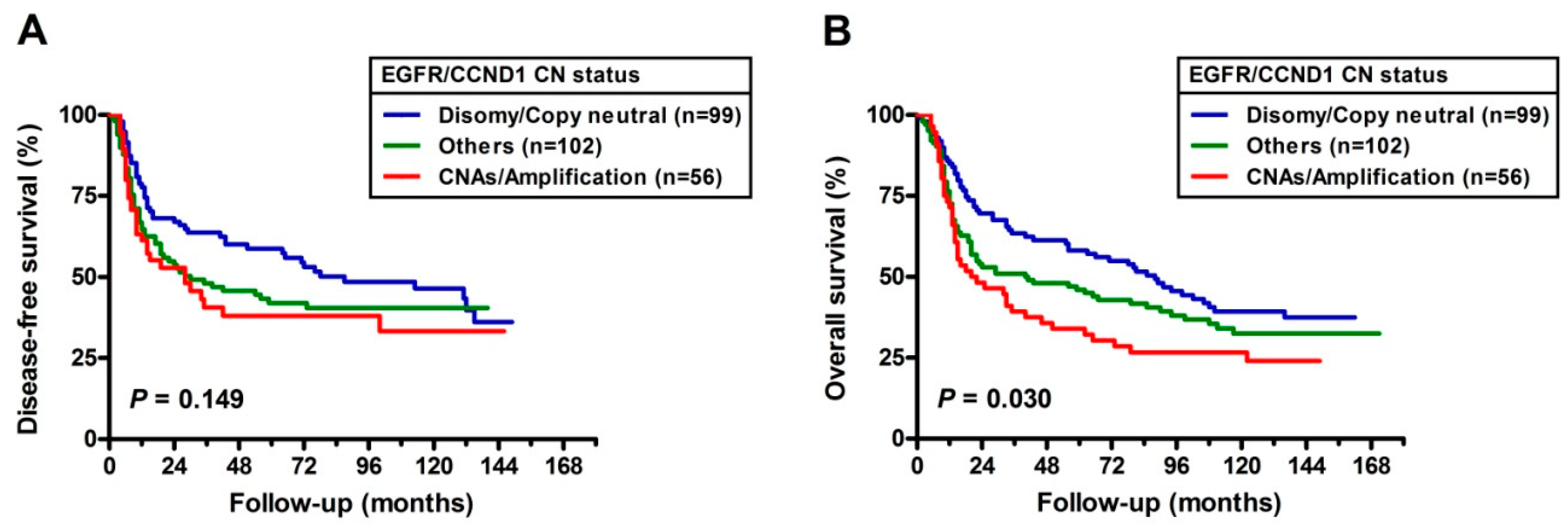
2.4. Prognostic Implications of CNA’s with EGFR and CCND1
3. Discussion
4. Materials and Methods
4.1. Patients, Specimens and Clinical Diagnosis
4.2. High-Resolution SNP Array and Data Analysis
4.3. Fluorescence In Situ Hybridization (FISH) for EGFR
4.4. TaqMan CN Assays via Quantitative Real-Time Polymerase Chain Reaction (qPCR) for CCND1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Frohling, S.; Dohner, H. Chromosomal Abnormalities in Cancer. N. Engl. J. Med. 2008, 359, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S. Global Epidemiology of Oral and Oropharyngeal Cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.C.; Huang, Y.L.; Lee, C.H.; Chen, M.J.; Lin, L.M.; Tsai, C.C. Betel Quid Chewing, Cigarette Smoking and Alcohol Consumption Related to Oral Cancer in Taiwan. J. Oral Pathol. Med. 1995, 24, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.J.; Jiang, R.S.; Wu, S.H.; Chen, F.J.; Liu, S.A. Smoking, Alcohol, and Betel Quid and Oral Cancer: A Prospective Cohort Study. J. Oncol. 2011, 2011, 525976. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Sandhu, J.; Kaur, B.; Juren, T.; Steward, W.P.; Segerback, D.; Farmer, P.B. Evaluation of the DNA Damaging Potential of Cannabis Cigarette Smoke by the Determination of Acetaldehyde Derived N2-Ethyl-2’-Deoxyguanosine Adducts. Chem. Res. Toxicol. 2009, 22, 1181–1188. [Google Scholar] [CrossRef]
- Lai, K.C.; Lee, T.C. Genetic Damage in Cultured Human Keratinocytes Stressed by Long-Term Exposure to Areca Nut Extracts. Mutat. Res. 2006, 599, 66–75. [Google Scholar] [CrossRef]
- Silverman, S., Jr. Demographics and Occurrence of Oral and Pharyngeal Cancers. The Outcomes, the Trends, the Challenge. J. Am. Dent. Assoc. 2001, 132, 7S–11S. [Google Scholar] [CrossRef]
- Dahabreh, I.J.; Linardou, H.; Kosmidis, P.; Bafaloukos, D.; Murray, S. Egfr Gene Copy Number as a Predictive Biomarker for Patients Receiving Tyrosine Kinase Inhibitor Treatment: A Systematic Review and Meta-Analysis in Non-Small-Cell Lung Cancer. Ann. Oncol. 2011, 22, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Kwei, K.A.; Kung, Y.; Salari, K.; Holcomb, I.N.; Pollack, J.R. Genomic Instability in Breast Cancer: Pathogenesis and Clinical Implications. Mol. Oncol. 2010, 4, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Young, B.D.; Lu, Y.J. The Application of Single Nucleotide Polymorphism Microarrays in Cancer Research. Curr. Genom. 2007, 8, 219–228. [Google Scholar]
- Ambatipudi, S.; Gerstung, M.; Gowda, R.; Pai, P.; Borges, A.M.; Schaffer, A.A.; Beerenwinkel, N.; Mahimkar, M.B. Genomic Profiling of Advanced-Stage Oral Cancers Reveals Chromosome 11q Alterations as Markers of Poor Clinical Outcome. PLoS ONE 2011, 6, e17250. [Google Scholar] [CrossRef]
