1. Introduction
The histone demethylase Ubiquitously Transcribed Tetratricopeptide Repeat Protein X-Linked (UTX, also known as Lysine demethylase 6A, gene name
KDM6A) demethylates K27me2/3 on histone 3 (H3), which is usually associated with gene activation [
1]. In most instances UTX acts in conjunction with the COMPASS complex [
2]. This complex, which mediates H3K4 methylation, comprises a core complex termed WRAD (WDR5, RBBP5, ASH2L and DPY30) and the MLL3 (KMT2C) or MLL2/4 (KMT2D) proteins [
3].
UTX is involved in many different human cancers in different ways [
1,
4]. In some cancer types, UTX exerts pro-tumorigenic effects. For instance, in estrogen receptor-dependent breast cancer UTX serves as a coactivator that facilitates gene activation by the estrogen-receptor α and its co-transcription factors [
5]. In other cancer types, UTX function is compromised or abolished by deleterious mutations in the
KDM6A gene. While the consequences of UTX inactivation are not fully understood, studies on hematological malignancies and pancreatic carcinoma, among others, suggest that it leads to a general increase in H3K27 trimethylation, a redistribution of H3K4 methylation, altered activity of enhancers, and ultimately gene transcription patterns [
6,
7]. The function of UTX may even differ between subtypes of one disease. For instance, UTX is firmly established as a tumor suppressor in T-ALL, but acts as a protumorigenic coactivator in the TAL1-driven subtype [
8,
9].
Among all cancer types, deleterious
KDM6A mutations are most frequent in urothelial carcinoma (UC), the most common histological type of urinary bladder cancer. UC is categorized into non-muscle-invasive and more aggressive muscle-invasive tumors, each of which comprise several molecular subtypes [
10]. Subtyping of UC is thought to contribute to improved selection of patients for chemotherapy and immunotherapeutic agents. While
KDM6A mutations are found across all UC stages and subtypes, their frequency varies and is particularly high in non-muscle-invasive low-grade papillary tumors, with up to 70% in female patients [
11,
12], whereas the frequency is around 25% in muscle-invasive tumors [
4]. A few previous publications have addressed the functional consequences of
KDM6A inactivation in UC, with partly discrepant results. Nickerson et al. [
13] observed no differences in monolayer growth of T-24-T cells with a homozygous
KDM6A nonsense mutation following transfection of a UTX expression plasmid. Likewise, shRNA-mediated downregulation of wild-type UTX in MGH-U3 cells did not affect short-term proliferation. However, colony formation was affected by both treatments. Ahn et al. [
14] found an increase in cell proliferation over nine days following
KDM6A knockout in two UC cell lines (HT-1197 and UM-UC-3). Ler et al. [
15] observed a slight increase in proliferation following
KDM6A knockout in the papillary UC cell line RT-4, whereas restoration of
KDM6A in the Ku-19-19 cell line, which has a homozygous
KDM6A mutation, did not appear to affect the basal proliferative activity of this cell line. Interestingly, UC cell lines with constitutive or engineered
KDM6A inactivation were more sensitive to inhibitors of the H3K27 methyltransferase EZH2, the UTX antagonist [
15].
In preliminary work, we had observed quite variable responses of UC cell lines to plasmid-based overexpression or knockdown of UTX, especially with respect to long-term clonogenicity. Given the heterogeneous character of UC, we were therefore interested in extending previous observations to further cell lines to map out the heterogeneity of UTX action in this cancer type. To this end, we used a lentiviral vector to transduce UTX fused C-terminally to TagGFP2 or TagGFP2 only into various UC cell lines (UCCs) with different endogenous KDM6A status, as well as into a newly engineered UC KDM6A knockout cell line and into a non-transformed immortalized urothelial cell line. Among these cell line pairs, we then compared their proliferation rates, clone formation ability, histone modifications and gene expression patterns. We report that responses to UTX expression vary widely among the cell lines and appear strongly contingent on their genetic constitution. In particular, we identify nuclear localization of UTX, which may depend on other COMPASS components, as a new critical point in its action.
3. Discussion
The high prevalence of evidently deleterious
KDM6A mutations throughout all stages and molecular subtypes of UC suggests an important tumor-suppressive function of UTX. Unfortunately, the nature of that function has remained rather elusive to date. Concordant with findings of previous publications [
13,
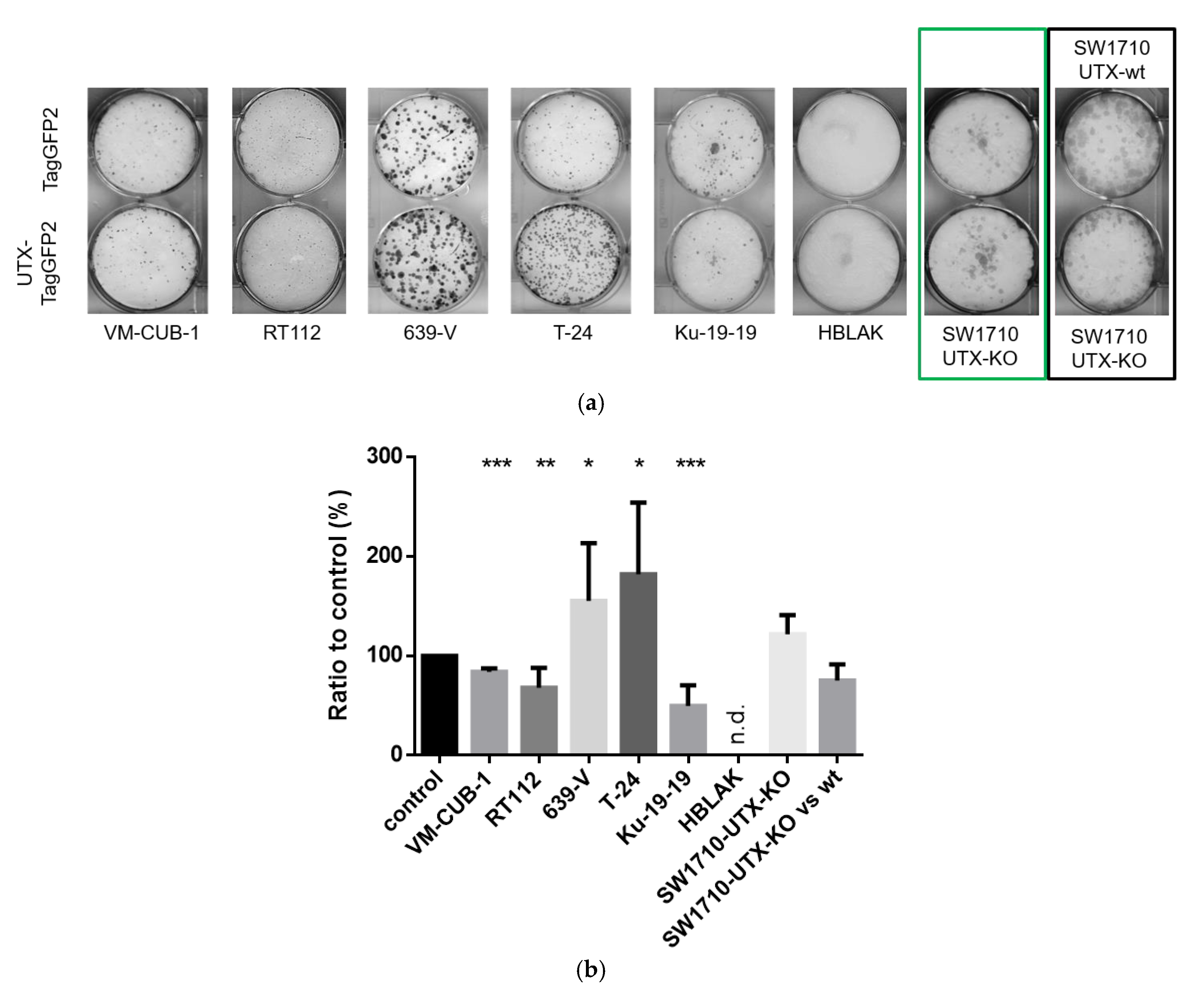
15], we observed that introduction of UTX did not affect short-term growth of UC cell lines, but in several cell lines impeded long-term proliferation as measured by clone formation assays. These repeated findings suggest that the tumor-suppressive function of UTX in UC is not exerted by fast effects on cell proliferation or survival, as observed during introduction of classical tumor suppressors like p53, but by long-term effects, as one might expect for an epigenetic regulator. We noted however considerable differences between individual UC cell lines in their response to introduced UTX. Obviously, its effects are contingent on several factors in the cell lines.
