A Case of Identity: HOX Genes in Normal and Cancer Stem Cells
Abstract
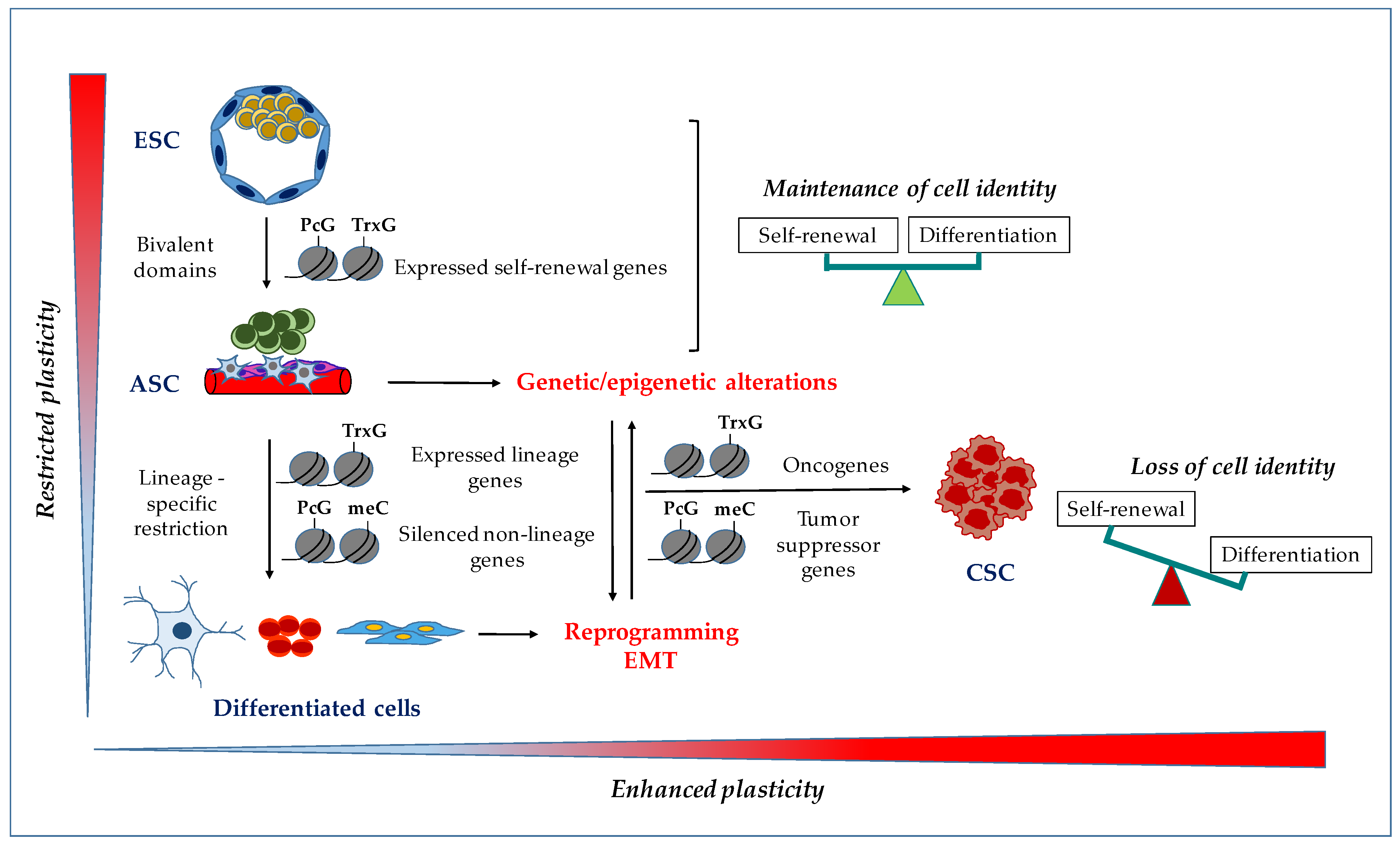
1. HOX Genes Are Master Regulators of Embryonic Development
2. HOX Genes and Cellular Identity
3. HOX Genes in Embryonic Stem Cells
4. HOX Genes in Adult Stem Cells
5. HOX Genes in Cancer Stem Cells
6. HOX Genes and CSC Targeted Therapies
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maconochie, M.; Nonchev, S.; Morrison, A.; Krumlauf, R. Paralogous Hox genes: Function and regulation. Annu. Rev. Genet. 1996, 30, 529–556. [Google Scholar] [CrossRef]
- Duboule, D. The rise and fall of Hox gene clusters. Development 2007, 134, 2549–2560. [Google Scholar] [CrossRef]
- Duboule, D.; Morata, G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994, 10, 358–364. [Google Scholar] [CrossRef]
- Young, T.; Deschamps, J. Hox, Cdx, and anteroposterior patterning in the mouse embryo. Curr. Top. Dev. Biol. 2009, 88, 235–255. [Google Scholar] [CrossRef] [PubMed]
- Montavon, T.; Soshnikova, N. Hox gene regulation and timing in embryogenesis. Semin. Cell Dev. Biol. 2014, 34, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Noordermeer, D.; Duboule, D. Chromatin architectures and Hox gene collinearity. Curr. Top. Dev. Biol. 2013, 104, 113–148. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef]
- Samadieh, Y.; Favaedi, R.; Ramezanali, F.; Afsharian, P.; Aflatoonian, R.; Shahhoseini, M. Epigenetic dynamics of HOXA10 gene in infertile women with endometriosis. Reprod. Sci. 2019, 26, 88–96. [Google Scholar] [CrossRef]
- Soshnikova, N.; Duboule, D. Epigenetic regulation of Hox gene activation: The waltz of methyls. Bioessays 2008, 30, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.-W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M.; Lacey, M. DNA methylation and differentiation: Silencing, upregulation and modulation of gene expression. Epigenomics 2013, 5, 553–568. [Google Scholar] [CrossRef]
- Shah, M.; Allegrucci, C. Stem cell plasticity in development and cancer: Epigenetic origin of cancer stem cells. Subcell. Biochem. 2013, 61, 545–565. [Google Scholar] [CrossRef]
- Ringrose, L.; Paro, R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 2007, 134, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.T.; Sano, M.; Kin, T.; Asai, K.; Hirose, T. Coordinated expression of ncRNAs and HOX mRNAs in the human HOXA locus. Biochem. Biophys. Res. Commun. 2007, 357, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Hombria, J.C.; Lovegrove, B. Beyond homeosis--HOX function in morphogenesis and organogenesis. Differentiation 2003, 71, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Rezsohazy, R.; Saurin, A.J.; Maurel-Zaffran, C.; Graba, Y. Cellular and molecular insights into Hox protein action. Development 2015, 142, 1212–1227. [Google Scholar] [CrossRef] [PubMed]
- Longobardi, E.; Penkov, D.; Mateos, D.; De Florian, G.; Torres, M.; Blasi, F. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev. Dyn. 2014, 243, 59–75. [Google Scholar] [CrossRef]