- Chiang, D.Y.; Villanueva, A.; Hoshida, Y.; Peix, J.; Newell, P.; Minguez, B.; LeBlanc, A.C.; Donovan, D.J.; Thung, S.N.; Sole, M.; et al. Focal Gains of Vegfa and Molecular Classification of Hepatocellular Carcinoma. Cancer Res. 2008, 68, 6779–6788. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, S.D.; Ferlito, A.; Takes, R.P.; Brakenhoff, R.H.; Valentin, M.D.; Woolgar, J.A.; Bradford, C.R.; Rodrigo, J.P.; Rinaldo, A.; Hier, M.P.; et al. Advances and Applications of Oral Cancer Basic Research. Oral Oncol. 2011, 47, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Leary, R.J.; Lin, J.C.; Cummins, J.; Boca, S.; Wood, L.D.; Parsons, D.W.; Jones, S.; Sjoblom, T.; Park, B.H.; Parsons, R.; et al. Integrated Analysis of Homozygous Deletions, Focal Amplifications, and Sequence Alterations in Breast and Colorectal Cancers. Proc. Natl. Acad. Sci. USA 2008, 105, 16224–16229. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Getz, G.; Nghiemphu, L.; Barretina, J.; Hsueh, T.; Linhart, D.; Vivanco, I.; Lee, J.C.; Huang, J.H.; Alexander, S.; et al. Assessing the Significance of Chromosomal Aberrations in Cancer: Methodology and Application to Glioma. Proc. Natl. Acad. Sci. USA 2007, 104, 20007–20012. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.H.; Liao, C.T.; Peng, S.C.; Chen, Y.J.; Cheng, A.J.; Juang, J.L.; Tsai, C.Y.; Chen, T.C.; Chuang, Y.J.; Tang, C.Y.; et al. A Novel Molecular Signature Identified by Systems Genetics Approach Predicts Prognosis in Oral Squamous Cell Carcinoma. PLoS ONE 2011, 6, e23452. [Google Scholar] [CrossRef]
- Sheu, J.J.; Hua, C.H.; Wan, L.; Lin, Y.J.; Lai, M.T.; Tseng, H.C.; Jinawath, N.; Tsai, M.H.; Chang, N.W.; Lin, C.F.; et al. Functional Genomic Analysis Identified Epidermal Growth Factor Receptor Activation as the Most Common Genetic Event in Oral Squamous Cell Carcinoma. Cancer Res. 2009, 69, 2568–2576. [Google Scholar] [CrossRef] [PubMed]
- Kalish, L.H.; Kwong, R.A.; Cole, I.E.; Gallagher, R.M.; Sutherland, R.L.; Musgrove, E.A. Deregulated Cyclin D1 Expression Is Associated with Decreased Efficacy of the Selective Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Gefitinib in Head and Neck Squamous Cell Carcinoma Cell Lines. Clin. Cancer Res. 2004, 10, 7764–7774. [Google Scholar] [CrossRef]
- Namazie, A.; Alavi, S.; Olopade, O.I.; Pauletti, G.; Aghamohammadi, N.; Aghamohammadi, M.; Gornbein, J.A.; Calcaterra, T.C.; Slamon, D.J.; Wang, M.B.; et al. Cyclin D1 Amplification and P16(Mts1/Cdk4i) Deletion Correlate with Poor Prognosis in Head and Neck Tumors. Laryngoscope 2002, 112, 472–481. [Google Scholar] [CrossRef]
