Mechanical Durotactic Environment Enhances Specific Glioblastoma Cell Responses
Abstract
1. Introduction
2. Results
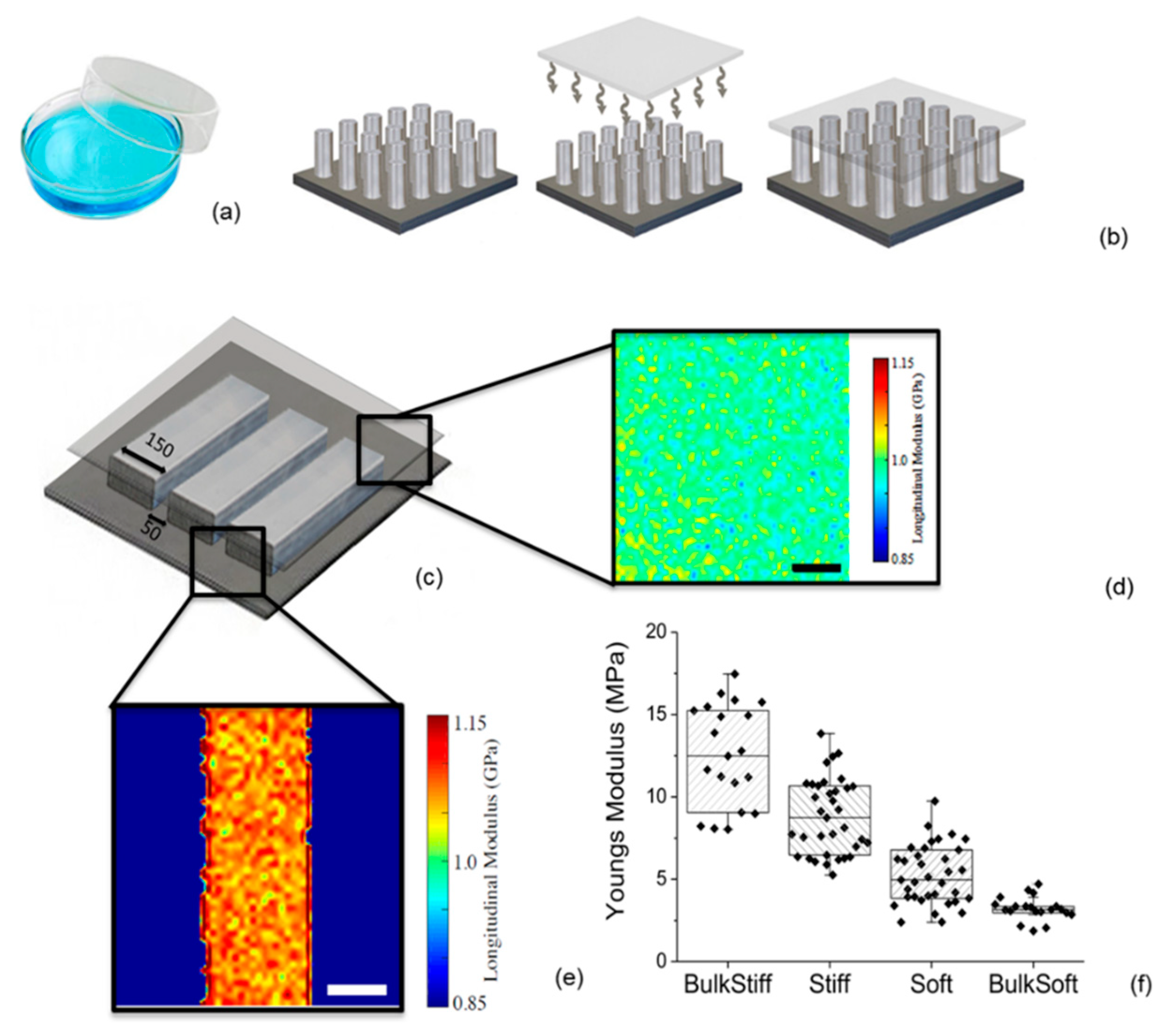
2.1. Substrate Characterization
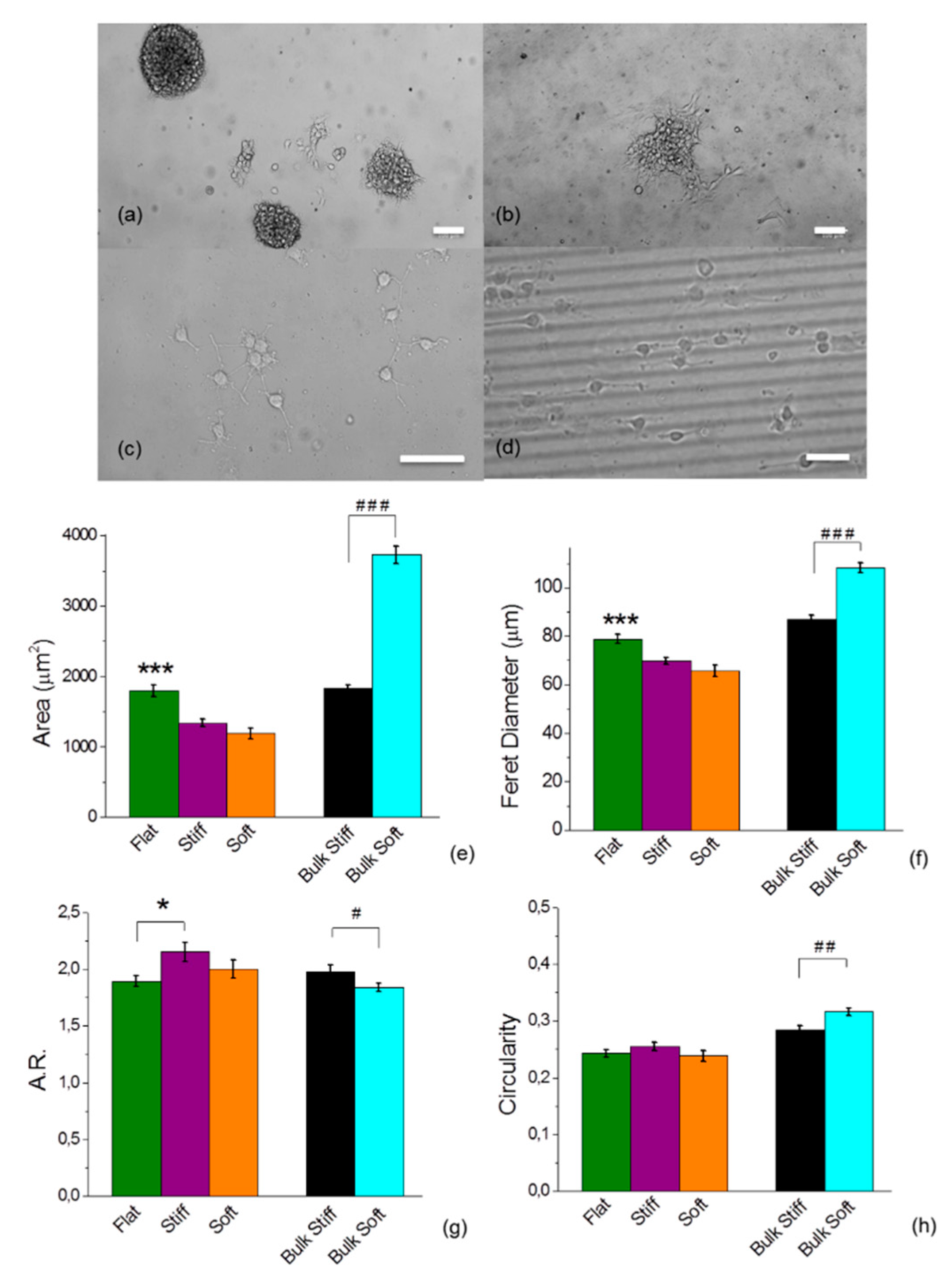
2.2. Glioblastoma Cell Morphology was Sensitive to Different Discrete Mechanical Stiffness in Particular to the Mechanically Uniform Durotactic Substrates
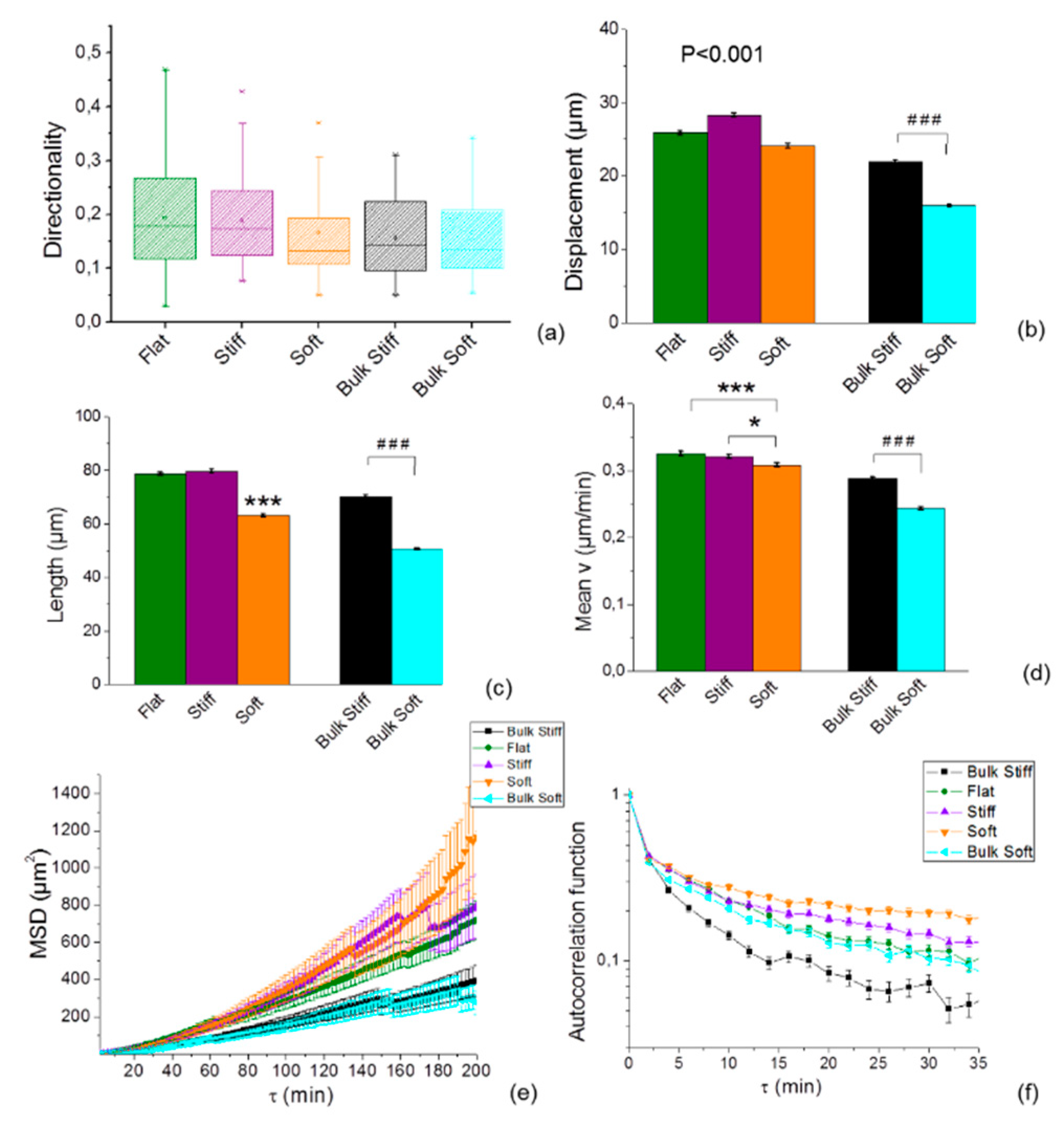
2.3. Cell Speed is Differentially Regulated by Substrate Stiffness
2.4. Cell Viability is not Influenced by Substrate Stiffness
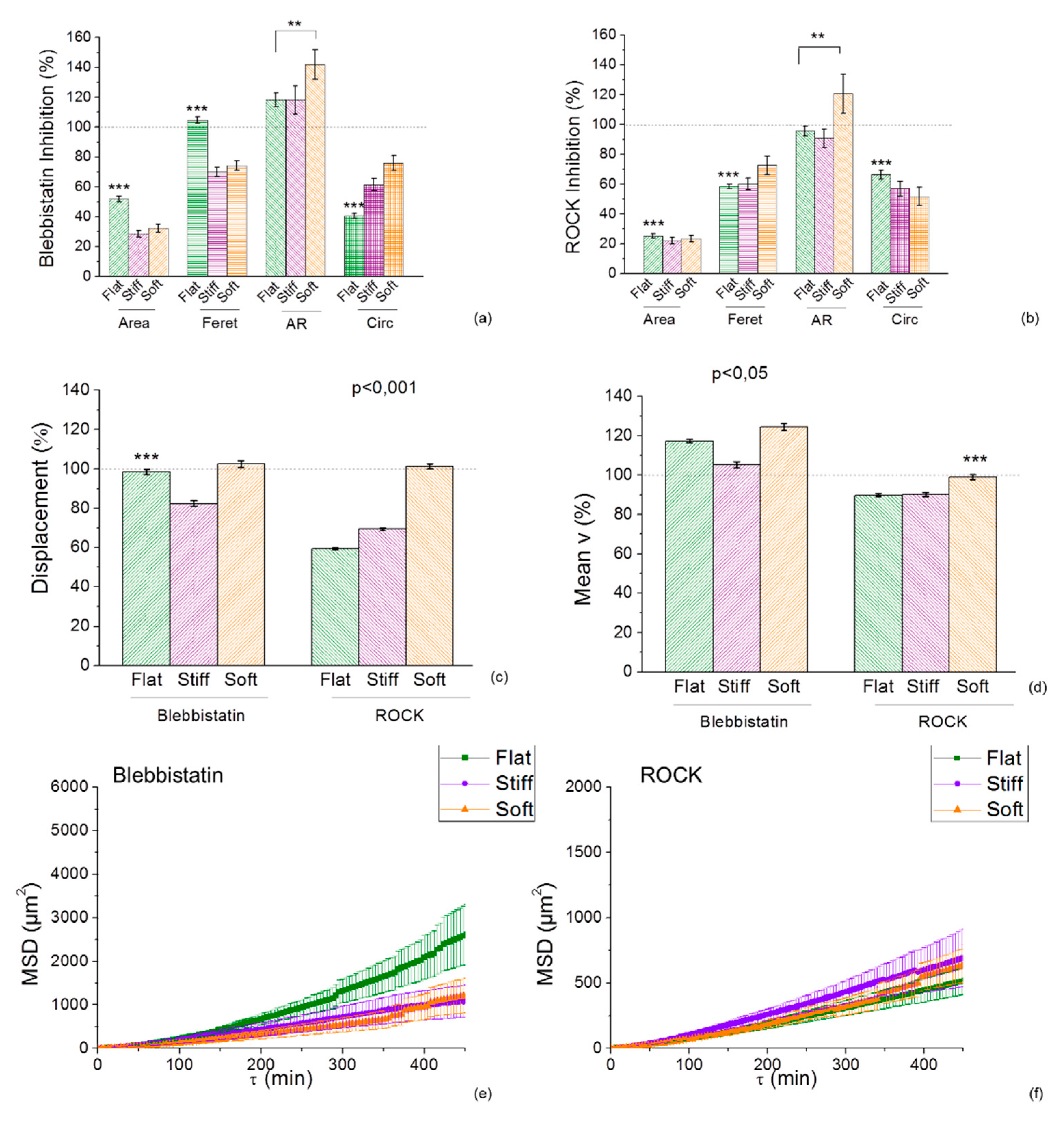
2.5. Inhibition of Myosin and RhoA is Morfologically Affected by the Durotactic Substrate Stiffness but not Dynamically
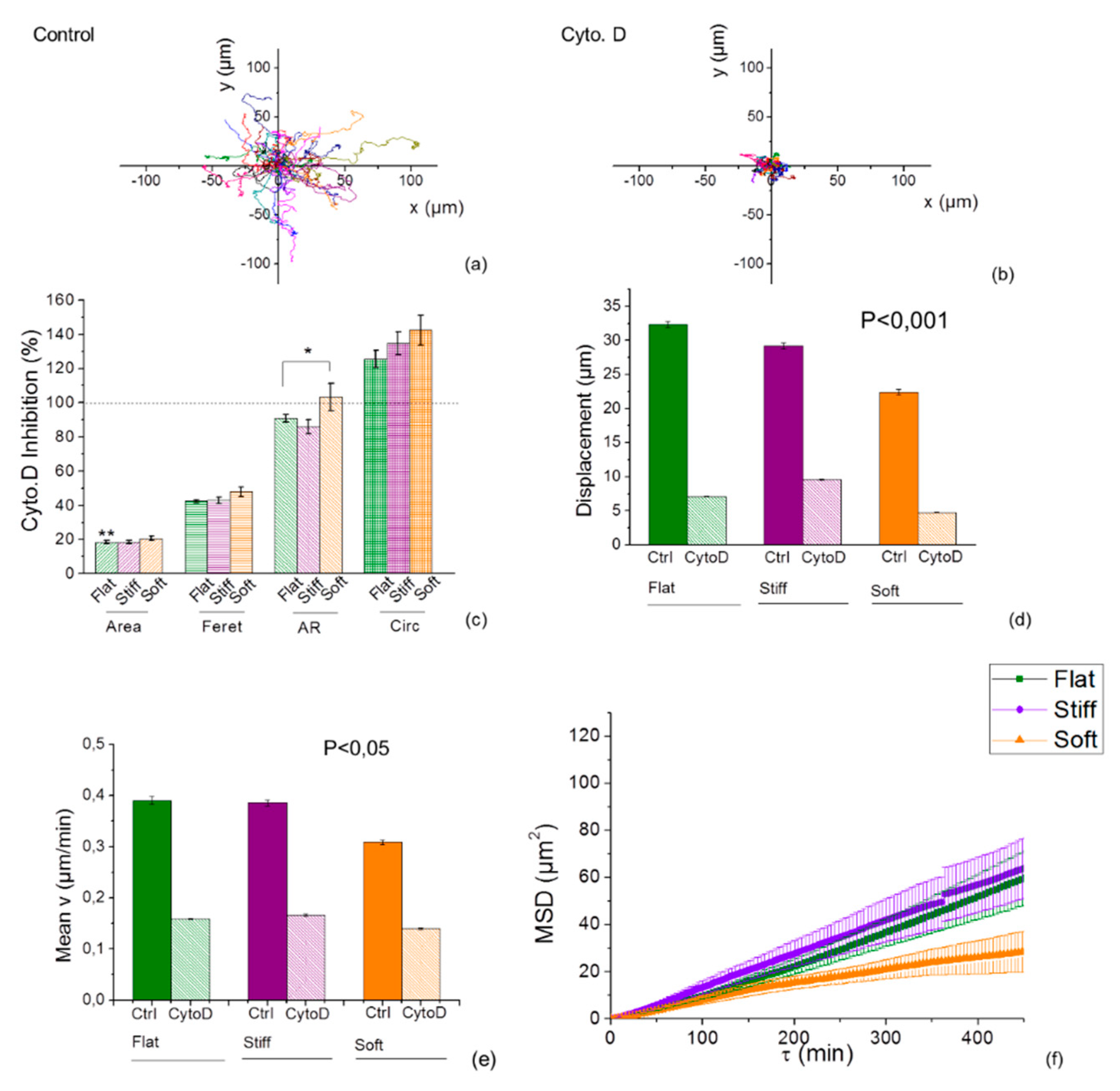
2.6. Inhibition of Actin Polymerization Increases Mechanotaxis of Cells
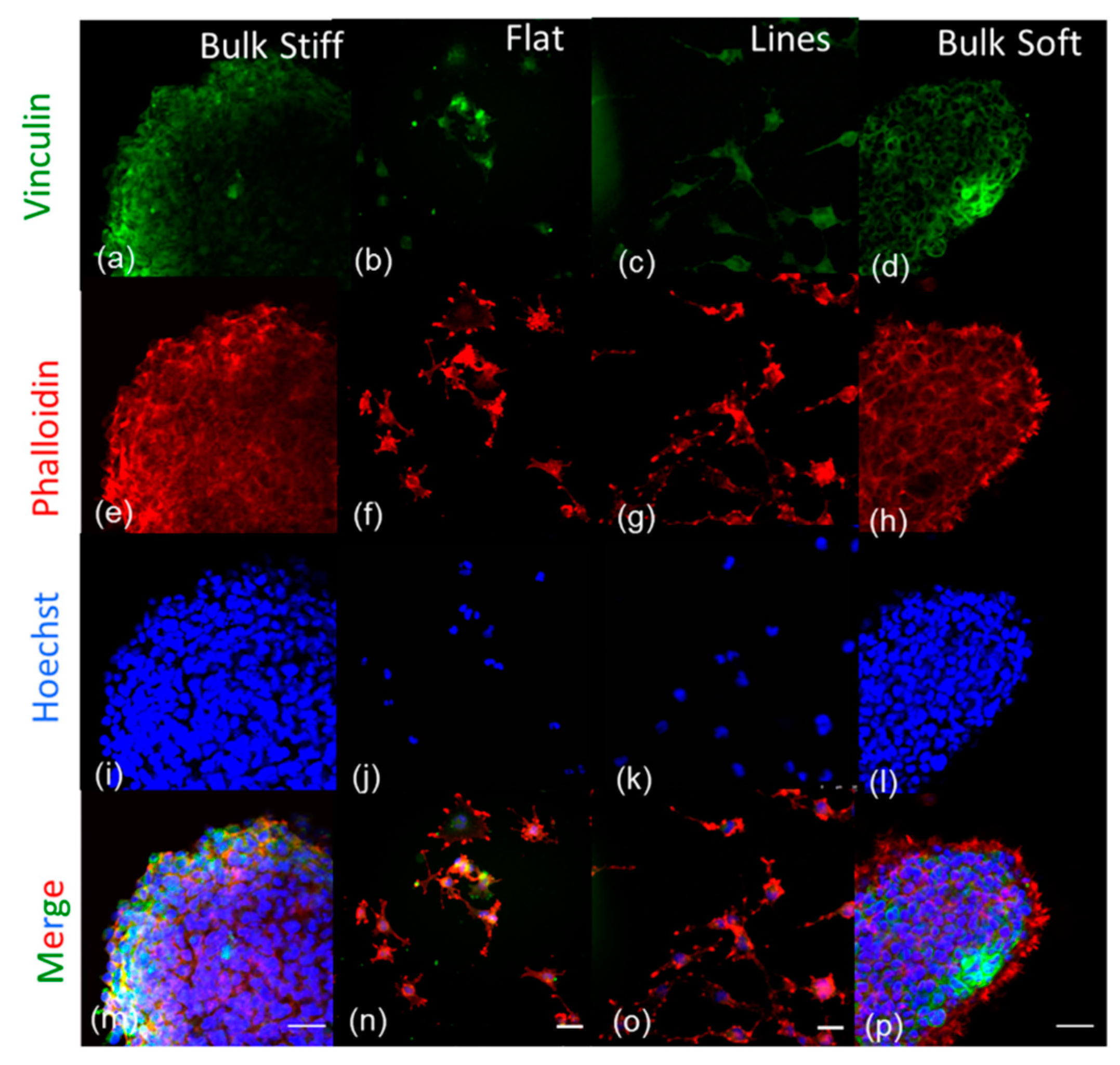
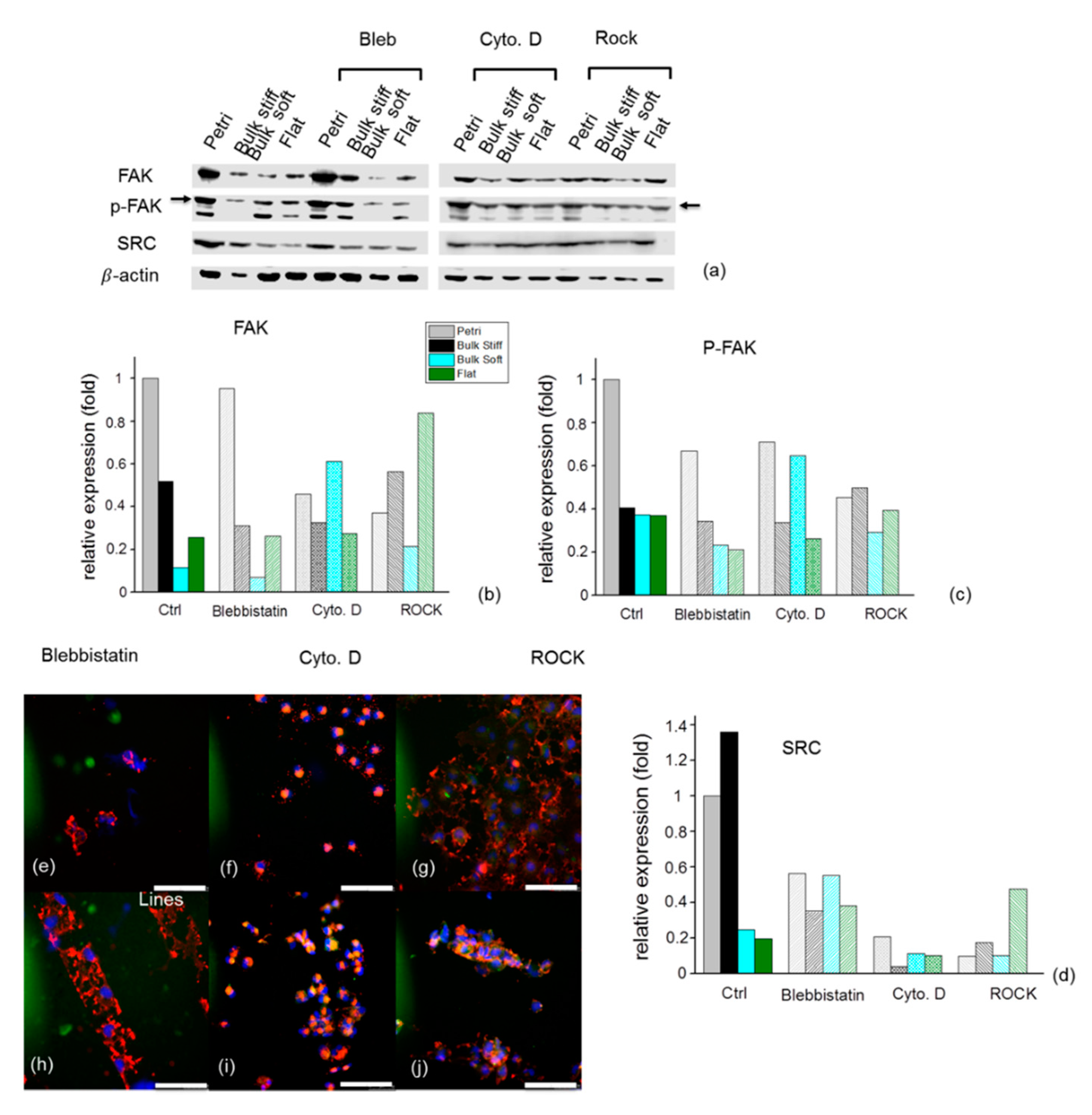
2.7. Effects of Inhibition on F- actin and Focal Adhesions Organization on GBM Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Cultures
4.3. Preparation of PDMS Uniform and Gradient Durotactic Substrates
4.4. Characterization of Substrates
4.4.1. Brillouin Measurements
4.4.2. AFM Measurements
4.5. Cell Morphological Analysis
4.6. Time-lapse Microscopy and Quantitative Analysis of Cell Migration
4.7. Cell Viability Assay
4.8. Apoptosis Analysis
4.9. Inhibition of Cell Contractility
4.10. Western Blotting Analysis
4.11. Immunofluorescence
4.12. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hou, L.C.; Veeravagu, A.; Hsu, A.R.; Tse, V.C. Recurrent glioblastoma multiforme: a review of natural history and management options. Neurosurg. Focus 2006, 20, E5. [Google Scholar] [CrossRef]
- Beauchesne, P. Extra-neural metastases of malignant gliomas: myth or reality? Cancers 2011, 3, 461–477. [Google Scholar] [CrossRef]