First, as one might expect, the effects of exogenous UTX appeared to depend on the status of its endogenous counterpart. Thus, the introduction of functional UTX in cell lines entirely lacking functional protein, such as Ku-19-19, or with hypomorphic genotypes, such as RT112 and VM-CUB-1, effectively diminished clonogenicity. Intriguingly, however, the ability of T-24 cells to form clones was not impaired by UTX. In contrast to this latter finding, Nickerson et al. [
13] reported a decreased long-term growth ability of T24-T cells, a T-24 subclone with the same homozygous
KDM6A nonsense mutation, following transient transfection with a UTX-expression plasmid. One explanation for our finding is that the original T-24 cell line may contain additional genetic and epigenetic changes that allow it to render the action of UTX irrelevant for its long-term growth ability.
A second contingency is the status of the
KMT2C and
KMT2D genes. In UC, mutations in either of these genes are almost mutually exclusive with those in
KDM6A [
4]. This observation indicates that the three proteins, MLL2/4 or MLL3 and UTX, cooperate in the maintenance of a normal state in the urothelium, likely as components of the COMPASS complex. In tumors with
KMT2C or
KMT2D inactivation, therefore, the level of UTX expression may be rather irrelevant. This situation is exemplified by the 639-V cell line, which has deleterious mutations in three out of four
KMT2C and
KMT2D alleles, but is wild-type for UTX. It is therefore not unexpected that introduction of additional UTX does not impede cell growth in this cell line, or may even promote it.
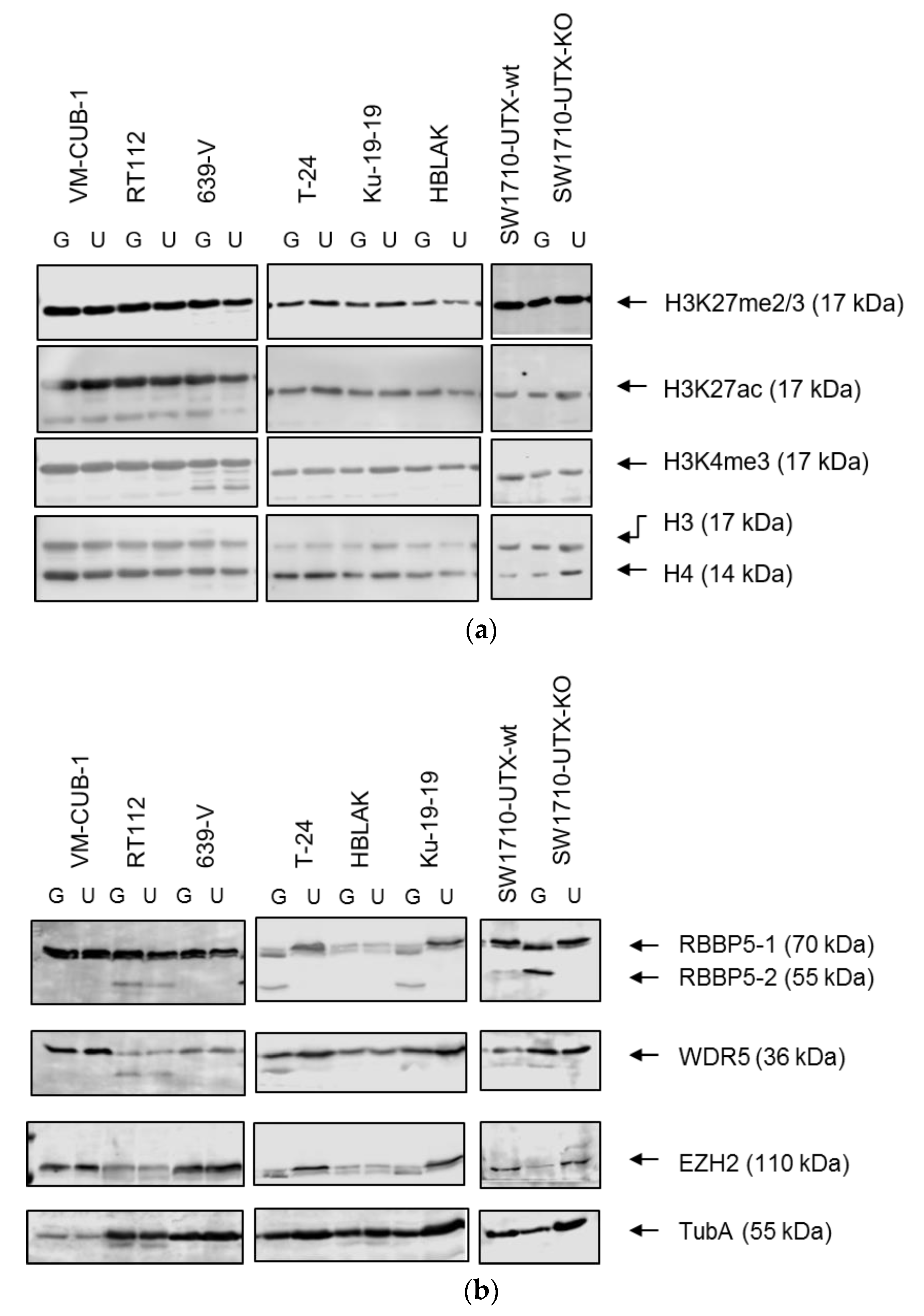
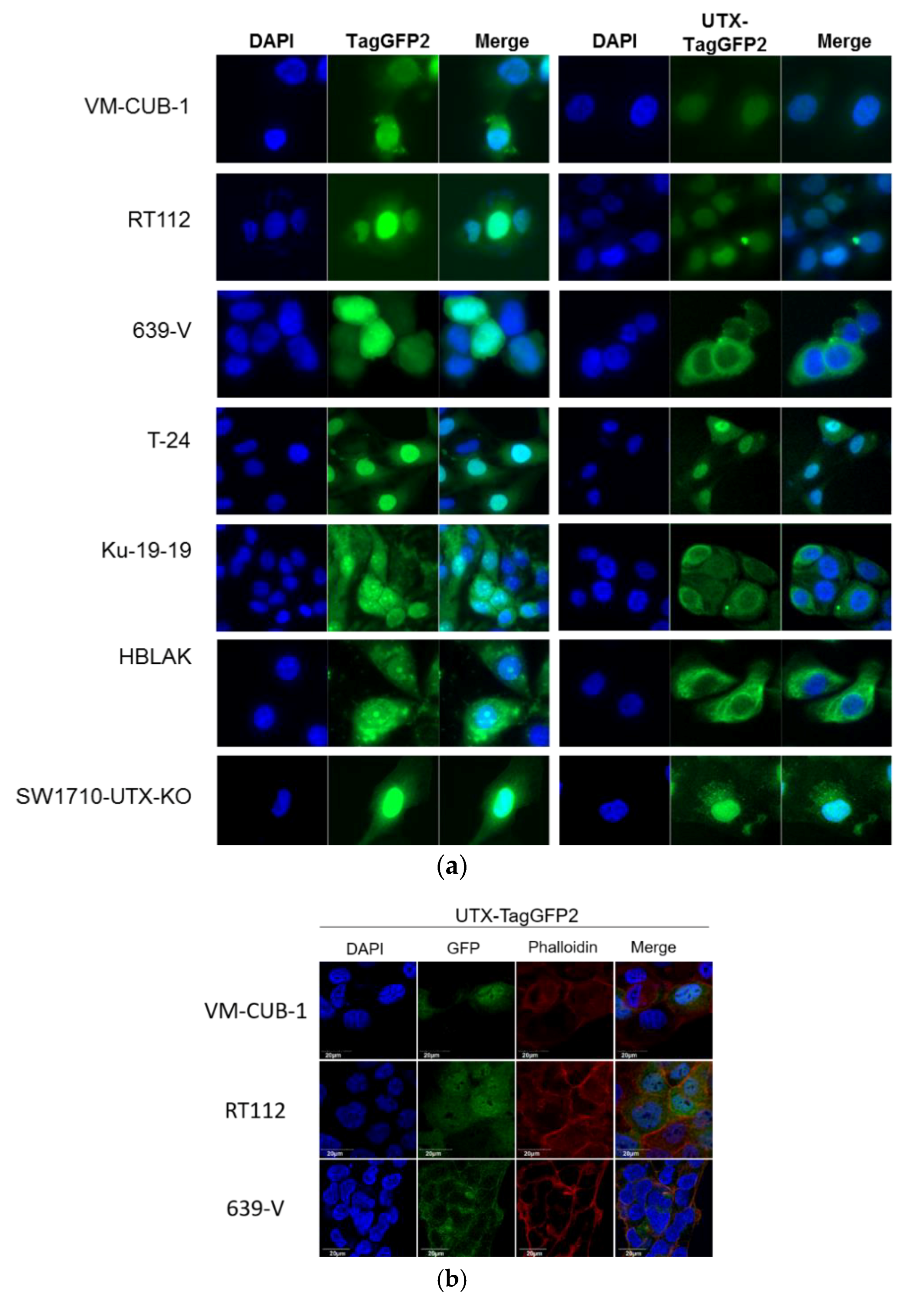
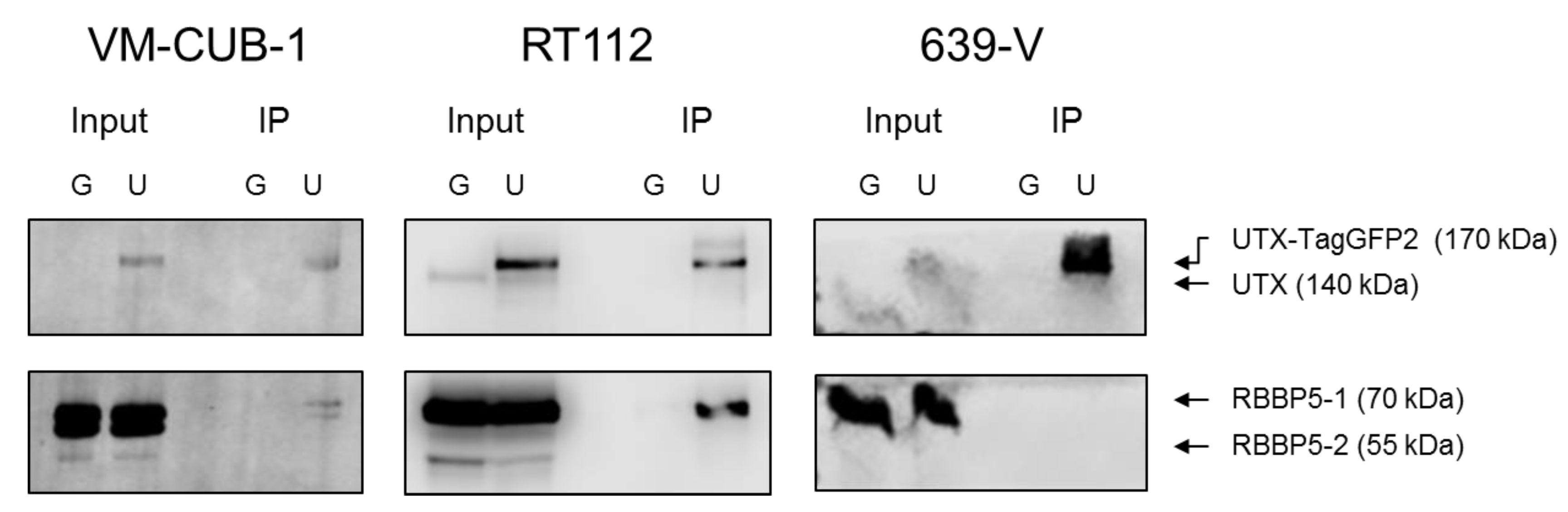