- Dard, A.; Reboulet, J.; Jia, Y.; Bleicher, F.; Duffraisse, M.; Vanaker, J.M.; Forcet, C.; Merabet, S. Human HOX Proteins Use Diverse and Context-Dependent Motifs to Interact with TALE Class Cofactors. Cell Rep. 2018, 22, 3058–3071. [Google Scholar] [CrossRef]
- Wang, K.C.; Helms, J.A.; Chang, H.Y. Regeneration, repair and remembering identity: The three Rs of Hox gene expression. Trends Cell Biol. 2009, 19, 268–275. [Google Scholar] [CrossRef]
- Chang, H.Y.; Chi, J.T.; Dudoit, S.; Bondre, C.; van de Rijn, M.; Botstein, D.; Brown, P.O. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. USA 2002, 99, 12877–12882. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Bondre, C.; Gladstone, H.B.; Brown, P.O.; Chang, H.Y. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006, 2, e119. [Google Scholar] [CrossRef]
- Donoghue, M.J.; Morris-Valero, R.; Johnson, Y.R.; Merlie, J.P.; Sanes, J.R. Mammalian muscle cells bear a cell-autonomous, heritable memory of their rostrocaudal position. Cell 1992, 69, 67–77. [Google Scholar] [CrossRef]
- Grieshammer, U.; Sassoon, D.; Rosenthal, N. A transgene target for positional regulators marks early rostrocaudal specification of myogenic lineages. Cell 1992, 69, 79–93. [Google Scholar] [CrossRef]
- Kamkar, F.; Xaymardan, M.; Asli, N.S. Hox-Mediated Spatial and Temporal Coding of Stem Cells in Homeostasis and Neoplasia. Stem Cells Dev. 2016, 25, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, J.; Duboule, D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 2017, 31, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Noordermeer, D.; Leleu, M.; Splinter, E.; Rougemont, J.; De Laat, W.; Duboule, D. The dynamic architecture of Hox gene clusters. Science (New York, N.Y.) 2011, 334, 222–225. [Google Scholar] [CrossRef]
- Bahrami, S.B.; Veiseh, M.; Dunn, A.A.; Boudreau, N.J. Temporal changes in Hox gene expression accompany endothelial cell differentiation of embryonic stem cells. Cell Adhes. Migr. 2011, 5, 133–141. [Google Scholar] [CrossRef]
- Riising, E.M.; Comet, I.; Leblanc, B.; Wu, X.; Johansen, J.V.; Helin, K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell 2014, 55, 347–360. [Google Scholar] [CrossRef]
- Di Croce, L.; Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013, 20, 1147–1155. [Google Scholar] [CrossRef]
- Neijts, R.; Amin, S.; van Rooijen, C.; Deschamps, J. Cdx is crucial for the timing mechanism driving colinear Hox activation and defines a trunk segment in the Hox cluster topology. Dev. Biol. 2017, 422, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Neijts, R.; Amin, S.; van Rooijen, C.; Tan, S.; Creyghton, M.P.; de Laat, W.; Deschamps, J. Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev. 2016, 30, 1937–1942. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, K.; Nakajima, M.; Kanokoda, M.; Kyba, M.; Dandapat, A.; Tolar, J.; Saito, M.K.; Toyoda, M.; Umezawa, A.; Hara, T. GSK3beta inhibition activates the CDX/HOX pathway and promotes hemogenic endothelial progenitor differentiation from human pluripotent stem cells. Exp. Hematol. 2016, 44, 68.e10–74.e10. [Google Scholar] [CrossRef] [PubMed]