- Hamilton, J.D.; Rapp, M.; Schneiderhan, T.; Sabel, M.; Hayman, A.; Scherer, A.; Kropil, P.; Budach, W.; Gerber, P.; Kretschmar, U.; et al. Glioblastoma multiforme metastasis outside the CNS: three case reports and possible mechanisms of escape. J. Clin. Oncol. 2014, 32, e80–e84. [Google Scholar] [CrossRef]
- Nagano, N.; Sasaki, H.; Aoyagi, M.; Hirakawa, K. Invasion of experimental rat brain tumor: early morphological changes following microinjection of C6 glioma cells. Acta Neuropathol. 1993, 86, 117–125. [Google Scholar] [CrossRef]
- Holash, J.; Maisonpierre, P.C.; Compton, D.; Boland, P.; Alexander, C.R.; Zagzag, D.; Yancopoulos, G.D.; Wiegand, S.J. Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF. Science 1999, 284, 1994–1998. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, M.; Stensjøen, A.L.; Berntsen, E.M.; Solheim, O.; Reinertsen, I. The Direction of Tumour Growth in Glioblastoma Patients. Sci. Rep. 2018, 8, 1199. [Google Scholar] [CrossRef] [PubMed]
- Zagzag, D.; Amirnovin, R.; Greco, M.A.; Yee, H.; Holash, J.; Wiegand, S.J.; Zabski, S.; Yancopoulos, G.D.; Grumet, M. Vascular apoptosis and involution in gliomas precede neovascularization: a novel concept for glioma growth and angiogenesis. Lab. Invest. 2000, 80, 837–849. [Google Scholar] [CrossRef]
- Grobben, B.; De Deyn, P.P.; Slegers, H. Rat C6 glioma as experimental model system for the study of glioblastoma growth and invasion. Cell and Tissue Res. 2002, 310, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Lugassy, C.; Haroun, R.I.; Brem, H.; Tyler, B.M.; Jones, R.V.; Fernandez, P.M.; Patierno, S.R.; Kleinman, H.K.; Barnhill, R.L. Pericytic-like angiotropism of glioma and melanoma cells. Am. J. Dermatopathol. 2002, 24, 473–478. [Google Scholar] [CrossRef]
- Farin, A.; Suzuki, S.O.; Weiker, M.; Goldman, J.E.; Bruce, J.N.; Canoll, P. Transplanted glioma cells migrate and proliferate on host brain vasculature: A dynamic analysis. Glia 2006, 53, 799–808. [Google Scholar] [CrossRef]
- Bellail, A.C.; Hunter, S.B.; Brat, D.J.; Tan, C.; Van Meir, E.G. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int. J. Biochem. Cell Biol. 2004, 36, 1046–1069. [Google Scholar] [CrossRef]