A novel finding in our study is that exogenous UTX in the 639-V cell line is not localized in the nucleus, but rather in the cytoplasm. In fact, biochemical fractionation demonstrated that the majority of endogenous UTX also localizes to the cytoplasm. Moreover, UTX in this cell line does not co-precipitate with RBBP5, a core component of the COMPASS complex. Taken together, these observations suggest that the COMPASS complex does not properly assemble in 639-V cells and UTX is mislocalized to the cytoplasm.
Recently, Wiedemuth et al. have drawn attention to the fact that extraneous UTX does not always localize to the nucleus, even in normal cells [
19]. We observed this phenomenon, too, in HBLAK cells, a spontaneously immortalized urothelial cell line with a few genomic changes related to immortalization [
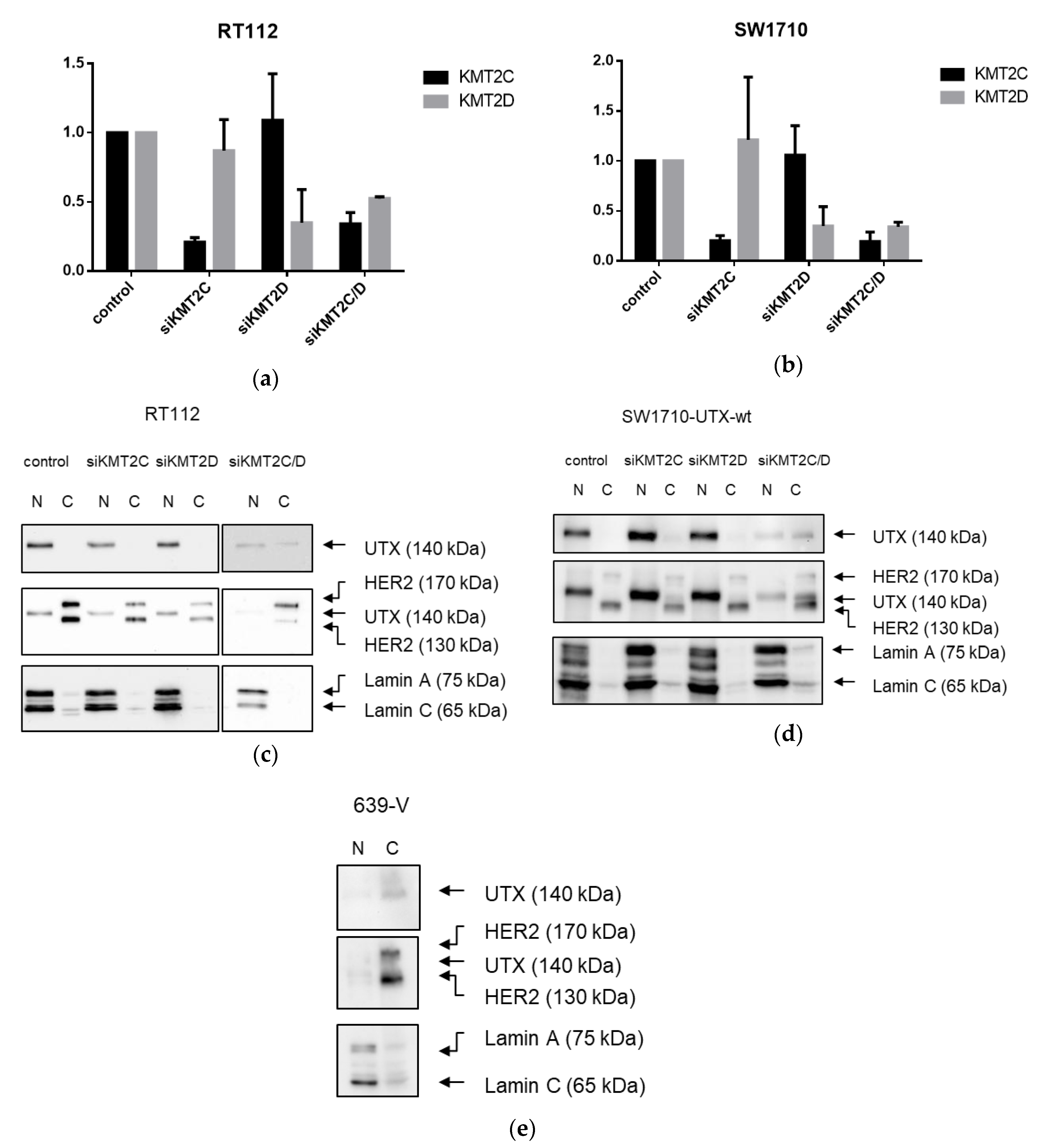
21], but without mutations in any COMPASS gene. We also note that the UTX protein sequence contains no obvious nuclear localization sequence. Therefore, UTX transport to and retention in the nucleus may depend on further interaction partners. Here, we provide evidence that either of the MLL proteins encoded by KMT2C and KMT2D may serve this function. In particular, knockdown of both factors, combined, but not individually, diminished the fraction of UTX in the nucleus and increased its amount in the cytoplasm. Unfortunately, we could not obtain antibodies specific and sensitive enough to detect endogenous levels of KMT2C and KMT2D proteins in the UCCs. Therefore, we could not evaluate the efficiency of the knockdowns at the protein level. Since the decrease in KMT2C and KMT2D mRNAs was significant, but not complete, it is possible that relevant amounts of these proteins remain after siRNA treatment and further decrease might lead to further exclusion of UTX from the nucleus, as suggested by the observations in the 639-V cell line.
