1. Introduction
Epithelial ovarian cancer (EOC) is the leading cause of gynecologic cancer mortality, and is difficult to detect at an early stage [
1,
2]. Over 70% of women with EOC are diagnosed with advanced stage disease with a 5-year relative survival of less than 50% [
2]. Current screening methods lack sufficient accuracy [
3,
4], and thus, there is an urgent need to develop new strategies for detection of early stage EOC to improve patient survival.
We previously identified a C4-binding protein with fully-sialylated N-glycans (FS-C4BP = A2160) as a novel EOC marker [
5]. The FS-C4BP particularly distinguished early-stage ovarian clear cell carcinoma (OCCC) from endometrioma more reliably than CA-125. In addition, A2160, as a FS-C4BP peptide with a fully sialylated and fucosylated triantennary glycan and fully sialylated biantennary glycan exhibited superior sensitivity and specificity as an EOC marker.
Common strategies for the discovery of new cancer biomarkers [
6] include metabolomics [
7,
8], identification of genetic and genomic markers and gene mutations by PCR and/or next generation sequencing, anomalous alternative mRNA splicing products of an expressed gene, as well gene re-expression following silencing during normal differentiation [
1,
9]. The use of liquid chromatography with mass spectrometry (LC–MS) to identify glycoproteins released into the circulation provides a new approach to the discovery of cancer markers [
5,
10]. With this novel technology, we investigated not only sugar chain alterations but also combinations of protein and glycan changes [
5].
The analysis of serum glycoproteins remains a difficult and exhausting procedure due to the large amount of data generated from the numerous peaks produced by the LC–MS method. High inter-individual variability and complexity in glycan and glycopeptide profiles makes the visual comparisons of these spectra impractical and multivariate analyses are required to determine consistent variations between data sets [
11,
12]. Some of the statistical methods required for the multivariate analysis of the instrumental spectral results involve bilinear and multiway models, cluster analysis, scatter plots, principal component analysis (PCA), linear and non-linear partial least squares, and pattern recognition computations through the newly developed application of orthogonal partial square discrimination analysis (OPLS-DA) in scatter and scores plots [
13,
14].
In our previous study, we identified the glycopeptide A2160 cancer marker from among more than 100,000 glycopeptide peaks detected in the blood sera of ovarian cancer patients [
5]. During the course of detecting, isolating and identifying the A2160 glycopeptide we developed filtering procedures to find thousands of MS glycopeptide peaks that showed statistically significant differences between ovarian cancer patients and control patients. In this study, we investigated the comprehensive glycopeptide spectra analysis (CSGSA) of serum between cancer and non-cancer patients using the basic
t-test, scatter plots, PCA, OPLS-DA, and heat maps in order to use the MS spectra as a possible novel approach for the early detection of ovarian cancer.
2. Materials and Methods
2.1. Patient Sample
Demographics are shown in
Supplementary Table S1. During the study period from January to December of 2016, consecutive serum samples were obtained from 39 EOC patients prior to the initiation of any treatment. In addition, serum samples were collected from 45 subjects in a non-cancer group. The mean age of EOC patients was 53.7 years. The ethnicity of the EOC group was Japanese (100%) and the four most common histologic subtypes of EOC were OCCC (
n = 15, 38.5%), endometrioid (
n = 14, 35.8%), serous (
n = 8, 20.5%), and mucinous adenocarcinoma (
n = 2, 5.1%). The majority of cases were early-stage (stage I–II; 58.9%).
The non-cancer group included Japanese women with the following gynecologic diseases and mean ages (years): Uterine myoma (n = 10) in women with an average age of 51.0 years, endometrioma (n = 18) with an average age of 43.78 years, and ovarian cysts (n = 17) in women with an average age of 54.41 years.
All blood samples were collected by venipuncture from patients before their surgery. Samples were centrifuged and stored at −80 °C until needed, with avoidance of repeated freeze–thaw cycles. Samples were excluded from the study based on the patient criteria listed in
Supplementary Table S2.
2.2. Quality Control Serum and Inter- and Intra-Assay Coefficients of Variability (CVs)
Quality control (QC) serum samples were prepared by separately pooling ten sera of ovarian cancer patients and ten sera of benign gynecologic disease patients. The pooled sera were preserved in −80 °C until they were analyzed. The QC samples were used during the process of glycoprotein profiling to calculate the inter- and intra-assay coefficients of variability (CVs) due to operator or laboratory generated variations and errors.