- Sahm, F.; Capper, D.; Jeibmann, A.; Habel, A.; Paulus, W.; Troost, D.; von Deimling, A. Addressing Diffuse Glioma as a Systemic Brain Disease with Single-Cell Analysis. Arch Neurol. 2012, 69, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Holland, E.C. Glioblastoma multiforme: The terminator. PNAS 2000, 97, 6242–6244. [Google Scholar] [CrossRef]
- Giese, A.; Bjerkvig, R.; Berens, M.E.; Westphal, M. Cost of migration: Invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 2003, 21, 1624–1636. [Google Scholar] [CrossRef]
- Hirata, E.; Yukinaga, H.; Kamioka, Y.; Arakawa, Y.; Miyamoto, S.; Okada, T.; Sahai, E.; Matsuda, M. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J. Cell Sci. 2012, 125, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Ivkovic, S.; Beadle, C.; Noticewala, S.; Massey, S.C.; Swanson, K.R.; Toro, L.N.; Bresnick, A.R.; Canoll, P.; Rosenfeld, S.S. Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol. Biol. Cell 2012, 23, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Beadle, C.; Assanah, M.C.; Monzo, P.; Vallee, R.; Rosenfeld, S.S.; Canoll, P. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell 2008, 19, 3357–3368. [Google Scholar] [CrossRef] [PubMed]
- Winkler, F.; Kienast, Y.; Fuhrmann, M.; Von Baumgarten, L.; Burgold, S.; Mitteregger, G.; Kretzschmar, H.; Herms, J. Imaging glioma cell invasion in vivo reveals mechanisms of dissemination and peritumoral angiogenesis. Glia 2009, 57, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, F.; Brotchi, J.; Kiss, R. Possible future issues in the treatment of glioblastomas: Special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J. Clin. Oncol. 2005, 23, 2411–2422. [Google Scholar] [CrossRef] [PubMed]
- Elkin, B.S.; Azeloglu, E.U.; Costa, K.D.; Morrison, B., 3rd. Mechanical heterogeneity of the rat hippocampus measured by atomic force microscope indentation. J. Neurotrauma 2007, 24, 812–822. [Google Scholar] [CrossRef]