The findings of Wiedemuth et al. [
19] and our own thus hint at the possibility that cells restrict the amount of UTX in their nuclei. Experimentally increased levels of UTX may only saturate the available interaction sites, and additional UTX protein may be retained in the cytoplasm, or become extruded from the nucleus. Taken together, these observations indicate the possibility that transport into the nucleus may constitute another layer of regulation of UTX.
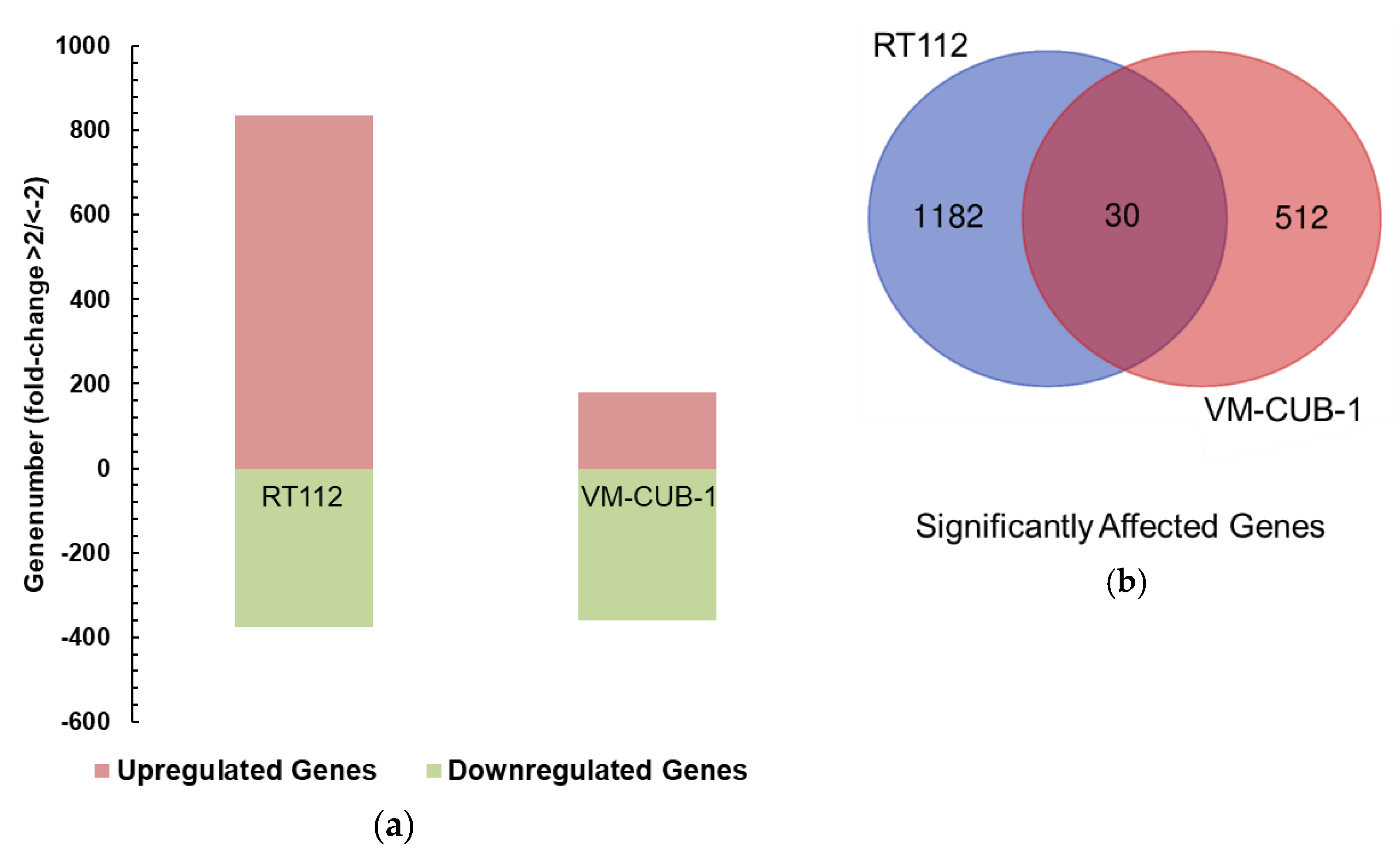
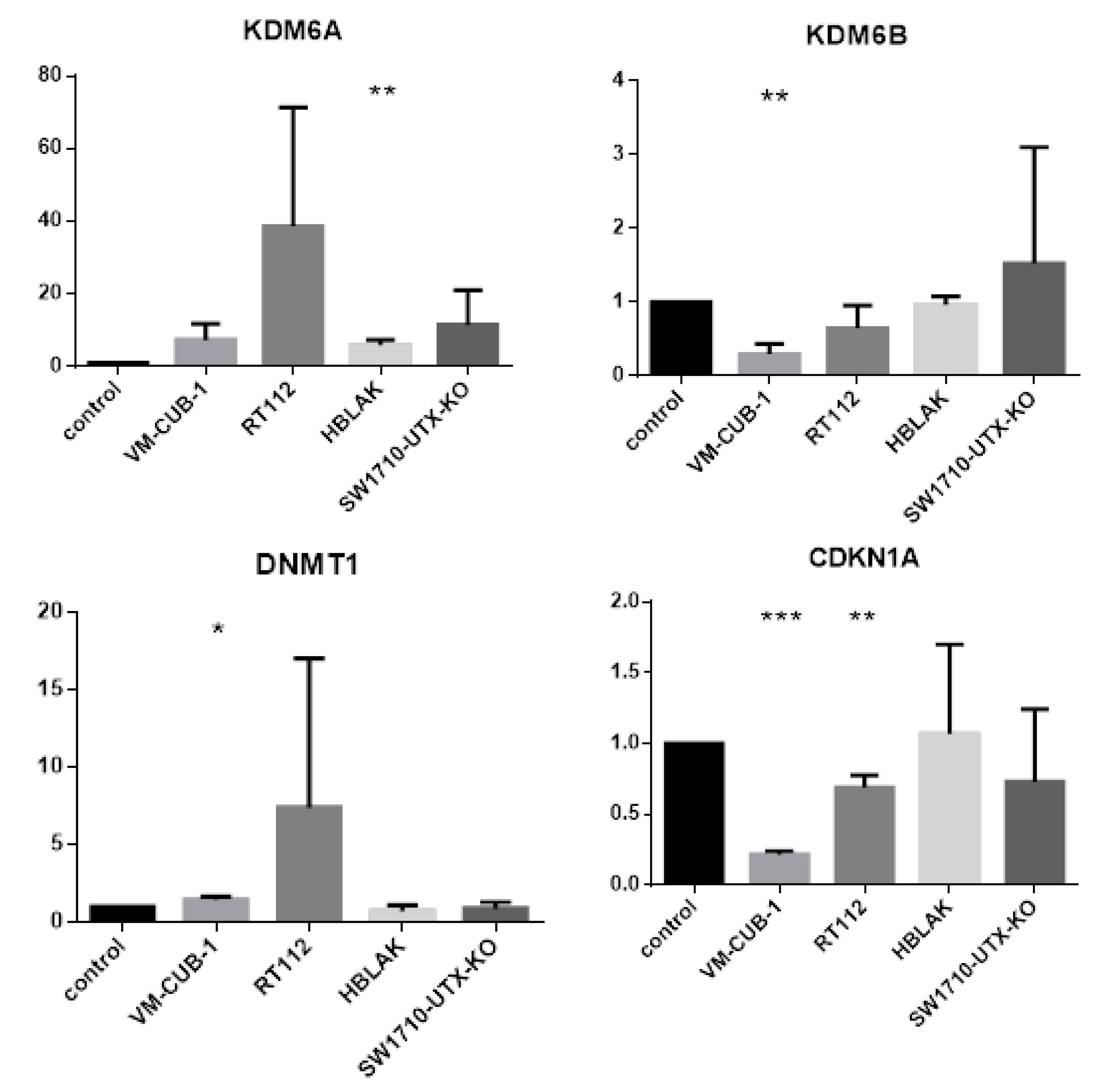
A limit to the amount of UTX in the nucleus in normal cells could in particular account for the observation that expression of additional UTX did not significantly alter gene expression in HBLAK cells. In contrast, significant effects on gene expression were observed in tumor cell lines with likely suboptimal levels of functional UTX, such as RT112 and VM-CUB-1, and upon reintroduction of UTX into SW-1710 KDM6A knockout cells. These changes were observed, even though the relevant global histone modification levels were not appreciably altered, suggesting that they were gene-specific.
Between the UCCs, the overlap between the differentially expressed gene sets was limited and not larger than expected by chance. Despite this reservation, the intersecting set between RT112 and VM-CUB-1 contained several genes that deserve further investigation as potential UTX target genes. For instance,
DNMT1 encodes the major cellular DNA maintenance methyltransferase and
CDKN1A encodes the crucial cell cycle inhibitor p21
CIP1. Inverse regulation of
KDM6A/UTX and the paralogous histone demethylase
KDM6B/JMJD3 has been observed in other cancers [
22]. Canonical HOX genes are classical, albeit cell type-specific targets of UTX during development [
23,
24]. Between the two cell lines, only one HOX gene (namely