- Sugahara, K.; Michikawa, Y.; Ishikawa, K.; Shoji, Y.; Iwakawa, M.; Shibahara, T.; Imai, T. Combination Effects of Distinct Cores in 11q13 Amplification Region on Cervical Lymph Node Metastasis of Oral Squamous Cell Carcinoma. Int. J. Oncol. 2011, 39, 761–769. [Google Scholar]
- Liu, H.S.; Lu, H.H.; Lui, M.T.; Yu, E.H.; Shen, W.; Chen, Y.P.; Chang, K.W.; Tu, H.F. Detection of Copy Number Amplification of Cyclin D1 (Ccnd1) and Cortactin (Cttn) in Oral Carcinoma and Oral Brushed Samples from Areca Chewers. Oral Oncol. 2009, 45, 1032–1036. [Google Scholar] [CrossRef] [PubMed]
- Pavelic, K.; Krizanac, S.; Cacev, T.; Hadzija, M.P.; Radosevic, S.; Crnic, I.; Levanat, S.; Kapitanovic, S. Aberration of Fhit Gene Is Associated with Increased Tumor Proliferation and Decreased Apoptosis-Clinical Evidence in Lung and Head and Neck Carcinomas. Mol. Med. 2001, 7, 442–453. [Google Scholar] [CrossRef]
- Nakaya, K.; Yamagata, H.D.; Arita, N.; Nakashiro, K.I.; Nose, M.; Miki, T.; Hamakawa, H. Identification of Homozygous Deletions of Tumor Suppressor Gene Fat in Oral Cancer Using Cgh-Array. Oncogene 2007, 26, 5300–5308. [Google Scholar] [CrossRef] [PubMed]
- Seethala, R.R.; Gooding, W.E.; Handler, P.N.; Collins, B.; Zhang, Q.; Siegfried, J.M.; Grandis, J.R. Immunohistochemical Analysis of Phosphotyrosine Signal Transducer and Activator of Transcription 3 and Epidermal Growth Factor Receptor Autocrine Signaling Pathways in Head and Neck Cancers and Metastatic Lymph Nodes. Clin. Cancer Res. 2008, 14, 1303–1309. [Google Scholar] [CrossRef]
- Lukas, J.; Aagaard, L.; Strauss, M.; Bartek, J. Oncogenic Aberrations of P16ink4/Cdkn2 and Cyclin D1 Cooperate to Deregulate G1 Control. Cancer Res. 1995, 55, 4818–4823. [Google Scholar] [PubMed]
- Chen, Y.J.; Lin, S.C.; Kao, T.; Chang, C.S.; Hong, P.S.; Shieh, T.M.; Chang, K.W. Genome-Wide Profiling of Oral Squamous Cell Carcinoma. J. Pathol. 2004, 204, 326–332. [Google Scholar] [CrossRef]
- Nakata, Y.; Uzawa, N.; Takahashi, K.; Sumino, J.; Michikawa, C.; Sato, H.; Sonoda, I.; Ohyama, Y.; Okada, N.; Amagasa, T. Egfr Gene Copy Number Alteration Is a Better Prognostic Indicator Than Protein Overexpression in Oral Tongue Squamous Cell Carcinomas. Eur. J. Cancer 2011, 47, 2364–2372. [Google Scholar] [CrossRef]
- Szabo, B.; Nelhubel, G.A.; Karpati, A.; Kenessey, I.; Jori, B.; Szekely, C.; Petak, I.; Lotz, G.; Hegedus, Z.; Hegedus, B.; et al. Clinical Significance of Genetic Alterations and Expression of Epidermal Growth Factor Receptor (Egfr) in Head and Neck Squamous Cell Carcinomas. Oral Oncol. 2011, 47, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging Functions of the Egfr in Cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Ely, K.; McGavran, L.; Varella-Garcia, M.; Parker, J.; Parker, N.; Jarrett, C.; Carter, J.; Murphy, B.A.; Netterville, J.; et al. Increased Epidermal Growth Factor Receptor Gene Copy Number Is Associated with Poor Prognosis in Head and Neck Squamous Cell Carcinomas. J. Clin. Oncol. 2006, 24, 4170–4176. [Google Scholar] [CrossRef]