- Shahhoseini, M.; Taghizadeh, Z.; Hatami, M.; Baharvand, H. Retinoic acid dependent histone 3 demethylation of the clustered HOX genes during neural differentiation of human embryonic stem cells. Biochem. Cell Biol. 2013, 91, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, J.; van Nes, J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 2005, 132, 2931–2942. [Google Scholar] [CrossRef] [PubMed]
- Langston, A.W.; Gudas, L.J. Retinoic acid and homeobox gene regulation. Curr. Opin. Genet. Dev. 1994, 4, 550–555. [Google Scholar] [CrossRef]
- Kashyap, V.; Laursen, K.B.; Brenet, F.; Viale, A.J.; Scandura, J.M.; Gudas, L.J. RARgamma is essential for retinoic acid induced chromatin remodeling and transcriptional activation in embryonic stem cells. J. Cell Sci. 2013, 126, 999–1008. [Google Scholar] [CrossRef]
- Kashyap, V.; Gudas, L.J.; Brenet, F.; Funk, P.; Viale, A.; Scandura, J.M. Epigenomic reorganization of the clustered Hox genes in embryonic stem cells induced by retinoic acid. J. Biol. Chem. 2011, 286, 3250–3260. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.S.; Lee, S.H.; Chen, K.; Zhu, G.; Oh, W.; Allton, K.; Gafni, O.; Kim, Y.Z.; Tomoiga, A.S.; Barton, M.C.; et al. An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res. 2016, 44, 3659–3674. [Google Scholar] [CrossRef] [PubMed]
- Gouti, M.; Gavalas, A. Hoxb1 controls cell fate specification and proliferative capacity of neural stem and progenitor cells. Stem Cells 2008, 26, 1985–1997. [Google Scholar] [CrossRef]
- Fan, R.; Bonde, S.; Gao, P.; Sotomayor, B.; Chen, C.; Mouw, T.; Zavazava, N.; Tan, K. Dynamic HoxB4-regulatory network during embryonic stem cell differentiation to hematopoietic cells. Blood 2012, 119, e139–e147. [Google Scholar] [CrossRef] [PubMed]
- Lengerke, C.; Grauer, M.; Niebuhr, N.I.; Riedt, T.; Kanz, L.; Park, I.H.; Daley, G.Q. Hematopoietic development from human induced pluripotent stem cells. Ann. N. Y. Acad. Sci. 2009, 1176, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Bowles, K.M.; Vallier, L.; Smith, J.R.; Alexander, M.R.; Pedersen, R.A. HOXB4 overexpression promotes hematopoietic development by human embryonic stem cells. Stem Cells 2006, 24, 1359–1369. [Google Scholar] [CrossRef]
- Larsen, B.M.; Marty-Santos, L.; Newman, M.; Lukacs, D.T.; Spence, J.R.; Wellik, D.M. Hox6 genes modulate in vitro differentiation of mESCs to insulin-producing cells. In Vitro Cell. Dev. Biol. Anim. 2016, 52, 974–982. [Google Scholar] [CrossRef]
- Sagi, B.; Maraghechi, P.; Urban, V.S.; Hegyi, B.; Szigeti, A.; Fajka-Boja, R.; Kudlik, G.; Nemet, K.; Monostori, E.; Gocza, E.; et al. Positional identity of murine mesenchymal stem cells resident in different organs is determined in the postsegmentation mesoderm. Stem Cells Dev. 2012, 21, 814–828. [Google Scholar] [CrossRef] [PubMed]