2.3. Sample Preparation for Glycoprotein Profiling
Mikami et al. [
5] previously presented a description of the techniques and the schema that were used for the glycoprotein profiling in this study. Briefly, after extracting and protease digestion of the glycoproteins, the glycopeptides were enriched by filtration and analyzed by liquid chromatography mass spectrometry (LC–MS). The detailed sample preparation methods are described in
Supplementary Method S1 and are slightly modified from the previous report [
5].
2.4. Liquid Chromatography and Mass Spectrometry
UPLC–MS/MS data were acquired on an UPLC system (Agilent HP1200; Agilent Technologies, Palo Alto, CA, USA) equipped with a C18 column (Inertsil ODS-4, 2 μm, 100 Å, 100 mm × 1.5 mm ID; GL Science, Tokyo, Japan) and coupled with an electrospray ionization quadrupole time-of-flight (Q-TOF) mass spectrometer (Agilent 6520, Agilent Technologies). Solvent A was 0.1% formic acid, and solvent B comprised 0.1% formic acid in 9.9% water and 90% acetonitrile. Glycopeptides were eluted at 40 °C with a flow rate of 0.15 mL/min, using the following gradient program: 0–7 min, 15–30% solvent B; 7–12 min, 30–50% solvent B; and an additional 2-min hold at 100% solvent B. The mass spectrometer was operated in negative mode with a capillary voltage of 4000 V. The nebulizing gas pressure was 30 psi, and the dry gas flow was 8 L/min at 350 °C. The injection volume was 5 μL.
2.5. Data Analysis
All mass spectral data were analyzed by using our original software, “Marker Analysis,” developed using R (R 3.2.2, R Foundation) and Excel VBA (Excel 2010, Microsoft, Washington, VA, USA) as previously reported [
15]. After LC–MS raw data were converted to CSV-type data using Mass Hunter Export (Agilent Technologies), Marker Analysis was used to distinguish peak curve shapes, smooth and differentiate the peaks, and recognize the beginning, top, and end of them (determined at the points where the differentiation curve changed from zero to positive, from positive to negative, and from negative to zero, respectively). The peak area was calculated by integrating curves from beginning to end. The error in retention time and m/z was corrected using internal standard (fetal calf fetuin) peaks. Peak alignment was performed in such a way that the error of each peak position (retention time and m/z) was within 0.3 min and 0.06 Da, respectively.
The data was normalized by calculating ratios between each peak area and the average peak areas of QCs. The mode establishing method with SIMCA software (version 13.0.3; Umetrics; Sartorius AG, Göttingen, Germany) was used to form the orthogonal partial least-squares discriminant analysis (OPLS-DA) model [
16]. Heat maps were prepared using the protocols developed for the software program Excel VBA.
2.6. Pattern Recognition (PR) Analysis and Cross-Validation
To establish a global overview of differences between the EOC patients and the non-cancer controls, multivariate analysis was applied to glycopeptide spectra data as previously described [
13,
14]. Normalized glycopeptide spectral data sets were unit variance scaled and mean-centered, then analyzed by PCA and OPLS-DA using the SIMCA-P+ program (version 14.1, Umetrics AB; Umeå, Sweden). Model quality was evaluated using R2Y and Q2 values, which reflect the explained fraction of variance and model predictability. PCA was utilized to overview an unsupervised pattern of samples, then OPLS-DA analysis was performed to distinguish two groups, EOC and non-EOC. Before OPLS-DA analysis, the data set was divided into two sets, a training set and a test set, including EOC and non-EOC samples respectively to evaluate the training model’s validity. Separation was evaluated using the first and second principal components taken by PCA or OPLS-DA. OPLS-DA analysis was conducted again for further verification of the differences between the comparison groups.
2.7. Study Approval
Institutional Review Board (IRB) approval was obtained at Tokai University (IRB registration number, 09R-082). Signed informed consents were obtained from all participants in the study.