- Unsgaard, G.; Rygh, O.M.; Selbekk, T.; Muller, T.B.; Kolstad, F.; Lindseth, F.; Hernes, T.A. Intra-operative 3D ultrasound in neurosurgery. Acta neurochir. 2006, 148, 235–253; discussion 253. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.I.; Kang, I.; You, W.K.; McDonald, D.M.; Weaver, V.M. In situ force mapping of mammary gland transformation. Integr. Bio. 2011, 3, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, T.A.; de Juan Pardo, E.M.; Kumar, S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009, 69, 4167–4174. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.N.; Jeibmann, A.; Halama, K.; Witte, H.T.; Walte, M.; Matzat, T.; Schillers, H.; Faber, C.; Senner, V.; Paulus, W.; et al. ECM stiffness regulates glial migration in Drosophila and mammalian glioma models. Development 2014, 141, 3233–3242. [Google Scholar] [CrossRef] [PubMed]
- Ananthanarayanan, B.; Kim, Y.; Kumar, S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials 2011, 32, 7913–7923. [Google Scholar] [CrossRef] [PubMed]
- Scherer, H.J. Structural Development in Gliomas. Am. J. Cancer 1938, 34, 18. [Google Scholar]
- Cortese, B.; Gigli, G.; Riehle, M. Mechanical Gradient Cues for Guided Cell Motility and Control of Cell Behavior on Uniform Substrates. Adv. Funct. Mater. 2009, 19, 2961–2968. [Google Scholar] [CrossRef]
- Palamà, I.E.D.A.S.; Cortese, B. Mechanical Guidance of Cell Migration; IAPC Publishing: Zagreb, Croatia, 2016. [Google Scholar]
- Palama, I.E.; Coluccia, A.M.; Gigli, G.; Riehle, M. Modulation of alignment and differentiation of skeletal myoblasts by biomimetic materials. Integr. Bio. 2012, 4, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ontanon, P.; Orgaz, J.L.; Aldaz, B.; Elosegui-Artola, A.; Martino, J.; Berciano, M.T.; Montero, J.A.; Grande, L.; Nogueira, L.; Diaz-Moralli, S.; et al. Cellular plasticity confers migratory and invasive advantages to a population of glioblastoma-initiating cells that infiltrate peritumoral tissue. Stem Cells 2013, 31, 1075–1085. [Google Scholar] [CrossRef]