- Ackema, K.B.; Charite, J. Mesenchymal stem cells from different organs are characterized by distinct topographic Hox codes. Stem Cells Dev. 2008, 17, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, S.; Buchheiser, A.; Bosch, J.; Bosse, F.; Kruse, F.; Zhao, X.; Santourlidis, S.; Kogler, G. The HOX Code as a “biological fingerprint” to distinguish functionally distinct stem cell populations derived from cord blood. Stem Cell Res. 2010, 5, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Leucht, P.; Kim, J.B.; Amasha, R.; James, A.W.; Girod, S.; Helms, J.A. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development 2008, 135, 2845–2854. [Google Scholar] [CrossRef]
- Alharbi, R.A.; Pettengell, R.; Pandha, H.S.; Morgan, R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 2013, 27, 1000–1008. [Google Scholar] [CrossRef]
- Collins, E.M.; Thompson, A. HOX genes in normal, engineered and malignant hematopoiesis. Int. J. Dev. Biol. 2018, 62, 847–856. [Google Scholar] [CrossRef]
- Hassan, M.Q.; Tare, R.; Lee, S.H.; Mandeville, M.; Weiner, B.; Montecino, M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol. Cell. Biol. 2007, 27, 3337–3352. [Google Scholar] [CrossRef]
- Myers, C.; Charboneau, A.; Cheung, I.; Hanks, D.; Boudreau, N. Sustained expression of homeobox D10 inhibits angiogenesis. Am. J. Pathol. 2002, 161, 2099–2109. [Google Scholar] [CrossRef]
- Seifert, A.; Werheid, D.F.; Knapp, S.M.; Tobiasch, E. Role of Hox genes in stem cell differentiation. World J. Stem Cells 2015, 7, 583–595. [Google Scholar] [CrossRef]
- Briscoe, J.; Wilkinson, D.G. Establishing neuronal circuitry: Hox genes make the connection. Genes Dev. 2004, 18, 1643–1648. [Google Scholar] [CrossRef][Green Version]
- Shah, N.; Sukumar, S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. Role of HOX Genes in Stem Cell Differentiation and Cancer. Stem Cells Int. 2018, 2018, 3569493. [Google Scholar] [CrossRef]
- Abate-Shen, C. Deregulated homeobox gene expression in cancer: Cause or consequence? Nat. Rev. Cancer 2002, 2, 777–785. [Google Scholar] [CrossRef]
- Eoh, K.J.; Kim, H.J.; Lee, J.Y.; Nam, E.J.; Kim, S.; Kim, S.W.; Kim, Y.T. Upregulation of homeobox gene is correlated with poor survival outcomes in cervical cancer. Oncotarget 2017, 8, 84396–84402. [Google Scholar] [CrossRef][Green Version]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. (Berl.) 2014, 92, 811–823. [Google Scholar] [CrossRef]
- Ben Khadra, Y.; Said, K.; Thorndyke, M.; Martinez, P. Homeobox genes expressed during echinoderm arm regeneration. Biochem. Genet. 2014, 52, 166–180. [Google Scholar] [CrossRef]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Pastushenko, I.; Skibinski, A.; Blanpain, C.; Kuperwasser, C. Phenotypic Plasticity: Driver of Cancer Initiation, Progression, and Therapy Resistance. Cell Stem Cell 2019, 24, 65–78. [Google Scholar] [CrossRef]