4. Discussion
In a previous study, we used LC–MS for comparative profiling of serum glycoproteins of ovarian cancer cases and non-cancer controls and found that there were more than 30,000 glycopeptide peaks that could be used potentially as serum biomarkers for the early diagnosis of ovarian cancer [
5]. The glycopeptide A2160 was one of the first glycopeptides that we identified as a potential cancer marker and peptide sequencing revealed that it was a component of the fully-sialylated alpha-chain of the complement 4-binding protein. Moreover, we showed that the serum levels of A2160 exhibited superior sensitivity and specificity to CA-125 as an EOC marker. However, many other glycopeptide peaks showed a statistically significant difference between ovarian cancer patients and control patients, suggesting that they also could be used as markers to identify cancer patients.
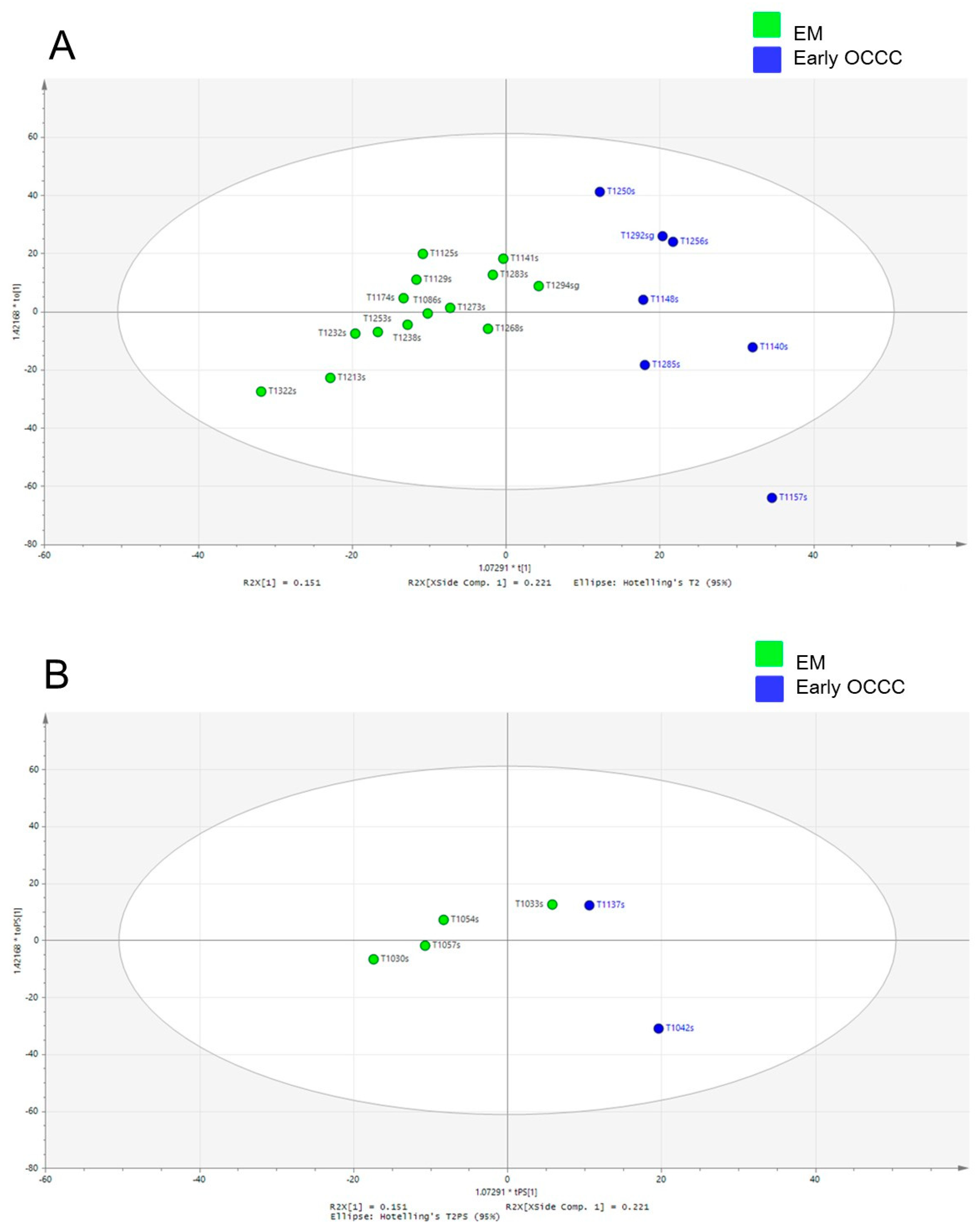
The aim of the present study was to validate a LC–MS profiling method that we had used previously to determine and visually quantitate multiple serum glycopeptides as a useful tool for the detection and diagnosis of EOC. We investigated the LC–MS quantitative profiling of 2281 glycopeptides selected on the basis of statistical significance following an initial t-test of 30,000 glycopeptides detected in the serum of cancer and non-cancer patients. We used the same validated statistical and visualization methods to quantitate the signature glycopeptides in the ovarian cancers compared to two separate non-cancer controls, the endometrioma control group as well as the leiomyoma and ovarian cyst control group. In both cases the signature glycopeptides in the cancer group were highly differentiated from the control groups using various measures including PCA, OPLS-DA scatter and scores plots, and heat maps.