- Wong, S.Y.; Ulrich, T.A.; Deleyrolle, L.P.; MacKay, J.L.; Lin, J.M.; Martuscello, R.T.; Jundi, M.A.; Reynolds, B.A.; Kumar, S. Constitutive activation of myosin-dependent contractility sensitizes glioma tumor-initiating cells to mechanical inputs and reduces tissue invasion. Cancer Res. 2015, 75, 1113–1122. [Google Scholar] [CrossRef]
- Grundy, T.J.; De Leon, E.; Griffin, K.R.; Stringer, B.W.; Day, B.W.; Fabry, B.; Cooper-White, J.; O’Neill, G.M. Differential response of patient-derived primary glioblastoma cells to environmental stiffness. Sci. Rep. 2016, 6, 23353. [Google Scholar] [CrossRef]
- Vicente-Manzanares, M.; Ma, X.; Adelstein, R.S.; Horwitz, A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Conti, M.A.; Adelstein, R.S. Nonmuscle myosin II moves in new directions. J. Cell Sci. 2008, 121, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Wolfenson, H.; Bershadsky, A.; Henis, Y.I.; Geiger, B. Actomyosin-generated tension controls the molecular kinetics of focal adhesions. J. Cell Sci. 2011, 124, 1425–1432. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.F. Applications for ROCK kinase inhibition. Curr. Opin. Cell Biol. 2008, 20, 242–248. [Google Scholar] [CrossRef]
- Burridge, K.; Chrzanowska-Wodnicka, M. Focal adhesions, contractility, and signaling. Annu. Rev. Cell Dev. Biol. 1996, 12, 463–518. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Cohen, M.; Addadi, L.; Geiger, B. Hierarchical assembly of cell-matrix adhesion complexes. Biochem. Soc. Trans. 2004, 32, 416–420. [Google Scholar] [CrossRef]
- Janoštiak, R.; Pataki, A.C.; Brábek, J.; Rösel, D. Mechanosensors in integrin signaling: The emerging role of p130Cas. Eur. J. Cell Biol. 2014, 93, 445–454. [Google Scholar] [CrossRef]
- Schwartz, M.A. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb. Perspect Biol. 2010, 2, a005066. [Google Scholar] [CrossRef]