- Campbell, L.L.; Polyak, K. Breast tumor heterogeneity: Cancer stem cells or clonal evolution? Cell Cycle 2007, 6, 2332–2338. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Twomey, D.; Feng, Z.; Stubbs, M.C.; Wang, Y.; Faber, J.; Levine, J.E.; Wang, J.; Hahn, W.C.; Gilliland, D.G.; et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006, 442, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Da Silva-Diz, V.; Lorenzo-Sanz, L.; Bernat-Peguera, A.; Lopez-Cerda, M.; Munoz, P. Cancer cell plasticity: Impact on tumor progression and therapy response. Semin. Cancer Biol. 2018, 53, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, E.N.; Scaffidi, P. Epigenetics and Cancer Stem Cells: Unleashing, Hijacking, and Restricting Cellular Plasticity. Trends Cancer 2017, 3, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.F.; Esteves, C.M.; Xavier, F.C.; Nunes, F.D. Methylation status of homeobox genes in common human cancers. Genomics 2016, 108, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Cardenas, R.; Wang, B.; Persson, J.; Mongan, N.P.; Grabowska, A.; Allegrucci, C. HOXC8 regulates self-renewal, differentiation and transformation of breast cancer stem cells. Mol. Cancer 2017, 16, 19. [Google Scholar] [CrossRef] [PubMed]
- Tabuse, M.; Ohta, S.; Ohashi, Y.; Fukaya, R.; Misawa, A.; Yoshida, K.; Kawase, T.; Saya, H.; Thirant, C.; Chneiweiss, H.; et al. Functional analysis of HOXD9 in human gliomas and glioma cancer stem cells. Mol. Cancer 2011, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Gallo, M.; Ho, J.; Coutinho, F.J.; Vanner, R.; Lee, L.; Head, R.; Ling, E.K.; Clarke, I.D.; Dirks, P.B. A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res. 2013, 73, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Qian, P.; De Kumar, B.; He, X.C.; Nolte, C.; Gogol, M.; Ahn, Y.; Chen, S.; Li, Z.; Xu, H.; Perry, J.M.; et al. Retinoid-Sensitive Epigenetic Regulation of the Hoxb Cluster Maintains Normal Hematopoiesis and Inhibits Leukemogenesis. Cell Stem Cell 2018, 22, 740.e7–754.e7. [Google Scholar] [CrossRef] [PubMed]
- Castro-Oropeza, R.; Melendez-Zajgla, J.; Maldonado, V.; Vazquez-Santillan, K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell. Oncol. (Dordr.) 2018, 41, 585–603. [Google Scholar] [CrossRef] [PubMed]
- Padua Alves, C.; Fonseca, A.S.; Muys, B.R.; de Barros, E.L.B.R.; Burger, M.C.; de Souza, J.E.; Valente, V.; Zago, M.A.; Silva, W.A., Jr. Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells 2013, 31, 2827–2832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014, 32, 2858–2868. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; An, J.; Wu, M.; Zheng, Q.; Gui, X.; Li, T.; Pu, H.; Lu, D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget 2015, 6, 27847–27864. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, F.; Xu, Y.; Wang, B.; Zhao, Y.; Xu, W.; Shi, L.; Lu, X.; Liu, Q. Epithelial-mesenchymal transition and cancer stem cells, mediated by a long non-coding RNA, HOTAIR, are involved in cell malignant transformation induced by cigarette smoke extract. Toxicol. Appl. Pharmacol. 2015, 282, 9–19. [Google Scholar] [CrossRef]
