The Role of MicroRNAs in Hepatoblastoma Tumors
Abstract
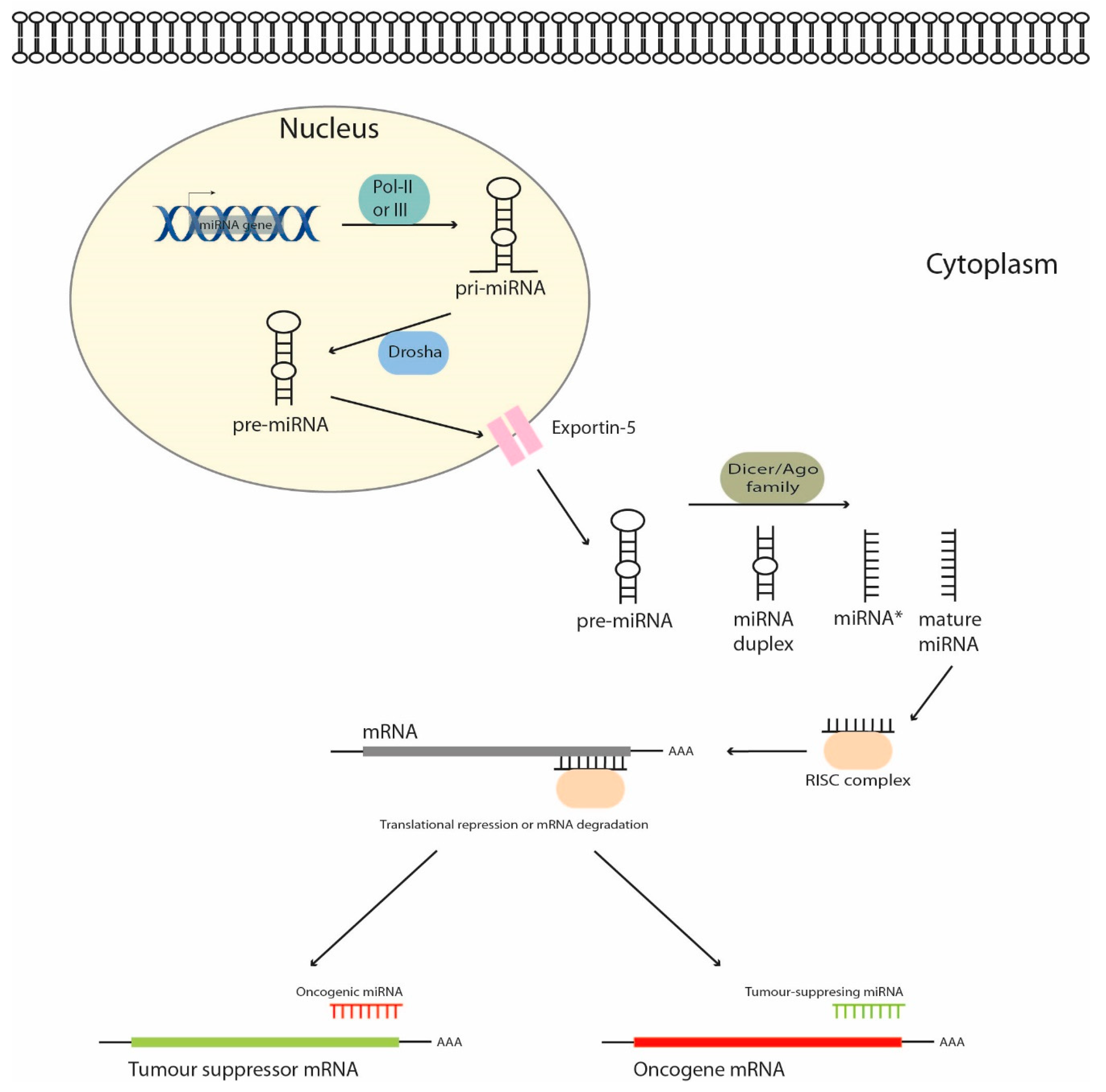
1. Introduction
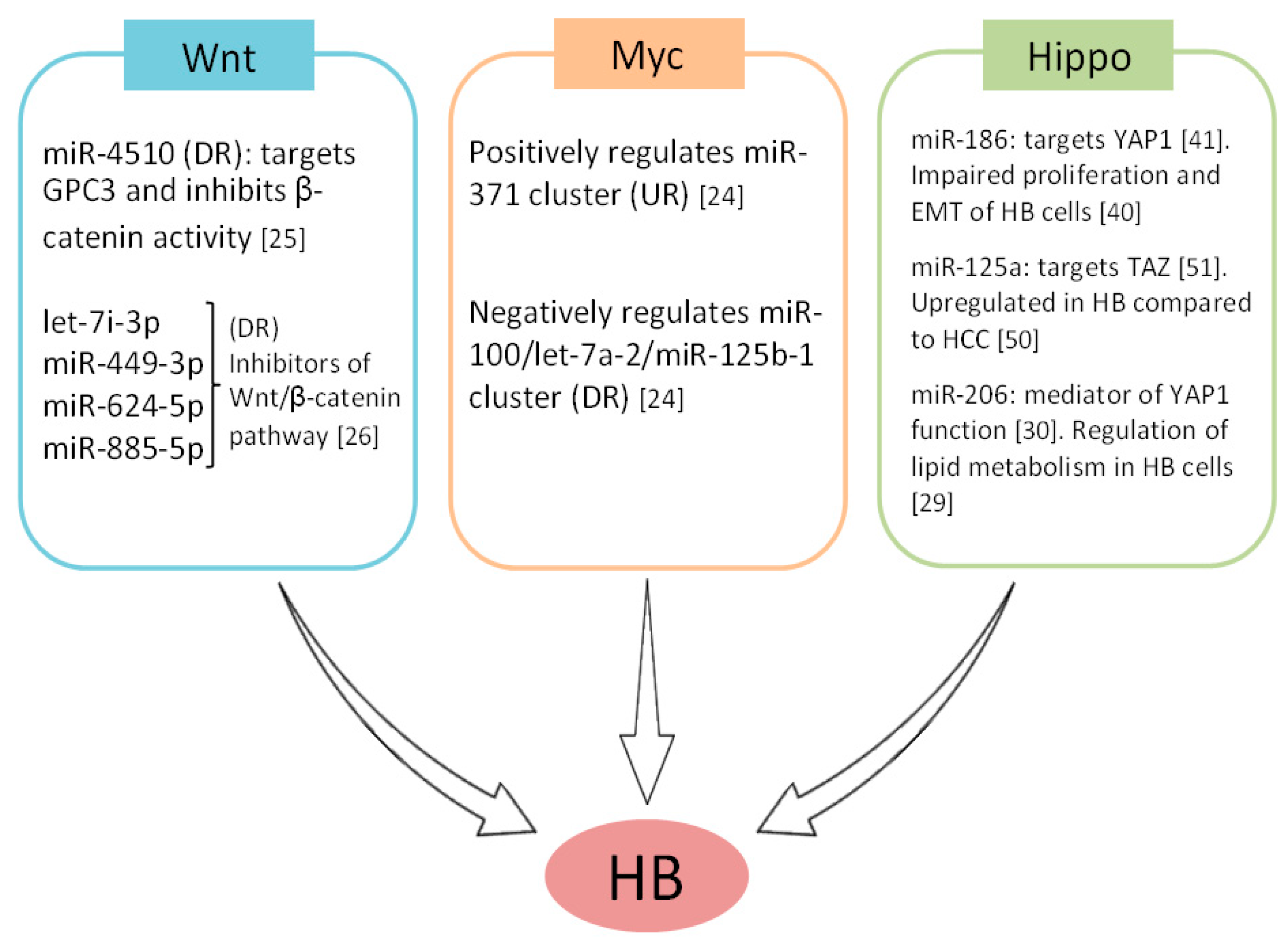
2. MicroRNAs as Regulators of Signaling Pathways in Hepatoblastoma Cells
3. MicroRNAs as Potential Therapeutic Targets
4. MicroRNA Expression Patterns and Their Potential Clinical Impact as Novel Biomarkers in Hepatoblastoma
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lim, I.I.P.; Bondoc, A.J.; Geller, J.I.; Tiao, G.M. Hepatoblastoma-The Evolution of Biology, Surgery, and Transplantation. Children 2018, 6, 1. [Google Scholar] [CrossRef]
- Kehm, R.D.; Osypuk, T.L.; Poynter, J.N.; Vock, D.M.; Spector, L.G. Do pregnancy characteristics contribute to rising childhood cancer incidence rates in the United States? Pediatr. Blood Cancer 2018, 65, e26888. [Google Scholar] [CrossRef]
- Meyers, R.L.; Maibach, R.; Hiyama, E.; Häberle, B.; Krailo, M.; Rangaswami, A.; Aronson, D.C.; Malogolowkin, M.H.; Perilongo, G.; von Schweinitz, D.; et al. Risk-stratified staging in paediatric hepatoblastoma: A unified analysis from the Children’s Hepatic tumors International Collaboration. Lancet Oncol. 2017, 18, 122–131. [Google Scholar] [CrossRef]
- Bell, D.; Ranganathan, S.; Tao, J.; Monga, S.P. Novel advances in understanding of molecular pathogenesis of hepatoblastoma: A Wnt/beta-catenin perspective. Gene Expr. 2017, 17, 141–154. [Google Scholar] [CrossRef]
- Sumazin, P.; Chen, Y.; Treviño, L.R.; Sarabia, S.F.; Hampton, O.A.; Patel, K.; Mistretta, T.A.; Zorman, B.; Thompson, P.; Heczey, A.; et al. Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology 2017, 65, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; El Jabbour, T.; Ainechi, S.; Gay, L.M.; Elvin, J.A.; Vergilio, J.A.; Suh, J.; Ramkissoon, S.H.; Ali, S.M.; Schrock, A. General paucity of genomic alteration and low tumor mutation burden in refractory and metastatic hepatoblastoma: Comprehensive genomic profiling study. Hum. Pathol. 2017, 70, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Cairo, S.; Armengol, C.; Buendía, M.A. Activation of Wnt and Myc signaling in hepatoblastoma. Front. Biosci. 2012, 4, 480–486. [Google Scholar] [CrossRef]
- Li, N.; Xie, C.; Lu, N. Crosstalk between Hippo signalling and miRNAs in tumour progression. FEBS J. 2017, 284, 1045–1055. [Google Scholar] [CrossRef]
- Kong, Y.W.; Ferland-McCollough, D.; Jackson, T.J.; Bushell, M. microRNAs in cancer management. Lancet Oncol. 2012, 13, e249–e258. [Google Scholar] [CrossRef]