HOXC4) was regulated in common by UTX-TagGFP2, whereas others were only affected in RT112 cells.
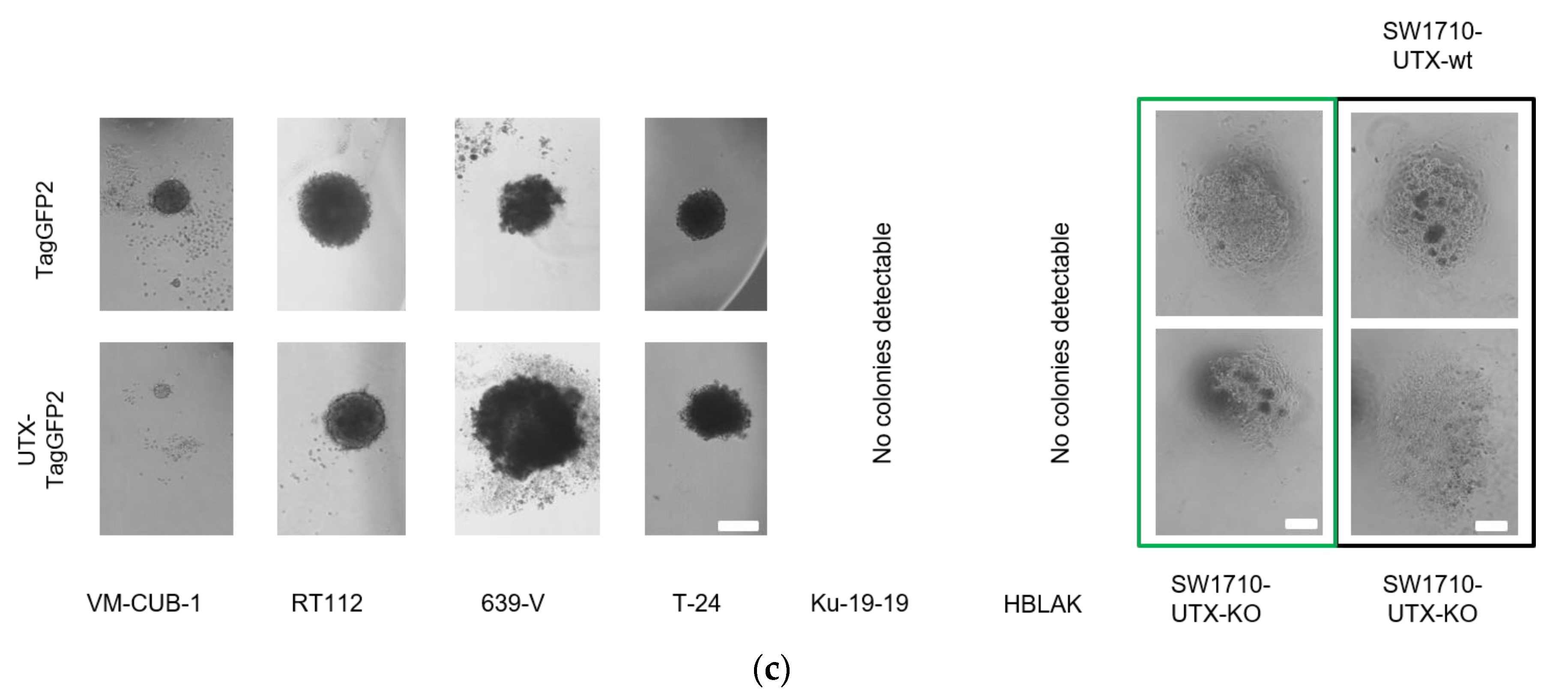
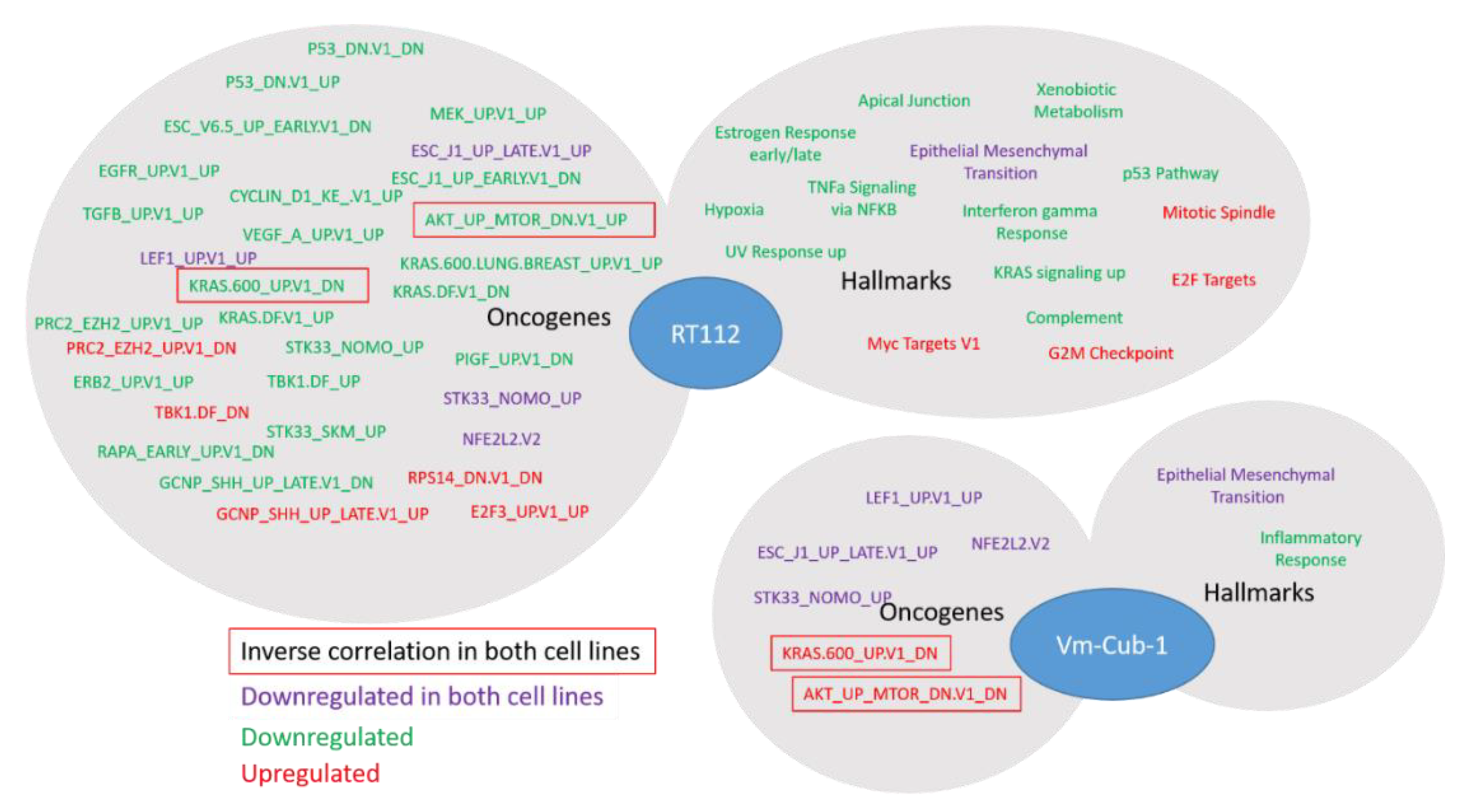
Whereas the gene expression changes at individual genes were mostly divergent, closer investigation by GSEA revealed several common biological functions. In particular, UTX appeared to negatively regulate genes associated with the epithelial-mesenchymal transition in both RT112 and VM-CUB-1. These changes in gene expression correspond to changes in morphology, namely the more compact colonies observed in 3D and 2D cultures, respectively. Accordingly, Nickerson et al. [
13] reported decreased invasion and migration following transfection of UTX into T24-T cells. Differentially expressed KEGG gene sets between
KDM6A-mutated and wildtype bladder tumors identified by Ler et al. [
15] also include prominently ECM_Receptor Interaction, Focal_Adhesion and Cell-Adhesion_Molecules. The possible regulation of cell morphology and adhesion in UC by UTX thus deserves further study. Another interesting observation in our study is that many gene sets, although not individual genes, upregulated by UTX in both RT112 and VM-CUB-1 relate to RNA processing. This may reflect an additional function of UTX during transcription which has been observed in some previous studies [
25] but is not well understood in the context of cancer.
The study by Ler et al. [
15] has highlighted the effects of the
KDM6A status of UC tissues and cell lines on genes repressed by EZH2 and the PRC2 polycomb complex, in keeping with the antagonism between EZH2 and UTX at the biochemical level and during development. In our study, this effect was also apparent in RT112 UTX-TagGFP2 transduced cells, but genes from the PRC2_EZH2 gene set were not uniformly