- Huang, S.F.; Cheng, S.D.; Chien, H.T.; Liao, C.T.; Chen, I.H.; Wang, H.M.; Chuang, W.Y.; Wang, C.Y.; Hsieh, L.L. Relationship between Epidermal Growth Factor Receptor Gene Copy Number and Protein Expression in Oral Cavity Squamous Cell Carcinoma. Oral Oncol. 2012, 48, 67–72. [Google Scholar] [CrossRef]
- Ulanovski, D.; Stern, Y.; Roizman, P.; Shpitzer, T.; Popovtzer, A.; Feinmesser, R. Expression of Egfr and Cerb-B2 as Prognostic Factors in Cancer of the Tongue. Oral Oncol. 2004, 40, 532–537. [Google Scholar] [CrossRef]
- Myo, K.; Uzawa, N.; Miyamoto, R.; Sonoda, I.; Yuki, Y.; Amagasa, T. Cyclin D1 Gene Numerical Aberration Is a Predictive Marker for Occult Cervical Lymph Node Metastasis in Tnm Stage I and Ii Squamous Cell Carcinoma of the Oral Cavity. Cancer 2005, 104, 2709–2716. [Google Scholar] [CrossRef]
- Saunders, W.S.; Shuster, M.; Huang, X.; Gharaibeh, B.; Enyenihi, A.H.; Petersen, I.; Gollin, S.M. Chromosomal Instability and Cytoskeletal Defects in Oral Cancer Cells. Proc. Natl. Acad. Sci. USA 2000, 97, 303–308. [Google Scholar] [CrossRef]
- Wight, A.J.; Ogden, G.R. Possible Mechanisms by Which Alcohol May Influence the Development of Oral Cancer—A Review. Oral Oncol. 1998, 34, 441–447. [Google Scholar] [CrossRef]
- Urashima, M.; Hama, T.; Suda, T.; Suzuki, Y.; Ikegami, M.; Sakanashi, C.; Akutsu, T.; Amagaya, S.; Horiuchi, K.; Imai, Y.; et al. Distinct Effects of Alcohol Consumption and Smoking on Genetic Alterations in Head and Neck Carcinoma. PLoS ONE 2013, 8, e80828. [Google Scholar] [CrossRef]
- Michikawa, C.; Uzawa, N.; Sato, H.; Ohyama, Y.; Okada, N.; Amagasa, T. Epidermal Growth Factor Receptor Gene Copy Number Aberration at the Primary Tumour Is Significantly Associated with Extracapsular Spread in Oral Cancer. Br. J. Cancer 2011, 104, 850–855. [Google Scholar] [CrossRef][Green Version]
- Huang, S.F.; Cheng, S.D.; Chuang, W.Y.; Chen, I.H.; Liao, C.T.; Wang, H.M.; Hsieh, L.L. Cyclin D1 Overexpression and Poor Clinical Outcomes in Taiwanese Oral Cavity Squamous Cell Carcinoma. World J. Surg. Oncol. 2012, 10, 40. [Google Scholar] [CrossRef]
- Lee, H.-H.; Wang, Y.-N.; Hung, M.-C. Non-Canonical Signaling Mode of the Epidermal Growth Factor Receptor Family. Am. J. Cancer Res. 2015, 5, 2944–2958. [Google Scholar]
- Takahashi, K.-I.; Uzawa, N.; Myo, K.; Okada, N.; Amagasa, T. Simultaneous Assessment of Cyclin D1 and Epidermal Growth Factor Receptor Gene Copy Number for Prognostic Factor in Oral Squamous Cell Carcinomas. Oral Sci. Int. 2009, 6, 8–20. [Google Scholar] [CrossRef]