- Xuan, Y.M.; Yang, H.; Zhao, L.; Lau, W.B.; Lau, B.; Ren, N.; Hu, Y.; Yi, T.; Zhao, X.; Zhou, S.; et al. MicroRNAs in colorectal cancer: Small molecules with big functions. Cancer Lett. 2015, 360, 89–105. [Google Scholar] [CrossRef]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204–1225. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Lorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Ageilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 103, 2464. [Google Scholar] [CrossRef] [PubMed]
- Cekaite, L.; Eide, P.W.; Lind, G.E.; Skotheim, R.I.; Lothe, R.A. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget 2016, 7, 6476–6505. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Wentzel, E.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Zeitels, L.R.; Hwang, H.-W.; Chivuluka, R.R.; Wentzel, E.A.; Dews, M.; Jung, J.; Gao, P.; Dang, C.V.; Beer, M.A.; et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl. Acad. Sci. USA 2009, 106, 3384–3389. [Google Scholar] [CrossRef]
- Boyerinas, B.; Park, S.M.; Hau, A.; Murmann, A.E.; Peter, M.E. The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer 2010, 17, F19–F36. [Google Scholar] [CrossRef]
- Watahiki, A.; Wang, Y.; Morris, J.; Dennis, K.; O´Dwyer, H.M.; Gleave, M.; Gout, P.W.; Wang, Y. MicroRNAs associated with metastatic prostate cancer. PLoS ONE 2011, 6, e24950. [Google Scholar] [CrossRef]
- Toyota, M.; Suzuki, H.; Sasaki, Y.; Maruyama, R.; Imai, K.; Shinomura, Y.; Tokino, T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008, 68, 4123–4132. [Google Scholar] [CrossRef]
- Chin, L.J.; Ratner, E.; Leng, S.; Zhai, R.; Nallur, S.; Babar, I.; Muller, R.U.; Straka, E.; Su, L.; Burki, E.A.; et al. A SNP in a let-7 microRNA complementary site in the KRAS 3 untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008, 68, 8535–8540. [Google Scholar] [CrossRef] [PubMed]
- Takebe, N.; Harris, P.J.; Warren, R.Q.; Ivy, S.P. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 2011, 8, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Lv, Q.; Ye, W.; Wong, C.K.; Cai, G.; Gu, D.; Ji, Y.; Zhao, C.; Wang, J.; Yang, B.B.; et al. Mirna-directed regulation of VEGF and other angiogenic under hypoxia. PLoS ONE 2006, 1, e116. [Google Scholar] [CrossRef] [PubMed]
- Cairo, S.; Wang, Y.; de Reyniès, A.; Duroure, K.; Dahan, J.; Redon, M.J.; Fabre, M.; McClelland, M.; Wang, X.W.; et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 20471–20476. [Google Scholar] [CrossRef] [PubMed]