up- or down-regulated. Another developmental gene set was commonly affected in RT112 and VM-CUB-1 cells, namely ESC_J1_UP-LATE.V1_UP; it contains genes upregulated during the late phase of embroid body formation from ESC cells. As for HOX genes, regulation of these genes may reflect the known developmental function of UTX. Finally, in both cell lines, a gene set regulated by the cytoprotective transcription factor NRF2 reacted to UTX-TagGFP2 expression. Generally, relatively little is known about epigenetic regulation of NRF2 action, but, interestingly, one paper reported that the transcription factor was repressed by EZH2 in lung cancer cells [
26]; an interaction with UTX has not been reported to the best of our knowledge.
Importantly, despite some similarities at the level of gene sets, our data do not support the idea that UTX affects a common set of genes across all UC. Gene expression scores for UTX have been suggested based on bioinformatical analyses of NGS data from UC tissues. For instance, like Ler et al. [
15], Dancik et al. [
27] derived an "UTX activity score" comparing
KDM6A wildtype and mutant cancers, which differs from that of Ler et al. and consists of much fewer, 27 genes. Only three of these gene were significantly changed in any of our cell line comparisons, namely
USP54 and
TNC (in RT112) as well as
C1orf21 (in SW-1710 and RT112). While cell lines may not fully reflect regulation of gene expression in vivo, this comparison suggests due caution in the application of such scores, especially in a tumor type like UC which is characterized by high heterogeneity. In particular, Ler et al. [
15] have provided evidence that
KDM6A mutations are predictive for a response of UC to inhibitors of EZH2. In that respect, our findings caution that this response might not be uniform, either.