- Fang, K.; Liu, P.; Dong, S.; Guo, Y.; Cui, X.; Zhu, X.; Li, X.; Jiang, L.; Liu, T.; Wu, Y. Magnetofection based on superparamagnetic iron oxide nanoparticle-mediated low lncRNA HOTAIR expression decreases the proliferation and invasion of glioma stem cells. Int. J. Oncol. 2016, 49, 509–518. [Google Scholar] [CrossRef]
- Fu, Z.; Chen, C.; Zhou, Q.; Wang, Y.; Zhao, Y.; Zhao, X.; Li, W.; Zheng, S.; Ye, H.; Wang, L.; et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017, 410, 68–81. [Google Scholar] [CrossRef]
- Bhatlekar, S.; Ertel, A.; Gonye, G.E.; Fields, J.Z.; Boman, B.M. Gene expression signatures for HOXA4, HOXA9, and HOXD10 reveal alterations in transcriptional regulatory networks in colon cancer. J. Cell. Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Goolsby, C.; Yaseen, N.R. NUP98-HOXA9 induces long-term proliferation and blocks differentiation of primary human CD34+ hematopoietic cells. Cancer Res. 2006, 66, 6628–6637. [Google Scholar] [CrossRef] [PubMed]
- Velu, C.S.; Chaubey, A.; Phelan, J.D.; Horman, S.R.; Wunderlich, M.; Guzman, M.L.; Jegga, A.G.; Zeleznik-Le, N.J.; Chen, J.; Mulloy, J.C.; et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J. Clin. Investig. 2014, 124, 222–236. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Fu, L.; Xie, C.; Zuo, W.S.; Liu, Y.S.; Zheng, M.Z.; Yu, J.M. miR-375 inhibits cancer stem cell phenotype and tamoxifen resistance by degrading HOXB3 in human ER-positive breast cancer. Oncol. Rep. 2017, 37, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Teo, W.W.; Merino, V.F.; Cho, S.; Korangath, P.; Liang, X.; Wu, R.C.; Neumann, N.M.; Ewald, A.J.; Sukumar, S. HOXA5 determines cell fate transition and impedes tumor initiation and progression in breast cancer through regulation of E-cadherin and CD24. Oncogene 2016, 35, 5539–5551. [Google Scholar] [CrossRef]
- Ordonez-Moran, P.; Dafflon, C.; Imajo, M.; Nishida, E.; Huelsken, J. HOXA5 Counteracts Stem Cell Traits by Inhibiting Wnt Signaling in Colorectal Cancer. Cancer Cell 2015, 28, 815–829. [Google Scholar] [CrossRef]
- Bhatlekar, S.; Viswanathan, V.; Fields, J.Z.; Boman, B.M. Overexpression of HOXA4 and HOXA9 genes promotes self-renewal and contributes to colon cancer stem cell overpopulation. J. Cell. Physiol. 2018, 233, 727–735. [Google Scholar] [CrossRef]
- Haria, D.; Naora, H. Homeobox Gene Deregulation: Impact on the Hallmarks of Cancer. Cancer Hallm. 2013, 1, 67–76. [Google Scholar] [CrossRef][Green Version]
- Wu, X.; Chen, H.; Parker, B.; Rubin, E.; Zhu, T.; Lee, J.S.; Argani, P.; Sukumar, S. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006, 66, 9527–9534. [Google Scholar] [CrossRef]