- Hsieh, L.L.; Chien, H.T.; Chen, I.H.; Liao, C.T.; Wang, H.M.; Jung, S.M.; Wang, P.F.; Chang, J.T.; Chen, M.C.; Cheng, A.J. The Xrcc1 399gln Polymorphism and the Frequency of P53 Mutations in Taiwanese Oral Squamous Cell Carcinomas. Cancer Epidemiol. Biomark. Prev. 2003, 12, 439–443. [Google Scholar]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. Gistic2.0 Facilitates Sensitive and Confident Localization of the Targets of Focal Somatic Copy-Number Alteration in Human Cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The Miqe Guidelines: Minimum Information for Publication of Quantitative Real-Time Pcr Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | 500 K Array (n = 26) | SNP 6.0 Array (n = 46) | 257 OSCC Cases for Further Analysis |
|---|---|---|---|
| Age (years) | |||
| Mean ± SD | 48.15 ± 11.39 | 50.41 ± 10.79 | 49.95 ± 11.23 |
| Range | 27–71 | 30–74 | 26–78 |
| Site of primary tumor [N (%)] | |||
| Tongue | 11 (42) | 26 (57) | 90 (35) |
| Bucca | 15 (58) | 20 (43) | 94 (37) |
| Others | - | - | 73 (28) |
| Clinical stage [N (%)] | |||
| I/II | 2 (8) | 0 (0) | 65 (25) |
| III/IV | 24 (92) | 46 (100) | 192 (75) |
| Primary tumor status [N (%)] | |||
| T1/T2 | 9 (35) | 18 (39) | 121 (47) |
| T3/T4 | 17 (65) | 28 (61) | 136 (53) |
| Lymph node metastasis [N (%)] | |||
| No | 8 (31) | 10 (22) | 132 (51) |
| Yes | 18 (69) | 36 (78) | 125 (49) |
| Extra-capsular spread [N (%)] a | |||
| Yes | 14 (78) | 26 (72) | 76 (61) |
| No | 4 (22) | 10 (28) | 48 (39) |
| Tumor differentiation [N (%)] | |||
| Well differentiated | 9 (35) | 25 (35) | 98 (38) |
| Moderately/Poorly differentiated | 17 (65) | 47 (65) | 159 (62) |
| AQ chewing [N (%)] | |||
| Yes | 15 (58) | 36 (78) | 223 (87) |
| No | 11 (42) | 10 (22) | 34 (13) |
| Cigarette smoking [N (%)] | |||
| Yes | 11 (42) | 35 (76) | 220 (86) |
| No | 15 (58) | 11 (24) | 37 (14) |
| Alcohol drinking [N (%)] | |||
| Yes | 14 (54) | 20 (43) | 138 (54) |
| No | 12 (46) | 26 (57) | 119 (46) |
| Cytogenetic Loci | GISTIC Wide Peak Region (Mb a) | Size (Mb) | Total Frequency (%, n = 72) | Cases with CNAs by 500-K Platform (n = 26) | Cases with CNAs by SNP 6.0 Platform (n = 46) | Candidate Genes |
|---|---|---|---|---|---|---|
| Gains | ||||||
| 7p11.2 b | 54.62–55.86 | 1.24 | 51 | 14 | 23 | SEC61G, EGFR |
| 11q13.3 b | 68.96–70.32 | 1.36 | 51 | 14 | 23 | MYEOV1, CCND1, ORAOV1, FADD, CTTN |
| Losses | ||||||
| 2q23.3–q24.2 b | 124.78–243.20 | 118.24 | 10 | 3 | 4 | ING5, LRP1B, DAPL1 |
| 3p14.2–p12.1 | 25.64–93.78 | 68.14 | 42 | 12 | 18 | FHIT, MLH1, BAP1, SETD2, PBRM1 |
| 4q35.2 b | 187.48–188.24 | 0.76 | 33 | 8 | 16 | FAT1 |
| 7q33–q34 b | 97.61–159.14 | 63.91 | 18 | 6 | 7 | MIR335, ING3 |
| 9p21.3 b | 21.56–22.00 | 0.44 | 26 | 4 | 15 | CDKN2A |