In this study, we did not characterize or identify individual glycopeptides. Instead we used them together as a single group to demonstrate the potential marker signature of ovarian cancer. It is clear from the present study that there are thousands of potential glycopeptide biomarkers for ovarian cancer, but it will take considerable time to isolate, sequence and identify all of them. In the meantime, we have devised an analytical method to measure the individual concentrations of thousands of glycopeptides using a single assay, which substantially reduces analytical time and sample size. In contrast to classical biochemical approaches using immunological methods such as ELISA, RIA and EIA that mainly focus on single targets, our CSGSA involves the collation of quantitative results for a broad series of glycopeptides to reveal an overall change in blood sera of ovarian cancer patients compared to controls.
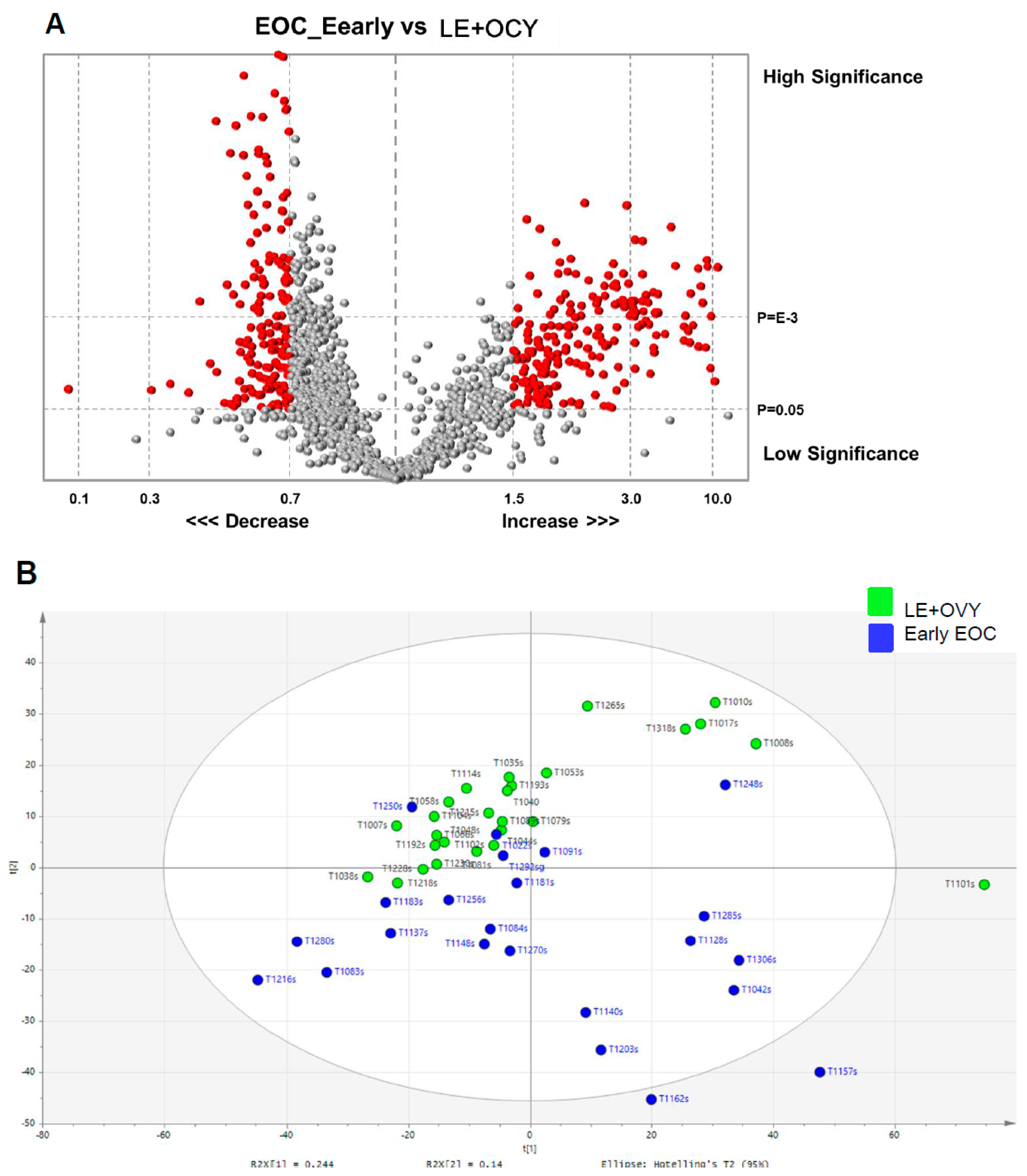
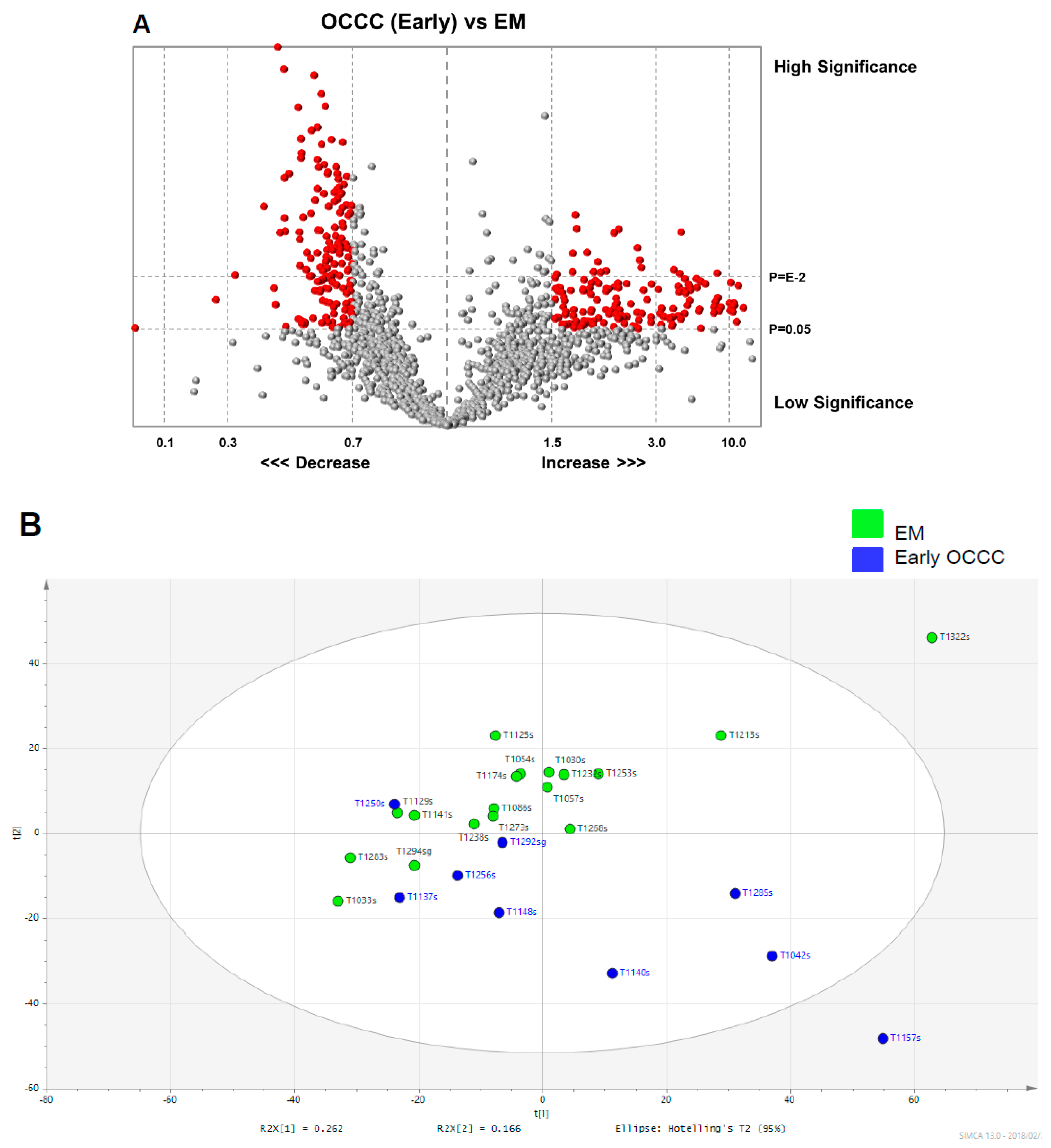
As part of our statistical and visual analysis to differentiate between the cancer and non-cancer groups, we used various measures including the volcano plot, PCA, OPLS-DA scatter and scores plots, and heat maps. The volcano plots were used initially to plot significance (t-statistic) versus fold-change on the Y and X axes