- Cai, J.Q.; Xu, X.W.; Mou, Y.P.; Chen, K.; Pan, Y.; Wu, D. Upregulation of HOXB7 promotes the tumorigenesis and progression of gastric cancer and correlates with clinical characteristics. Tumour Biol. 2016, 37, 1641–1650. [Google Scholar] [CrossRef]
- Liao, W.T.; Jiang, D.; Yuan, J.; Cui, Y.M.; Shi, X.W.; Chen, C.M.; Bian, X.W.; Deng, Y.J.; Ding, Y.Q. HOXB7 as a prognostic factor and mediator of colorectal cancer progression. Clin. Cancer Res. 2011, 17, 3569–3578. [Google Scholar] [CrossRef] [PubMed]
- Topisirovic, I.; Kentsis, A.; Perez, J.M.; Guzman, M.L.; Jordan, C.T.; Borden, K.L. Eukaryotic translation initiation factor 4E activity is modulated by HOXA9 at multiple levels. Mol. Cell. Biol. 2005, 25, 1100–1112. [Google Scholar] [CrossRef]
- Rubin, E.; Wu, X.; Zhu, T.; Cheung, J.C.; Chen, H.; Lorincz, A.; Pandita, R.K.; Sharma, G.G.; Ha, H.C.; Gasson, J.; et al. A role for the HOXB7 homeodomain protein in DNA repair. Cancer Res. 2007, 67, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Kong, X.; Shah, T.; Penet, M.F.; Wildes, F.; Sgroi, D.C.; Ma, X.J.; Huang, Y.; Kallioniemi, A.; Landberg, G.; et al. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 2736–2741. [Google Scholar] [CrossRef]
- Raman, V.; Martensen, S.A.; Reisman, D.; Evron, E.; Odenwald, W.F.; Jaffee, E.; Marks, J.; Sukumar, S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature 2000, 405, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.S.; Li, B.; Xu, G.; Yang, S.Q.; Wang, P.; Tang, H.H.; Liu, Y.Y. Long noncoding RNA LINC00483/microRNA-144 regulates radiosensitivity and epithelial-mesenchymal transition in lung adenocarcinoma by interacting with HOXA10. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef]
- Yoshida, H.; Broaddus, R.; Cheng, W.; Xie, S.; Naora, H. Deregulation of the HOXA10 homeobox gene in endometrial carcinoma: Role in epithelial-mesenchymal transition. Cancer Res. 2006, 66, 889–897. [Google Scholar] [CrossRef]
- Dang, C.V.; Reddy, E.P.; Shokat, K.M.; Soucek, L. Drugging the ‘undruggable’ cancer targets. Nat. Rev. Cancer 2017, 17, 502–508. [Google Scholar] [CrossRef]
- Lappin, T.R.; Grier, D.G.; Thompson, A.; Halliday, H.L. HOX genes: Seductive science, mysterious mechanisms. Ulster Med. J. 2006, 75, 23–31. [Google Scholar] [PubMed]
- Luo, Z.; Rhie, S.K.; Farnham, P.J. The Enigmatic HOX Genes: Can We Crack Their Code? Cancers 2019, 11, 323. [Google Scholar] [CrossRef]
- Morgan, R.; El-Tanani, M.; Hunter, K.D.; Harrington, K.J.; Pandha, H.S. Targeting HOX/PBX dimers in cancer. Oncotarget 2017, 8, 32322–32331. [Google Scholar] [CrossRef] [PubMed]
- Northcott, J.M.; Northey, J.J.; Barnes, J.M.; Weaver, V.M. Fighting the force: Potential of homeobox genes for tumor microenvironment regulation. Biochim. Biophys. Acta 2015, 1855, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, K.; Arderiu, G.; Charboneau, A.; Hansen, S.L.; Hoffman, W.; Boudreau, N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat. Res. Biol. 2005, 3, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Carrio, M.; Arderiu, G.; Myers, C.; Boudreau, N.J. Homeobox D10 induces phenotypic reversion of breast tumor cells in a three-dimensional culture model. Cancer Res. 2005, 65, 7177–7185. [Google Scholar] [CrossRef]