| 11q22.3–q24.3 b | 72.39–135.01 | 62.62 | 38 | 10 | 17 | ATM, MRE11A, CHEK1, H2AFX |
| 16q23.1 | 75.46–79.63 | 4.17 | 11 | 2 | 6 | WWOX |
| 18q11.2–q22.3 b | 18.69–78.08 | 59.39 | 40 | 9 | 20 | DCC |
| 21q21.1 | 10.19–29.10 | 18.91 | 33 | 7 | 17 | CHODL |
| 21q22.3 b | 44.30–48.13 | 3.83 | 33 | 6 | 18 | NDUFV3 |
| EGFR | CCND1 | EGFR/CCND1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Di | Poly | Amp | P Value | N | Amp | P Value | Di/Neu | Others | Amp/CNA | P Value | |
| Age (years) | |||||||||||
| <50 | 90 (64) | 17 (12) | 34 (24) | 0.11 | 63 (45) | 78 (55) | 0.32 | 51 (36) | 51 (36) | 39 (28) | 0.04 |
| ≥50 | 88 (76) | 10 (9) | 18 (16) | 0.04a | 59 (51) | 57 (49) | 48 (41) | 51 (44) | 17 (15) | 0.06 a | |
| Subsites | |||||||||||
| Tongue | 63 (70) | 9 (10) | 18 (20) | 0.97 | 41 (46) | 49 (54) | 0.90 | 38 (42) | 28 (31) | 24 (27) | 0.24 |
| Bucca | 63 (67) | 10 (11) | 21 (22) | 0.82 a | 46 (49) | 48 (51) | 35 (37) | 39 (41) | 20 (21) | 0.77 a | |
| Others | 52 (71) | 8 (11) | 13 (18) | 35 (48) | 38 (52) | 26 (36) | 35 (48) | 12 (16) | |||
| Clinical stage | |||||||||||
| I/II | 52 (80) | 8 (12) | 5 (8) | 0.01 | 38 (58) | 27 (44) | 0.04 | 33 (51) | 24 (37) | 8 (12) | 0.03 |
| III/IV | 126 (66) | 19 (10) | 47 (24) | <0.01 a | 84 (44) | 108 (56) | 66 (34) | 78 (41) | 48 (25) | <0.01a | |
| Primary tumor status | |||||||||||
| T1/T2 | 90 (74) | 12 (10) | 19 (16) | 0.20 | 60 (50) | 61 (50) | 0.52 | 51 (42) | 48 (40) | 22 (18) | 0.34 |
| T3/T4 | 88 (65) | 15 (11) | 33 (24) | 0.07 a | 62 (46) | 74 (54) | 48 (35) | 54 (40) | 34 (25) | 0.15 a | |
| LNM c | |||||||||||
| No | 103 (78) | 14 (11) | 15 (11) | <0.01 | 80 (61) | 52 (39) | <0.01 | 67 (51) | 49 (37) | 16 (12) | <0.01 |
| Yes | 75 (60) | 13 (10) | 37 (30) | <0.01 a | 42 (34) | 83 (66) | 32 (26) | 53 (42) | 40 (32) | <0.01a | |
| ECSd | |||||||||||
| Yes | 45 (59) | 9 (12) | 22 (29) | 0.82 | 32 (42) | 44 (58) | 0.02 | 23 (30) | 31 (41) | 22 (29) | 0.33 |
| No | 29 (60) | 4 (8) | 15 (31) | 0.95 a | 10 (21) | 38 (79) | 9 (19) | 21 (44) | 18 (38) | 0.15 a | |
| Tumor differentiation | |||||||||||
| Well | 77 (79) | 9 (9) | 12 (12) | 0.03 | 63 (64) | 36 (36) | <0.01 | 57 (58) | 27 (27) | 15 (15) | <0.01 |
| Mod/Poor | 101 (64) | 18 (11) | 40 (25) | <0.01 a | 59 (38) | 98 (62) | 42 (27) | 74 (47) | 41 (26) | <0.01 a | |
| AQ e chewing | |||||||||||
| Yes | 156 (70) | 21 (9) | 46 (21) | 0.34 | 110 (49) | 113 (51) | 0.13 | 89 (40) | 88 (39) | 46 (21) | 0.39 |
| No | 22 (65) | 6 (18) | 6 (18) | 0.88 a | 12 (35) | 22 (65) | 10 (29) | 14 (41) | 10 (29) | 0.17 a | |
| Cigarette smoking | |||||||||||
| Yes | 153 (70) | 23 (10) | 44 (20) | 0.97 | 107 (49) | 113 (51) | 0.36 | 86 (39) | 88 (40) | 46 (21) | 0.70 |