respectively to compare the cancer and non-cancer samples and identify changes in the large glycopeptide data set generated by the LC–MS. From these volcano plots, we identified glycopeptides candidate EOC glycopeptide markers. The unsupervised PCA identified a set of unique spectral patterns that generally separated the cancer and non-cancer data into relatively distinct clusters. However, some of the cancer spectral data overlapped with the non-cancer data.
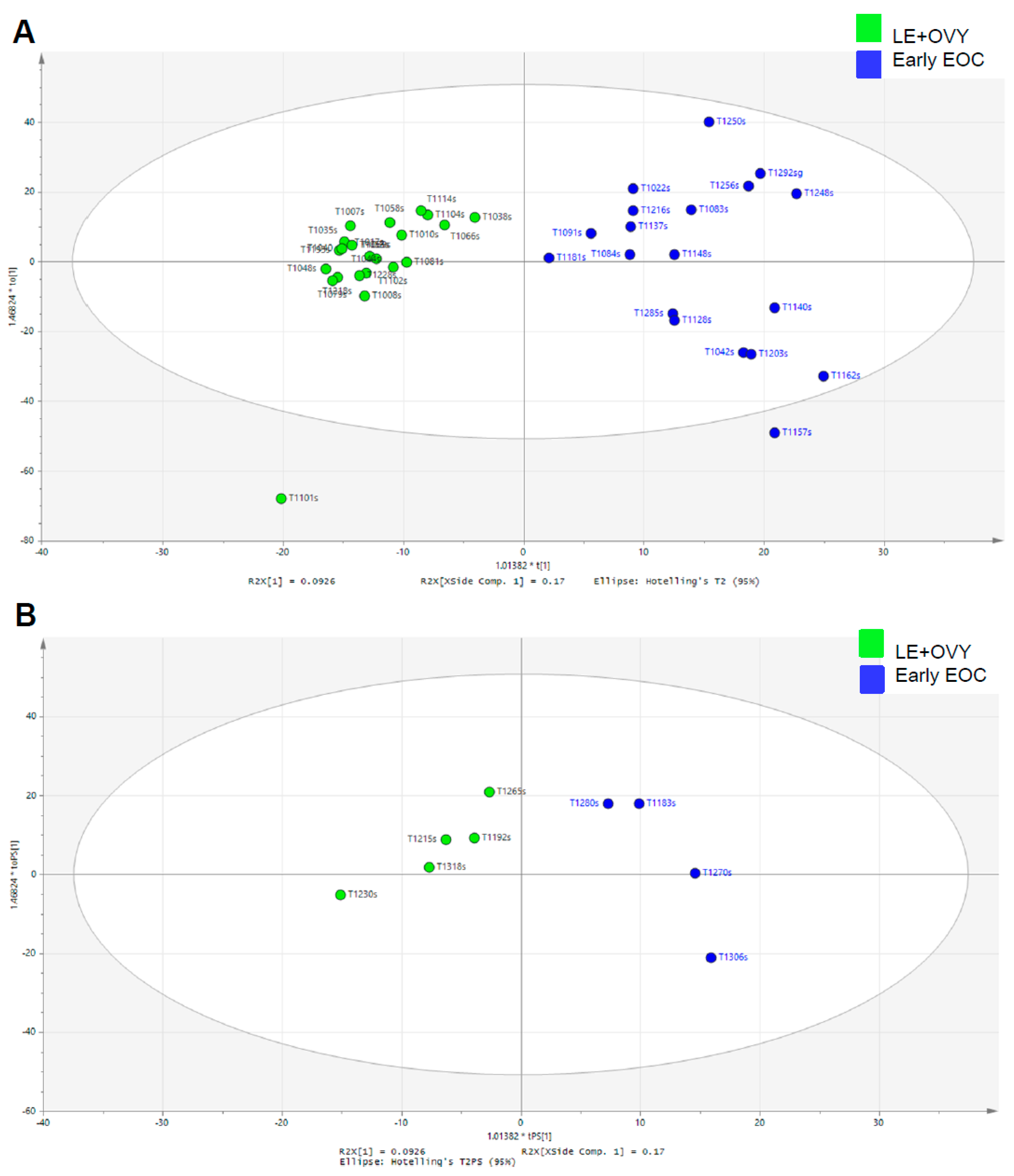
In comparison to PCA, the supervised OPLS-DA scatter plots clearly forced a greater scores-space separation that grouped the cancer and non-cancer data into two significantly distinct clusters. Worley and Powers [
13] used Monte Carlo simulations to demonstrate that the PCA and the OPLS-DA analyses should be performed together on the same samples to confirm reliability because the OPLS-DA analysis alone can yield statistically unreliable group separation. They found that when the PCA failed to expose group separation, OPLS-DA continued to separate the groups at the expense of model reliability. Therefore, we used both the PCA and the OPLS-DA analyses to test and confirm the reliability of our results. We further validated the reliability of our results by computing OPLS-DA scatter and scores plots in association with the OPLS-DA prediction model.