- Cartier, F.; Indersie, E.; Lesjean, S.; Charpentier, J.; Hooks, K.B.; Ghousein, A.; Desplat, A.; Dugot-Senant, N.; Trézéguet, V.; Sagliocco, F.; et al. New tumor suppressor microRNAs target glypican-3 in human liver cancer. Oncotarget 2017, 8, 41211–41226. [Google Scholar] [CrossRef]
- Indersie, E.; Lesjean, S.; Hooks, K.B.; Sagliocco, F.; Ernault, T.; Cairo, S.; Merched-Sauvage, M.; Rullier, A.; Le Bail, B.; Taque, S.; et al. MicroRNA therapy inhibits hepatoblastoma growth in vivo by targeting β-catenin and Wnt signaling. Hepatol. Commun. 2017, 1, 168–183. [Google Scholar] [CrossRef]
- Zhang, N.; Lei, J.; Lei, H.; Ruan, X.; Liu, Q.; Chen, Y.; Huang, W. MicroRNA-101 overexpression by IL-6 and TNF-α inhibits cholesterol efflux by suppressing ATP-binding cassette transporter A1 expression. Exp. Cell Res. 2015, 336, 33–42. [Google Scholar] [CrossRef]
- Awortwe, C.; Kaehler, M.; Rosenkranz, B.; Cascorbi, I.; Bruckmueller, H. MicroRNA-655-3p regulates Echinacea purpurea mediated activation of ABCG2. Xenobiotica 2018, 48, 1050–1058. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhao, C.; Zhang, N.; Kang, W.; Lu, R.; Wu, H.; Geng, Y.; Zhao, Y.; Xu, X. Serum microRNA miR-206 is decreased in hyperthyroidism and mediates thyroid hormone regulation of lipid metabolism in HepG2 human hepatoblastoma cells. Mol. Med. Rep. 2018, 17, 5635–5641. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Del Re, D.P.; Nakano, N.; Sciarretta, S.; Zhai, P.; Park, J.; Sabed, D.; Shirakabe, A.; Matsushima, S.; Park, Y.; et al. miR-206 mediates YAP-induced cardiac hypertrophy and survival. Circ. Res. 2015, 117, 891–904. [Google Scholar] [CrossRef]
- Pei, Y.; Yao, Q.; Yuan, S.; Xie, B.; Liu, Y.; Ye, C.; Zhuo, H. GATA4 promotes hepatoblastoma cell proliferation by altering expression of miR125b and DKK3. Oncotarget 2016, 7, 77890–77901. [Google Scholar] [CrossRef]
- von Frowein, J.; Pagel, P.; Kappler, R.; von Schweinitz, D.; Roscher, A.; Schmid, I. MicroRNA-492 is processed from the keratin 19 gene and up-regulated in metastatichepatoblastoma. Hepatology 2011, 53, 833–842. [Google Scholar] [CrossRef]
- von Frowein, J.; Hauck, S.M.; Kappler, R.; Pagel, P.; Fleischmann, K.K.; Magg, T.; Cairo, S.; Roscher, A.; von Schweinitz, D.; Schmid, I. MiR-492 regulates metastatic properties of hepatoblastoma via CD44. Liver Int. 2018, 38, 1280–1291. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Li, Z.; Chen, R.; Guo, N.; Zhou, J.; Zhou, Q.; Lin, Q.; Cheng, D.; Liao, Q.; Zheng, L.; et al. Epigenetic regulation of miR-124 by hepatitis C virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett. 2012, 586, 3271–3278. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Liao, Y.J.; Cai, M.Y.; Liu, Y.H.; Liu, T.H.; Chen, S.P.; Bian, X.W.; Guan, X.Y.; Lin, M.C.; Zeng, Y.X.; et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 2012, 61, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Liu, H.; Wei, Z.; Jia, H.; Liu, Y.; Liu, J. Systematic analysis of the molecular mechanism of microRNA-124 in hepatoblastoma cells. Oncol. Lett. 2017, 14, 7161–7170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Wu, J.; Liangpunsakul, S.; Niu, J.; Wang, L. MicroRNA-26-5p functions as a new inhibitor of hepatoblastoma by repressing lin-28 homolog B and aurora kinase a expression. Hepatol. Commun. 2018, 2, 861–871. [Google Scholar] [CrossRef]
- Dong, R.; Liu, G.B.; Liu, B.H.; Chen, G.; Li, K.; Zheng, S.; Dong, K.R. Targeting long non-coding RNA-TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis. 2016, 7, e2278. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.C.; Yuan, L.Q.; Zhang, T.; Yan, X.M.; Zhou, Y.; Xia, H.L.; Wu, Y.; Xu, L.X.; Cao, X.; Wang, J. Nuclear paraspeckle assembly transcript 1 promotes the metastasis and epithelial-mesenchymal transition of hepatoblastoma cells by inhibiting miR-129-5p. Oncol. Lett. 2017, 14, 5773–5778. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, F.; Yang, F.; Liu, Y. Kockdown of OIP5-AS1 expression inhibits proliferation, metastasis and EMT progress in hepatoblastoma cells through up-regulating miR-186a-5p and down-regulating ZEB1. Biomed. Pharmacother. 2018, 101, 14–23. [Google Scholar] [CrossRef]
- Ruan, T.; He, X.; Yu, J.; Hang, Z. MicroRNA-186 targets Yes-associated protein 1 to inhibit Hippo signaling and tumorigenesis in hepatocellular carcinoma. Oncol. Lett. 2016, 11, 2941–2945. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Calvisi, D.F.; Ranganathan, S.; Cigliano, A.; Zhou, L.; Singh, S.; Jiang, L.; Fan, B.; Terracciano, L.; Armeanu-Ebinger, S.; et al. Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 2014, 147, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Min, Q.; Molina, L.; Li, J.; Adebayo Michael, A.O.; Russell, J.O.; Preziosi, M.E.; Singh, S.; Poddar, M.; Matz-Soja, M.; Ranganathan, S.; et al. β-Catenin and yes-associated protein 1 cooperate in hepatoblastoma pathogenesis. Am. J. Pathol. 2019, in press. [Google Scholar] [CrossRef] [PubMed]