- Harris, W.J.; Huang, X.; Lynch, J.T.; Spencer, G.J.; Hitchin, J.R.; Li, Y.; Ciceri, F.; Blaser, J.G.; Greystoke, B.F.; Jordan, A.M.; et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012, 21, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Nowak, D.; Stewart, D.; Koeffler, H.P. Differentiation therapy of leukemia: 3 decades of development. Blood 2009, 113, 3655–3665. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.; El-Tanani, M. HOX Genes as Potential Markers of Circulating Tumour Cells. Curr. Mol. Med. 2016, 16, 322–327. [Google Scholar] [CrossRef]
| HOX Genes and lncRNA | CSC Type | Function |
|---|---|---|
| HOXA5 | Breast | Silencing of HOXA5 induces loss of differentiation [85] |
| Colon | Silencing of HOXA5 maintains the pool of CSC [86] | |
| HOXA4 | Colon | Expression of HOXA4 induces CSC self-renewal [87] |
| HOXA9 | AML | HOXA9 and NUP98-HOXA9 fusion protein sustain self-renewal and impair differentiation of CSC [82,83] |
| Colon | Expression of HOXA9 induces CSC self-renewal [87] | |
| HOXA10 | Glioblastoma | Activation of HOXA10 by the TrxG protein MLL induces tumorigenicity of CSC [72] |
| HOXB cluster | Leukemia | Expression of HOXB genes induces expansion of CSC [73] |
| HOXB3 | Breast | Expression of HOXB3 sustains proliferation of drug resistance of CSC [84] |
| HOXC8 | Breast | Silencing of HOXC8 sustains self-renewal and impairs differentiation of CSC [70] |
| HOXD9 | Glioma | Silencing of HOXD9 induces self-renewal and survival of CSC [71] |
| HOTAIR | Colon | Expression of HOTAIR induces EMT and stemness [75] |
| Breast | Expression of HOTAIR induces EMT and stemness through activation of HOXD10 [76] | |
| Lung | Expression of HOTAIR induces EMT and stemness [78] | |
| Liver | Expression of HOTAIR induces EMT and stemness [77] | |
| Glioma | Expression of HOTAIR induces proliferation and invasion of CSC [79] | |
| HOTTIP | Pancreas | Expression of HOTTIP induces CSC proliferation by induction of HOXA9 [80] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, J.; Zyoud, A.; Allegrucci, C. A Case of Identity: HOX Genes in Normal and Cancer Stem Cells. Cancers 2019, 11, 512. https://doi.org/10.3390/cancers11040512
Smith J, Zyoud A, Allegrucci C. A Case of Identity: HOX Genes in Normal and Cancer Stem Cells. Cancers. 2019; 11(4):512. https://doi.org/10.3390/cancers11040512
Chicago/Turabian StyleSmith, Jessica, Ahmad Zyoud, and Cinzia Allegrucci. 2019. "A Case of Identity: HOX Genes in Normal and Cancer Stem Cells" Cancers 11, no. 4: 512. https://doi.org/10.3390/cancers11040512
APA StyleSmith, J., Zyoud, A., & Allegrucci, C. (2019). A Case of Identity: HOX Genes in Normal and Cancer Stem Cells. Cancers, 11(4), 512. https://doi.org/10.3390/cancers11040512