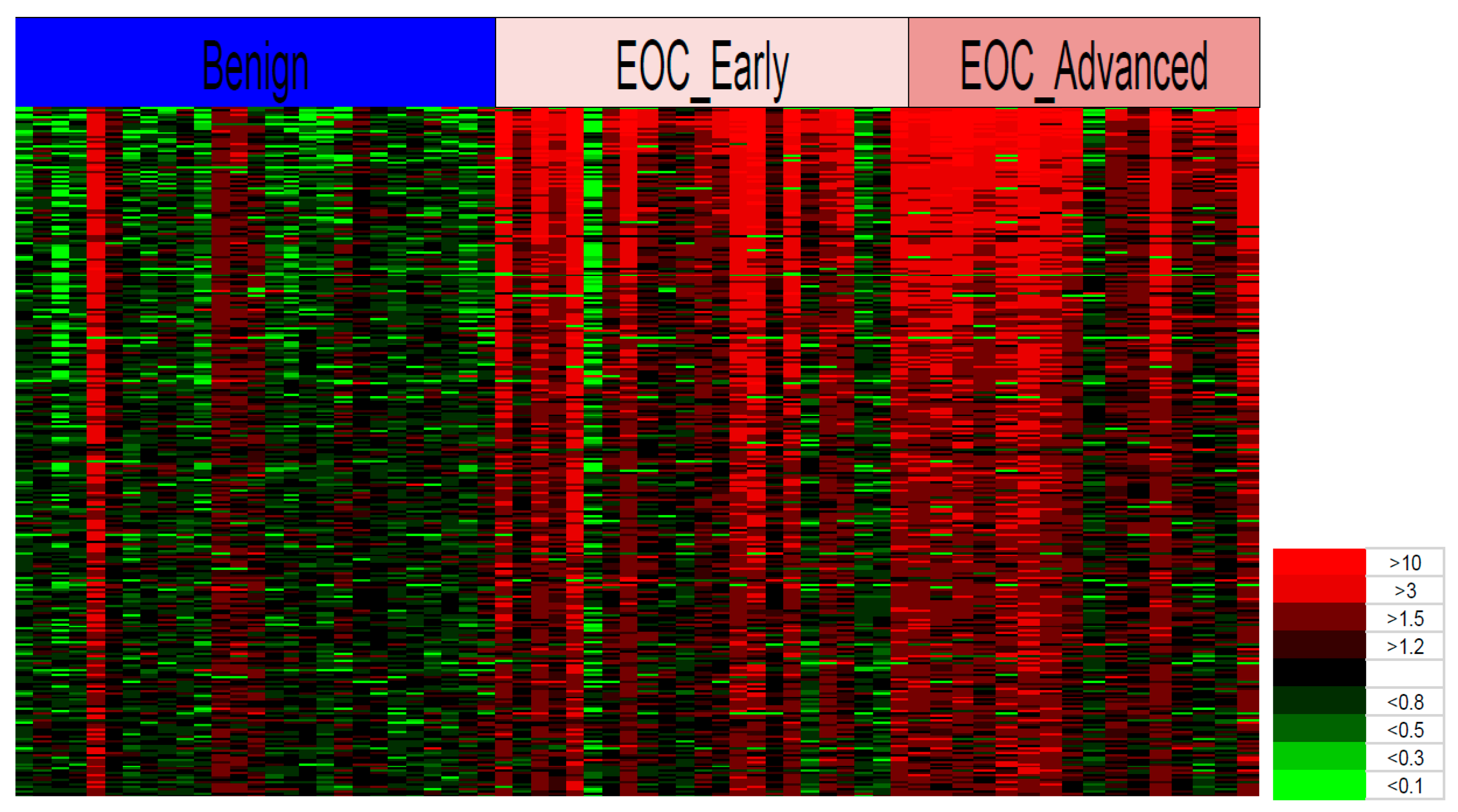
Heat maps are another efficient way to visualize complex data sets organized as matrices. Our study is the first to report on the MS-based glycopeptides quantitative levels in ovarian cancer and non-cancer individual patients using statistical comparative analysis with heat maps. We used heat maps to visualize the quantitative glycopeptide data in individual patients as a single graphic that allowed us to easily identify the signature patterns of many glycopeptides and their quantities across multiple subjects in highly complex MS datasets comparing cancer and non-cancer patients. The relative concentrations of the glycopeptides were statistically filtered between the patients and controls and the quantitative results were displayed for each individual to reveal the most effective glycopeptide signatures. Although most of the statistically significant glycopeptides showed higher levels in the patients with cancer, some individuals had levels similar to those in the non-cancer group. These inconsistencies may have arisen from the fact that individual ratios were based on the average glycopeptide levels in the non-cancer group and not on the potentially variable glycopeptide base-line levels of each individual. However, the heat