- Molina, L.; Yang, H.; Adebayo Michael, A.O.; Oertel, M.; Bell, A.; Singh, S.; Chen, X.; Tao, J.; Monga, S.P.S. mTOR inhibition affects Yap1-β-catenin-induced hepatoblastoma growth and development. Oncotarget 2019, 10, 1475–1490. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Tang, H.F.; Xiang, Q.; Yu, J.; Yang, X.Y.; Hu, N.; Lei, X.Y. MiR-122 increases sensitivity of drug-resistant BEL-7402/5-FU cells to 5-fluorouracil via down-regulation of bcl-2 family proteins. Pharmazie 2011, 66, 975–981. [Google Scholar] [PubMed]
- Yu, G.; Wang, J.; Xu, K.; Dong, J. Dynamic regulation of uncoupling protein 2 expression by microRNA-214 in hepatocellular carcinoma. Biosci. Rep. 2016, 36, 3. [Google Scholar] [CrossRef] [PubMed]
- Gyugos, M.; Lendvai, G.; Kenessey, I.; Schlachter, K.; Halász, J.; Nagy, P.; Garami, M.; Jakab, Z.; Schaff, Z.; Kiss, A. MicroRNA expression might predict prognosis of epithelial hepatoblastoma. Virchows Arch. 2014, 464, 419–427. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, J.; Pan, T.; Ai, S.; Tang, L.; Wang, F. miR-378a enhances the sensitivity of liver cancer to sorafenib by targeting VEGFR, PDGFRβ and c-Raf. Mol. Med. Rep. 2018, 17, 4581–4588. [Google Scholar] [CrossRef]
- Liu, B.H.; Zhang, B.B.; Liu, X.Q.; Zheng, S.; Dong, K.R.; Dong, R. Expression Profiling Identifies Circular RNA Signature in Hepatoblastoma. Cell. Physiol. Biochem. 2018, 45, 706–719. [Google Scholar] [CrossRef]
- Magrelli, A.; Azzalin, G.; Salvatore, M.; Viganotti, M.; Tosto, F.; Colombo, T.; Devito, R.; Di Masi, A.; Antoccia, A.; Lorenzetti, S.; et al. Altered microRNA Expression Patterns in Hepatoblastoma Patients. Transl. Oncol. 2009, 2, 157–163. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, C.; Zhu, X.; Zhu, X.; Xian, H.; Huang, Z. Suppression of microRNA-125a-5p upregulates the TAZ-EGFR signaling pathway and promotes retinoblastoma proliferation. Cell Signal. 2016, 28, 850–860. [Google Scholar] [CrossRef]
- Zatkova, A.; Rouillard, J.M.; Hartmann, W.; Lamb, B.J.; Kuick, R.; Eckart, M.; von Schweinitz, D.; Koch, A.; Fonatsch, C.; Pietsch, T.; et al. Amplification and overexpression of the IGF2 regulator PLAG1 in hepatoblastoma. Genes Chromosom. Cancer 2004, 39, 126–137. [Google Scholar] [CrossRef]