4. Materials and Methods
4.1. Cell Lines and Cell Culture
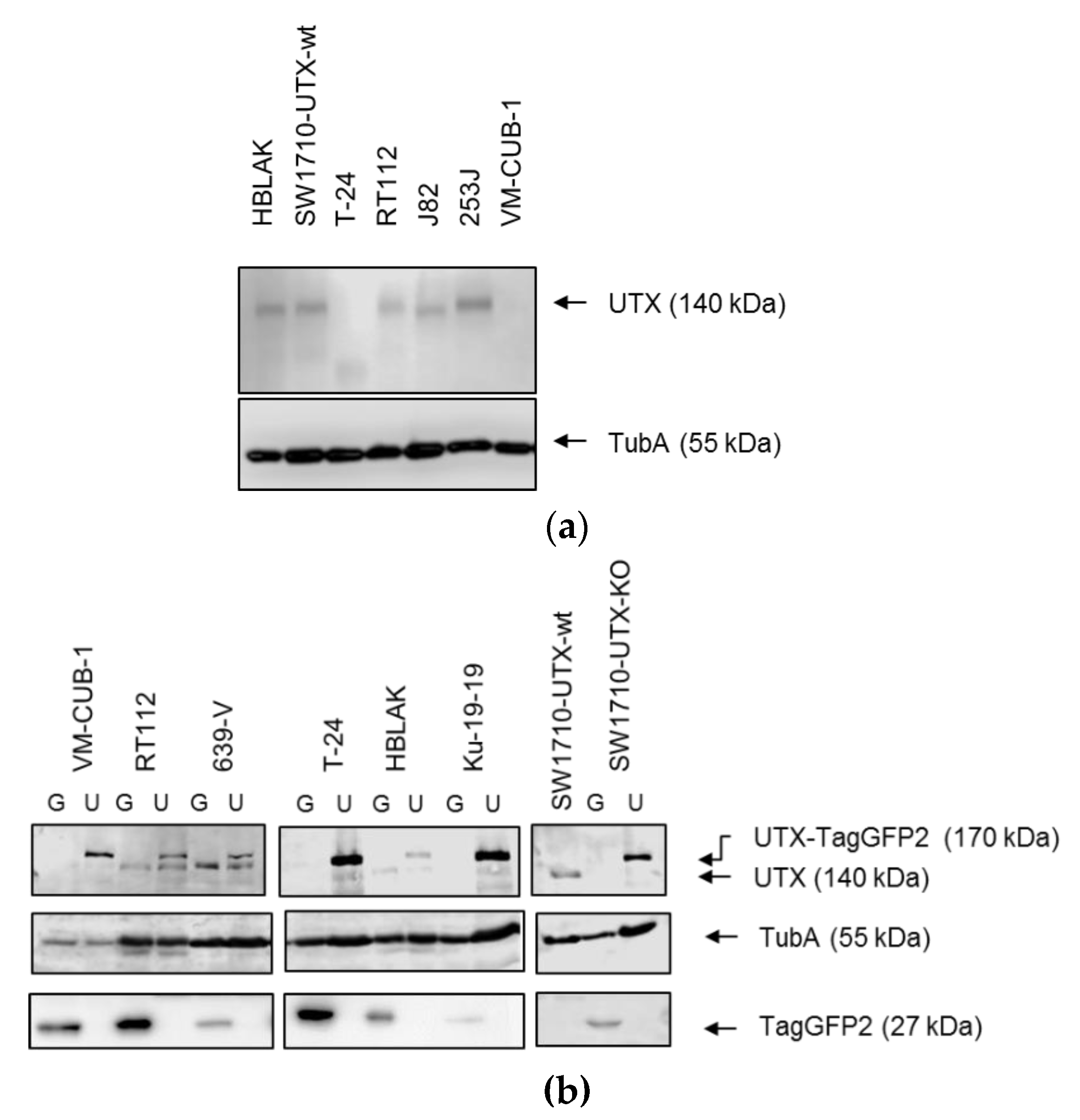
For most experiments, six different UCCs overexpressing TagGFP2 or UTX-TagGFP2 (VM-CUB1, RT112, SW1710, T24, 639-V and KU-19-19) were used. Parental UCCs were obtained from the DSMZ (Braunschweig, Germany) and Dr. H.B. Grossmann (Houston, TX, USA). For comparison, we investigated the spontaneously immortalized normal human urothelial cell line HBLAK (provided by CELLnTEC, Bern, Switzerland) [
21]. Cells were cultured and treated in DMEM GlutaMAX-I (Gibco, Darmstadt, Germany) supplemented with 10% fetal calf serum (Biochrom, Berlin, Germany), except for HBLAK cultured in CnT-Prime Epithelial Culture Medium (CELLnTEC, Bern, Switzerland; HBLAK) and KU-19-19 in RPMI-1640 (Gibco), at 37 °C and 5% CO
2. STR (short tandem repeat) profiling via DNA fingerprint analysis was performed for all cell lines.
KDM6A and
KMT2C/D genotypes were obtained from the CCLE database and ascertained by targeted PCR and Sanger sequencing. For individual cell lines, such as HBLAK, whole-exome sequencing data from other own projects [
21] was used to elucidate the mutation status.
4.2. Plasmids
KDM6A cDNA from the pLenti-C-mGFP-KDM6A plasmid purchased from OriGene (Herford, Germany, RC210861L2) was cloned into the lentiviral vector puc2CL12IPwo using standard techniques, thereby creating the vector puc2CL12IPwo-KDM6A-TagGFP2. Integrity of the KDM6A coding sequence was verified by sequencing.
4.3. Generation of Cell Lines by Lentiviral Transduction
Lentivirus production and cell transduction was performed as previously described [
28,
29]. In brief, to produce replication-deficient lentiviruses HEK-293T cells were transfected with helper plasmid expression construct (pCD/NL-BH [
30]), envelope vector (pczVSV-G [
31]) and the vector plasmids puc2CL12IPwo-TagGFP2 or puc2CL12IPwo-KDM6A-TagGFP2. Viral particles were harvested 48 h after transfection and used to infect UC cells. 24 h after transduction, the supernatant containing viral particles was removed and the transduced cells were selected with 1 µg/mL (2 µg/mL for RT112) puromycin (Invitrogen, Carlsbad, CA, USA) for seven days. Stable overexpression of UTX was confirmed by Western blot analysis of cells from several different passages. Experiments were in general conducted 2–5 passages after transduction. Cells reused after thawing were first retreated with puromycin.
4.4. Generation of SW1710 KDM6A Knockout Cells
Cells were transfected using X-tremeGENE 9 DNA Transfection Reagent (Roche, Mannheim, Germany) with KDM6A Double Nickase Plasmid (sc-402761-NIC, SantaCruz Biotechnology, Heidelberg, Germany) encoding a GFP-marker, Puromycin resistance, two different sgRNAs targeting KDM6A and Cas9 (Nickase). Transfected cells were selected with 1 µg/mL Puromycin for five days before single-cell seeding into 96-well plates. Genomic DNA was extracted from single-cell clones using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). An amplicon spanning the sgRNA binding sites was amplified using HotStarTaq polymerase (Qiagen), PCR products were Sanger sequenced. Mutant sequences were compared to the NCBI KDM6A reference sequence (NG_016260.1). Successful knockout was verified by western blot analysis for UTX.
4.5. Cell Viability and Clone Formation Assays
Viability of cells was measured after 24 h, 48 h and 72 h of seeding by 3-(4,5-dimetylthiazol-2-yl)-2,5-diphenyltetrazolium bromide dye reduction assay (MTT, M2128-G, Sigma Aldrich, St. Louis, MO, USA).
For colony forming assays, cells were seeded into six-well plates at a density of 500 cells/well. After 8–15 days, cells were fixed in methanol and stained with Giemsa (Merck, Darmstadt, Germany). Colony forming units were counted by Fiji (1.52b) with the BioVoxxel Toolbox [
32] using the green channel of a 300 dpi RGB picture. One well of a six-well plate was sectioned (makeOval) and a filter ("Mean...", "radius = 2") was used. Via Threshold (Triangle) a binary picture was generated and fused colonies were separated (Watershed). Binary particles were measured by the Analyze Particles tool.
For hanging-drop culture cells were seeded into the lids of six-well plates at a density of 5000 cells/drop in a volume of 20 µL of culture media. The wells were filled with sterile phosphate buffered saline (PBS) to prevent desiccation. After 4 days colonies in the drops were photographed using an inverted microscope.