- Schlaepfer, D.D.; Hanks, S.K.; Hunter, T.; van der Geer, P. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature 1994, 372, 786–791. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 1992, 69, 11–25. [Google Scholar] [CrossRef]
- Georges, P.C.; Janmey, P.A. Cell type-specific response to growth on soft materials. J. Appl. Physiol. (1985) 2005, 98, 1547–1553. [Google Scholar] [CrossRef] [PubMed]
- Tilghman, R.W.; Cowan, C.R.; Mih, J.D.; Koryakina, Y.; Gioeli, D.; Slack-Davis, J.K.; Blackman, B.R.; Tschumperlin, D.J.; Parsons, J.T. Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS ONE 2010, 5, e12905. [Google Scholar] [CrossRef] [PubMed]
- Palamà, I.E.; D’Amone, S.; Coluccia, A.M.; Biasiucci, M.; Gigli, G. Cell self-patterning on uniform PDMS-surfaces with controlled mechanical cues. Integr. Biol. (Camb.) 2012, 4, 228–236. [Google Scholar] [CrossRef]
- Gorelik, R.; Gautreau, A. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 2014, 9, 1931–1943. [Google Scholar] [CrossRef]
- Howe, A.; Aplin, A.E.; Alahari, S.K.; Juliano, R.L. Integrin signaling and cell growth control. Curr. Opin. Cell Biol. 1998, 10, 220–231. [Google Scholar] [CrossRef]
- LeDuc, P.P.; LeDuc, P.R.; Bellin, R.R.; Bellin, R.M. Nanoscale intracellular organization and functional architecture mediating cellular behavior. Ann. Biomed. Eng. 2006, 34, 102–113. [Google Scholar] [CrossRef]
- Kolch, W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 2005, 6, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Pogoda, K.; Bucki, R.; Byfield, F.J.; Cruz, K.; Lee, T.; Marcinkiewicz, C.; Janmey, P.A. Soft Substrates Containing Hyaluronan Mimic the Effects of Increased Stiffness on Morphology, Motility, and Proliferation of Glioma Cells. Biomacromolecules 2017, 18, 3040–3051. [Google Scholar] [CrossRef]
- Pathak, A.; Kumar, S. Independent regulation of tumor cell migration by matrix stiffness and confinement. PNAS 2012, 109, 10334–10339. [Google Scholar] [CrossRef]