- Ding, S.J.; Li, Y.; Tan, Y.X.; Jiang, M.R.; Tian, B.; Liu, Y.K.; Shao, X.X.; Ye, S.L.; Wu, J.R.; Zeng, R.; et al. From proteomic analysis to clinical significance: Overexpression of cytokeratin 19 correlates with hepatocellular carcinoma metastasis. Mol. Cell. Proteom. 2004, 3, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Kamohara, Y.; Haraguchi, N.; Mimori, K.; Tanaka, F.; Inoue, H.; Mori, M.; Kanematsu, T. The search for cancer stem cells in hepatocellular carcinoma. Surgery 2008, 144, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Finch, M.L.; Marquardt, J.U.; Yeoh, G.C.; Callus, B.A. Regulation of microRNAs and their role in liver development, regeneration and disease. Int. J. Biochem. Cell Biol. 2014, 54, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Ecevit, Ç.Ö.; Aktaş, S.; Tosun Yildirim, H.; Demirağ, B.; Erbay, A.; Karaca, İ.; Çelik, A.; Demir, A.B.; Erçetin, A.P.; Olgun, N. MicroRNA-17, MicroRNA-19b, MicroRNA-146a, MicroRNA-302d Expressions in Hepatoblastoma and Clinical Importance. J. Pediatr. Hematol. Oncol. 2019, 41, 7–12. [Google Scholar] [CrossRef]
- Liu, W.; Chen, S.; Liu, B. Diagnostic and prognostic values of serum exosomal microRNA-21 in children with hepatoblastoma: A Chinese population-based study. Pediatr. Surg. Int. 2016, 32, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Zhu, A.; Jiao, X.; Ge, J.; Xu, X. Combined low miR-34s are associated with unfavorable prognosis in children with hepatoblastoma: A Chinese population-based study. J. Pediatr. Surg. 2016, 51, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Jiao, X.; Zhu, A.; Ge, J.; Xu, X. Exosomal miR-34s panel as potential novel diagnostic and prognostic biomarker in patients with hepatoblastoma. J. Pediatr. Surg. 2017, 52, 618–624. [Google Scholar] [CrossRef]
- Chatterjee, A.; Leichter, A.L.; Fan, V.; Tsai, P.; Purcell, R.V.; Sullivan, M.J.; Eccles, M.R. A cross comparison of technologies for the detection of microRNAs in clinical FFPE samples of hepatoblastoma patients. Sci. Rep. 2015, 5, 10438. [Google Scholar] [CrossRef]
- Leichter, A.L.; Purcell, R.V.; Sullivan, M.J.; Eccles, M.R.; Chatterjee, A. Multi-platform microRNA profiling of hepatoblastoma patients using formalin fixed paraffin embedded archival samples. Gigascience 2015, 4, 54. [Google Scholar] [CrossRef]
| MicroRNA | Role | Therapeutic Impact | References |
|---|---|---|---|
| miR-100/let-7a-2/miR-125b-1 cluster | Tumor supressor | In vivo cooperation. OE of the first cluster concomitant with inhibition of the second one blocked tumor formation | Cairo et al., 2010 [24] |
| miR-371 cluster | Oncogenic | ||
| miR-122 | Tumor supressor | Sensitizes to 5-FU via Bcl-2 and Bcl-XL DR and P53 activation | Yin et al., 2011 [45] |