4.6. Western Blot Analysis
Total protein extraction, purification of histones and Western blot analysis were performed as previously described [
33]. Briefly, cells were incubated for 30 minutes on ice in RIPA-buffer (150 mM NaCl, 1% Triton X-100, 0.5% desoxycholate, 1% Nonidet P-40, 0.1% SDS (sodium dodecyl sulfate), 1 mM EDTA, 50 mM TRIS (pH 7.6)) containing 10 µL/mL protease inhibitor cocktail (#P-8340, Sigma Aldrich). Histones were extracted by a modified published protocol employing sulphuric acid extraction and TCA-precipitation [
34]. Concentrations of total protein and histones were determined by BCA protein assay (Thermo Fisher Scientific, Carlsbad, CA, USA). Subsequently, total cell proteins (50 µg) or extracted histones (2 µg) were separated by SDS-PAGE (total proteins 10% gels, histones 15% gels), transferred to PVDF membranes (Merck Millipore, Berlin, Germany) and were incubated with primary antibodies at RT for 1 h or 4°C overnight (see
Table S2) following blocking with 5% non-fat milk in TBST (150 mM NaCl, 10 mM TRIS, pH 7.4 and 0.1% Tween-20). For signal detection membranes were incubated with a suitable horseradish peroxidase-conjugated secondary antibody (see
Table S1) at RT for 1 h and signals were visualized by Clarity™ Western ECL Substrate (Bio-Rad Laboratories, Munich, Germany) and WesternBright Quantum kit (Biozym, Hessisch Oldendorf, Germany).
4.7. RNA Extraction and Reverse Transcription
Total cell RNA was isolated by the Qiagen RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol and cDNA was synthesized using QuantiTect Reverse Transcription Kit (Qiagen) with an extended incubation time of 30 min at 42 °C as previously described [
33]. Target mRNA expression was measured by qRT-PCR with QuantiTect SYBR Green RT-PCR Kit (Qiagen) on the LightCycler® 96 Real-Time PCR system with software version 1.1 (Roche Diagnostics, Rotkreuz, Switzerland). All used primers, comprising QuantiTect Primer assays (Qiagen), self-designed target primers and primers for the reference housekeeping gene
TBP (TATA-box binding protein), are listed in
Table S2.
4.8. RNA-Seq and Data Analysis
Total RNA samples used for transcriptome analyses were quantified (Qubit RNA HS Assay, Thermo Fisher Scientific) and quality measured by capillary electrophoresis using the Fragment Analyzer and the Total RNA Standard Sensitivity Assay (Agilent Technologies, Inc. Santa Clara, CA, USA). All samples in this study showed high quality RNA Quality Numbers (RQN; mean = 9.9). The library preparation was performed according to the manufacturer’s protocol using the ‘VAHTS™ Stranded mRNA-Seq Library Prep Kit’ for Illumina®. Briefly, 300 ng total RNA were used for mRNA capturing, fragmentation, the synthesis of cDNA, adapter ligation and library amplification. Bead purified libraries were normalized and finally sequenced on the HiSeq 3000/4000 system (Illumina Inc. San Diego, CA, USA) with a read setup of 1 × 150 bp. The bcl2fastq tool was used to convert the bcl files to fastq files as well for adapter trimming and demultiplexing.
Data analyses on fastq files were conducted with CLC Genomics Workbench (version 10.1.1, QIAGEN, Venlo, The Netherlands). The reads of all probes were adapter trimmed (Illumina TruSeq) and quality trimmed (using the default parameters: bases below Q13 were trimmed from the end of the reads, ambiguous nucleotides maximal 2). Mapping was done against the Homo sapiens (hg38) (Mai 25, 2017) genome sequence. After grouping of samples (three biological replicates each) according to their respective experimental condition, multi-group comparisons were made and statistically determined using the Empirical Analysis of DGE (version 1.1, cutoff = 5). The Resulting p values were corrected for multiple testing by FDR and Bonferroni-correction. A p value of ≤ 0.05 was considered significant.
4.9. Gene Set Enrichment Analysis (GSEA)
GSEA [
18] was performed with the software provided by
https://software.broadinstitute.org/gsea/index.jsp, and performed with the MSigDB database v6.2. The datasets generated by RNA-Seq were split into two based on up- and downregulated genes upon UTX-TagGFP2 overexpression compared to TagGFP2 overexpression and were statistically significant for multiple testing by FDR. Each dataset was used to correlate the gene set to GSEA Hallmarks, Oncogenes and Gene Ontology (GO) gene sets. A group was considered upon an enrichment of at least 15 genes and a statistical significance of FDR < 25%.
4.10. Immunocytochemistry
Analysis of UTX-TagGFP2 localization was performed in stably transduced UCCs. As previously described [
33], after fixation with 4% formaldehyde, cells were permeabilized using 0.2% Triton X100 in PBS for 10 min at RT, blocked with 1% BSA in PBS, for 30 min at RT and subsequently incubated for 1 h at RT with 14 nM Rhodamine Phalloidin in blocking solution. Following counter-staining of nuclei with 1 µg/mL DAPI (4´,6-diamidino-2-phenylindole) cells were mounted with fluorescence mounting medium (DAKO, Glostrup, Denmark). Imaging was performed using ZEISS Axio Observer.Z1 / 7; Plan-Apochromat 40x/1.4 Oil DIC (UV) VIS-IR M27; 90 HE DAPI/ GFP/ Cy3 /Cy5; LED-module "wavelength" nm (Colibri 7); Axiocam 512 mono (ZEISS, Jena, Germany)
Higher resolution images were obtained using an inverted confocal FluoView1000 laser scanning microscope with a 60xW UPLSAPO objective NA1.2 in a sequential DAPI-GFP-Phalloidin scanning mode (Olympus, Hamburg, Germany). Resolution was 1024 × 1024.
4.11. Immunopurification
Cells were harvested at a cell count of ≈1 × 10
7 cells and lysed as described in
Section 4.6. The immunopurification was performed with 2 mg cleared cell lysate, from which an aliquot was used as input control, and 10 µL magnetic GFP-beads (GFP-Trap®_MA ChromoTek, Martinsried, Germany) of 50% slurry (washed three times in lysis buffer) overnight at 4 °C under permanent rotation. Then, beads were washed three times with lysis buffer, incubated with 1× Laemmli denaturing solution and heated at 95 °C for 5 min. The entire beads fraction and 50 µg input protein each were loaded on a Tris-Glycin-SDS-PAGE. Subsequent steps were performed as described in
Section 4.6.
4.12. Transient siRNA Transfection
For transient transfection of siRNA the Lipofectamine™ RNAiMAX Transfection Reagent (13778030, Invitrogen) was used. Cells were seeded at a number of 2 × 105 (RT112) or 1.5 × 105 (SW1710) per well of 6-well plates and cultured for one day at 37 °C and 5% CO2. Then, cells were transfected with 4 nM of siRNA as described in the RNAiMAX datasheet. Media were changed one day after transfection. Cells were harvested after 3 days and RNA and proteins were isolated as described above or below, respectively.
4.13. Nuclear and Cytoplasmic Fractionation
Fractionation of cytoplasmatic and nuclear proteins was performed using three buffers: HB-Buffer (10 mM Tris-HCl, pH 8, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM 2-mercaptoethanol, protease inhibitor [P8340, Merck; 4 µL per 5 mL]); Lysis-Buffer: HB-Buffer + 0.4% NP-40); Buffer C (20 mM HEPES, 400 mM NaCl, 1 mM EDTA, 1 mM DTT, protease inhibitor [P8340, Merck; 4 µL per 1 mL]). 1–5 × 106 cells were harvested, pelleted at 200× g for 5 min and washed at RT. The cell pellet was resuspended in 350 µL HB-Buffer and centrifuged at 200× g for 1 min at 4 °C. The supernatant was removed and the pellet was resuspended in 100 µL Lysis-Buffer followed by incubation for 15 min at 4 °C. The supernatant containing the cytosolic fraction was harvested by centrifugation at 15,000× g for 5 min at 4 °C. The pellet was washed twice with 75 µL Lysis-Buffer, resuspended in 60 µL Buffer C and incubated for 15 min at 4 °C. Following centrifugation at 15,000× g for 5 min at 4 °C the supernatant containing the nuclear fraction was harvested. Protein concentrations were then determined using the Protein Assay Kit I (Bio Rad, 5000001).
4.14. Statistical Methods
p-values between different groups were determined by the Student´s t-test; asterisks denote significant (* < 0.05) differences; error bars indicate SD. The RNA-Seq data is available from the authors upon reasonable request.