- Torka, R.; Thuma, F.; Herzog, V.; Kirfel, G. ROCK signaling mediates the adoption of different modes of migration and invasion in human mammary epithelial tumor cells. Exp. Cell Res. 2006, 312, 3857–3871. [Google Scholar] [CrossRef]
- Sen, S.; Ng, W.P.; Kumar, S. Contractility dominates adhesive ligand density in regulating cellular de-adhesion and retraction kinetics. Ann. Biomed. Eng. 2011, 39, 1163–1173. [Google Scholar] [CrossRef][Green Version]
- Malchinkhuu, E.; Sato, K.; Maehama, T.; Mogi, C.; Tomura, H.; Ishiuchi, S.; Yoshimoto, Y.; Kurose, H.; Okajima, F. S1P(2) receptors mediate inhibition of glioma cell migration through Rho signaling pathways independent of PTEN. Biochem. Biophys. Res. Commun. 2008, 366, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Khalil, B.D.; Hanna, S.; Saykali, B.A.; El-Sitt, S.; Nasrallah, A.; Marston, D.; El-Sabban, M.; Hahn, K.M.; Symons, M.; El-Sibai, M. The regulation of RhoA at focal adhesions by StarD13 is important for astrocytoma cell motility. Exp. Cell Res. 2014, 321, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, S.V.; Pasapera, A.M.; Sabass, B.; Waterman, C.M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 2012, 151, 1513–1527. [Google Scholar] [CrossRef]
- Kiss, A.; Horvath, P.; Rothballer, A.; Kutay, U.; Csucs, G. Nuclear motility in glioma cells reveals a cell-line dependent role of various cytoskeletal components. PLoS ONE 2014, 9, e93431. [Google Scholar] [CrossRef][Green Version]
- Chintala, S.K.; Sawaya, R.; Aggarwal, B.B.; Majumder, S.; Giri, D.K.; Kyritsis, A.P.; Gokaslan, Z.L.; Rao, J.S. Induction of matrix metalloproteinase-9 requires a polymerized actin cytoskeleton in human malignant glioma cells. J. Biol. Chem. 1998, 273, 13545–13551. [Google Scholar] [CrossRef] [PubMed]
- Mierke, C.T.; Rösel, D.; Fabry, B.; Brábek, J. Contractile forces in tumor cell migration. Eur. J. Cell Biol. 2008, 87, 669–676. [Google Scholar] [CrossRef]
- Gingras, D.; Page, M.; Annabi, B.; Beliveau, R. Rapid activation of matrix metalloproteinase-2 by glioma cells occurs through a posttranslational MT1-MMP-dependent mechanism. Biochim. Biophys. Acta 2000, 1497, 341–350. [Google Scholar] [CrossRef]