| miR-214 | Tumor supressor | Sensitizes to gemcitabine via UCP2 targeting | Yu et al., 2016 [46] |
| miR-624-5p | Tumor supressor | Targets β-catenin and impairs tumor growth in vivo | Indersie et al., 2017 [26] |
| miR-4510 | Tumor supressor | Decreases proliferation and induces apoptosis in vitro through Wnt/β-catenin inhibition | Cartier et al., 2017 [25] |
| miR-34a-5p | Tumor supressor | Reduces tumor growth and microvascular density | Dong et al., 2016 [38] |
| miR-125b | Tumor supressor | Decreases cell growth, migration and invasion | Pei et al., 2016 [31] |
| miR-492 | Oncogenic | Targets CD44 and enhances anchorage-independent growth, migration and invasion | von Frowein et al., 2018 [33] |
| miR-378a | Tumor supressor | Sensitizes to sorafenib and inhibits cell proliferation and invasion | Fu et al., 2018 [48] |
| miR-26a-5p | Tumor supressor | Inhibits proliferation and colony formation | Zhang et al., 2018 [37] |
| miR-1250-3p | Tumor supressor | Decreases cell growth and invasion | Liu et al., 2018 [49] |
| MicroRNA | Expression | Clinical Significance | References |
|---|---|---|---|
| miR-492 | High | Metastatic disease | [30] |
| miR-21 | High | Independent markers of increased survival | [41] |
| miR-222 and miR-224 | Low | ||
| Combined miR-34a/b/c | Low | Independent unfavourable prognostic factor | [53] |
| Exosomal miR-21 | High | Independent predictor of larger EFS 1 | [52] |
| Exosomal miR-34a/b/c | Low | Independent unfavourable prognostic factor | [54] |
| miR-492 | High | Correlation with aggressive tumors and reduced EFS 1 | [31] |
| miR-17 | Low | Worse prognosis | [51] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cristóbal, I.; Sanz-Álvarez, M.; Luque, M.; Caramés, C.; Rojo, F.; García-Foncillas, J. The Role of MicroRNAs in Hepatoblastoma Tumors. Cancers 2019, 11, 409. https://doi.org/10.3390/cancers11030409
Cristóbal I, Sanz-Álvarez M, Luque M, Caramés C, Rojo F, García-Foncillas J. The Role of MicroRNAs in Hepatoblastoma Tumors. Cancers. 2019; 11(3):409. https://doi.org/10.3390/cancers11030409
Chicago/Turabian StyleCristóbal, Ion, Marta Sanz-Álvarez, Melani Luque, Cristina Caramés, Federico Rojo, and Jesús García-Foncillas. 2019. "The Role of MicroRNAs in Hepatoblastoma Tumors" Cancers 11, no. 3: 409. https://doi.org/10.3390/cancers11030409
APA StyleCristóbal, I., Sanz-Álvarez, M., Luque, M., Caramés, C., Rojo, F., & García-Foncillas, J. (2019). The Role of MicroRNAs in Hepatoblastoma Tumors. Cancers, 11(3), 409. https://doi.org/10.3390/cancers11030409