maps seem to reflect the trends of the PCA results more closely than the OPLS-DA. These sample outliers would need to be followed up in greater detail to better understand the inconsistencies between the cancer and non-cancer groups and between the PCA and OPLS-DA results.
We have used the mass spectra profiles of serum glycopeptides as multi-biomarkers for the purpose of cancer screening. Kim et al. [
17] previously had undertaken a serum multi-biomarker MS screening of ovarian cancers, but they screened for N-glycans instead of glycopeptides. No study has previously used the CSGSA for the detection of ovarian cancer. However, like Kim et al [
17], we focused on diagnosis rather than marker discovery and even though at this stage we do not understand the biological context of the changes, the MS signals exhibited differences between the cancer and non-cancer groups that could be exploited to improve current cancer screening procedures.
Our study shows that CSGSA is a potentially new tool for the early detection of ovarian cancer using glycopeptides within blood serum samples that could also be applied for the detection and diagnosis of other cancers. We believe that the use of multiple biomarkers can lead to far better diagnostic classification than current single-marker approaches [
11,
17] that depend on CA-125 or HE4. Our analysis is early and many more comprehensive studies are required to determine if the over-expressed blood serum glycopeptide markers are specific to certain cancers such as EOC or if they also markers for various other cancer and pre-cancer types. Thus, we will need to ascertain the specificity of our markers to ovarian cancer by comparing our current results in patients with ovarian cancer to those with other cancer types. We must also further address how these markers change with different stages of precancerous and cancerous lesions.
Another limitation in our study is small sample size. Generally, high-grade serous ovarian cancer is the most common histology type of ovarian cancer, and therefore, ovarian cancer screening to distinguish stage I high-grade serous ovarian cancer from benign ovarian tumors is the most clinically meaningful comparison. However, this comparison was not feasible as there were too few cases of this subgroup to analyze in our dataset. Moreover, comparisons between histology types were also not feasible. In Japan, OCCC is a particularly common histology, presenting typically at an early-stage [
18]. As women with endometrioma possess an increased risk of OCCC [
19], comparison of early-stage OCCC to endometrioma is more relevant to the Japanese population.