| No | 25 (68) | 4 (11) | 8 (22) | 0.80 a | 15 (41) | 22 (59) | 13 (35) | 14 (38) | 10 (27) | 0.46 a | |
| Alcohol drinking | |||||||||||
| Yes | 96 (70) | 14 (10) | 28 (20) | 0.98 | 57 (41) | 81 (59) | 0.03 | 46 (33) | 61 (44) | 31 (22) | 0.16 |
| No | 82 (69) | 13 (11) | 24 (20) | 0.96a | 65 (55) | 54 (45) | 53 (45) | 41 (34) | 25 (21) | 0.18a | |
| Disease-Free Survival | Overall Survival | |||
|---|---|---|---|---|
| Characteristic | HR (95% CI) | P Value | HR (95% CI) | P Value |
| Age (yrs) | ||||
| <50 | 1 | 1 | ||
| > = 50 | 0.901 (0.641–1.265) | 0.55 | 1.198 (0.883–1.626) | 0.25 |
| Tumor differentiation | ||||
| Well differentiated | 1 | 1 | ||
| Moderately/poorly differentiated | 1.282 (0.903–1.821) | 0.17 | 1.258 (0.915–1.729) | 0.16 |
| Primary tumor status | ||||
| T1/T2 | 1 | 1 | ||
| T3/T4 | 1.104 (0.788–1.546) | 0.57 | 1.487 (1.092–2.025) | 0.01 |
| Lymph node status | ||||
| LNM a−, ECS b− | 1 | 1 | ||
| LNM+, ECS− | 1.256 (0.777–2.030) | 0.35 | 1.643 (1.083–2.493) | 0.02 |
| LNM+, ECS+ | 2.354 (1.621–3.420) | <0.01 | 2.360 (1.673–3.329) | <0.01 |
| EGFR copy number | ||||
| Disomy | 1 | 1 | ||
| Polysomy | 1.160 (0.682–1.973) | 0.58 | 1.358 (0.819–2.253) | 0.24 |
| Amplification | 1.231 (0.802–1.891) | 0.34 | 1.264 (0.827–1.931) | 0.28 |
| CCND1 copy number | ||||
| Neutral | 1 | 1 | ||
| Amplification | 1.402 (0.998–1.971) | 0.05 | 1.485 (1.091–2.021) | 0.01 |
| EGFR and CCND1 CN status | ||||
| EGFR disomy/CCND1 copy neutral | 1 | 1 | ||
| Others | 1.332 (0.905–1.961) | 0.15 | 1.368 (0.926–2.022) | 0.12 |
| EGFR CNAs c and CCND1 amplification | 1.501 (0.960–2.347) | 0.08 | 1.750 (1.126–2.719) | 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chien, H.-T.; Cheng, S.-D.; Liao, C.-T.; Wang, H.-M.; Huang, S.-F. Amplification of the EGFR and CCND1 Are Coordinated and Play Important Roles in the Progression of Oral Squamous Cell Carcinomas. Cancers 2019, 11, 760. https://doi.org/10.3390/cancers11060760
Chien H-T, Cheng S-D, Liao C-T, Wang H-M, Huang S-F. Amplification of the EGFR and CCND1 Are Coordinated and Play Important Roles in the Progression of Oral Squamous Cell Carcinomas. Cancers. 2019; 11(6):760. https://doi.org/10.3390/cancers11060760
Chicago/Turabian StyleChien, Huei-Tzu, Sou-De Cheng, Chun-Ta Liao, Hung-Ming Wang, and Shiang-Fu Huang. 2019. "Amplification of the EGFR and CCND1 Are Coordinated and Play Important Roles in the Progression of Oral Squamous Cell Carcinomas" Cancers 11, no. 6: 760. https://doi.org/10.3390/cancers11060760
APA StyleChien, H.-T., Cheng, S.-D., Liao, C.-T., Wang, H.-M., & Huang, S.-F. (2019). Amplification of the EGFR and CCND1 Are Coordinated and Play Important Roles in the Progression of Oral Squamous Cell Carcinomas. Cancers, 11(6), 760. https://doi.org/10.3390/cancers11060760