- Antonacci, G.; Braakman, S. Biomechanics of subcellular structures by non-invasive Brillouin microscopy. Sci. Rep. 2016, 6, 37217. [Google Scholar] [CrossRef] [PubMed]
- Basilico, B.; Cortese, B.; Ratano, P.; Di Angelantonio, S.; Ragozzino, D. Time-lapse Whole-field Fluorescence Imaging of Microglia Processes Motility in Acute Mouse Hippocampal Slices and Analysis. Bio-protocol 2019, 9, e3220. [Google Scholar] [CrossRef]
- Stokes, C.L.; Lauffenburger, D.A.; Williams, S.K. Migration of individual microvessel endothelial cells: stochastic model and parameter measurement. J. Cell Sci. 1991, 99, 419–430. [Google Scholar] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palamà, I.E.; D’Amone, S.; Ratano, P.; Donatelli, A.; Liscio, A.; Antonacci, G.; Testini, M.; Di Angelantonio, S.; Ragozzino, D.; Cortese, B. Mechanical Durotactic Environment Enhances Specific Glioblastoma Cell Responses. Cancers 2019, 11, 643. https://doi.org/10.3390/cancers11050643
Palamà IE, D’Amone S, Ratano P, Donatelli A, Liscio A, Antonacci G, Testini M, Di Angelantonio S, Ragozzino D, Cortese B. Mechanical Durotactic Environment Enhances Specific Glioblastoma Cell Responses. Cancers. 2019; 11(5):643. https://doi.org/10.3390/cancers11050643
Chicago/Turabian StylePalamà, Ilaria Elena, Stefania D’Amone, Patrizia Ratano, Amato Donatelli, Andrea Liscio, Giuseppe Antonacci, Mariangela Testini, Silvia Di Angelantonio, Davide Ragozzino, and Barbara Cortese. 2019. "Mechanical Durotactic Environment Enhances Specific Glioblastoma Cell Responses" Cancers 11, no. 5: 643. https://doi.org/10.3390/cancers11050643
APA StylePalamà, I. E., D’Amone, S., Ratano, P., Donatelli, A., Liscio, A., Antonacci, G., Testini, M., Di Angelantonio, S., Ragozzino, D., & Cortese, B. (2019). Mechanical Durotactic Environment Enhances Specific Glioblastoma Cell Responses. Cancers, 11(5), 643. https://doi.org/10.3390/cancers11050643







