MicroRNAs and Long Non-Coding RNAs and Their Hormone-Like Activities in Cancer
Abstract
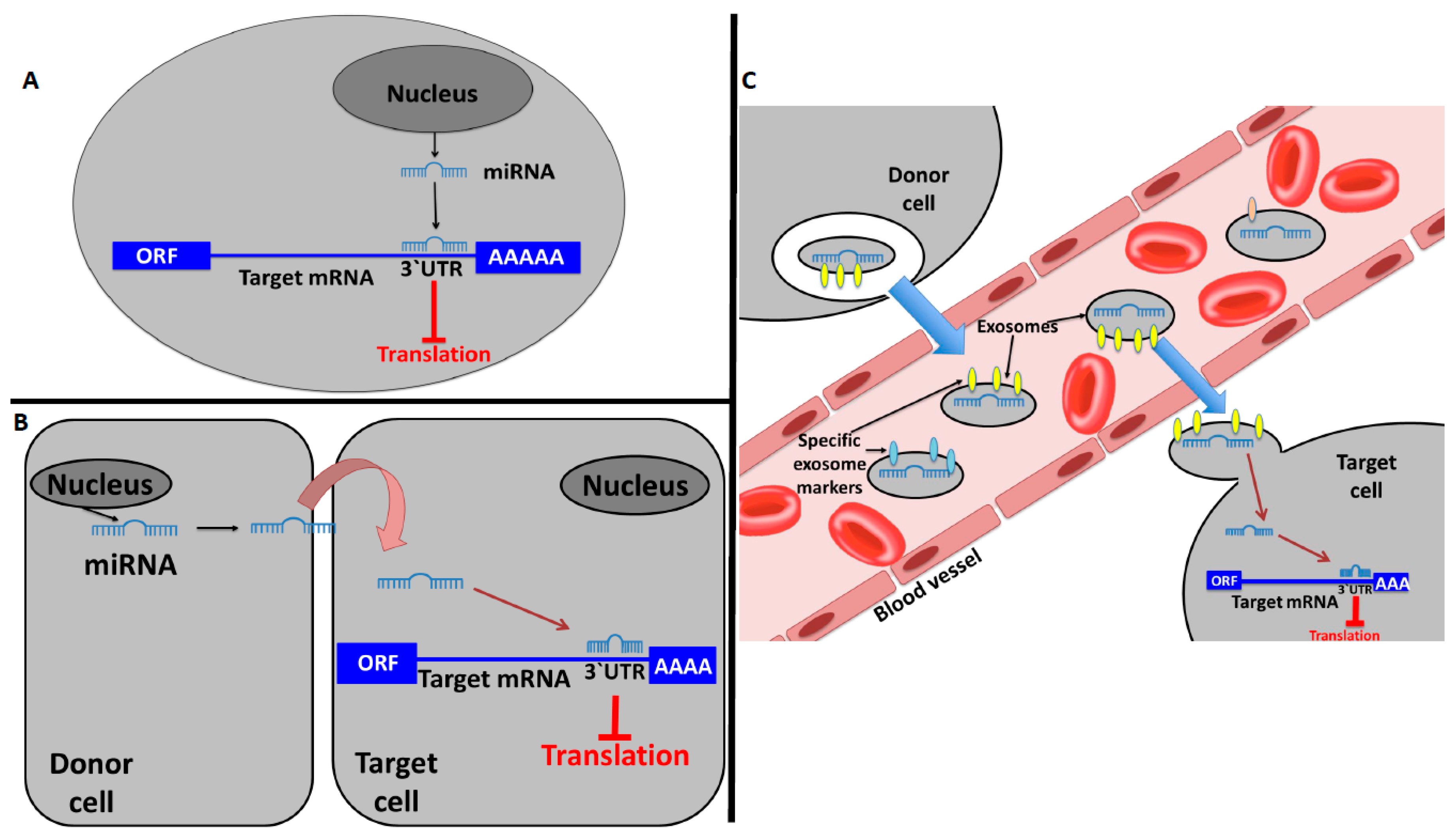
1. Introduction
2. miRNAs and Their Hormone-Like Activity in Cancer
3. Long Non-Coding RNAs Acting as Hormones
4. Conclusions
Funding
Conflicts of Interest
References
- Bayliss, W.M.; Starling, E.H. On the relation of enterokinase to trypsin. J. Physiol. 1905, 32, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J. Ernest Starling and ‘Hormones’: An historical commentary. J. Endocrinol. 2005, 184, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Litwack, G. Hormones and transport systems. Preface. Vitam. Horm. 2015, 98, xvii–xviii. [Google Scholar] [CrossRef]
- White, A. The interaction of enzymes and hormones. Pediatrics 1960, 26, 476–481. [Google Scholar]
- Ameri, P.; Ferone, D. Diffuse Endocrine System, Neuroendocrine Tumors and Immunity: What’s New? Neuroendocrinology 2012, 95, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K.; Calin, G.A. Cell-to-cell communication: MicroRNAs as hormones. Mol. Oncol. 2017, 11, 1673–1686. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Thery, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Drummen, G.P.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [PubMed]
- Gangoda, L.; Boukouris, S.; Liem, M.; Kalra, H.; Mathivanan, S. Extracellular vesicles including exosomes are mediators of signal transduction: Are they protective or pathogenic? Proteomics 2015, 15, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Cieslik, M.; Chinnaiyan, A.M. Cancer transcriptome profiling at the juncture of clinical translation. Nat. Rev. Genet. 2018, 19, 93–109. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Munker, R.; Calin, G.A. MicroRNA profiling in cancer. Clin. Sci. (Lond.) 2011, 121, 141–158. [Google Scholar] [CrossRef]
- Sevignani, C.; Calin, G.A.; Nnadi, S.C.; Shimizu, M.; Davuluri, R.V.; Hyslop, T.; Demant, P.; Croce, C.M.; Siracusa, L.D. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc. Natl. Acad. Sci. USA 2007, 104, 8017–8022. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. microRNA Therapeutics in Cancer—An Emerging Concept. EBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef]
- Inui, M.; Martello, G.; Piccolo, S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gou, X.; Jiang, T.; Ouyang, J. The effects of microRNAs on glucocorticoid responsiveness. J. Cancer Res. Clin. Oncol. 2017, 143, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Abramov, R.; Fu, G.; Zhang, Y.; Peng, C. Expression and regulation of miR-17a and miR-430b in zebrafish ovarian follicles. Gen. Comp. Endocrinol. 2013, 188, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Prizant, H.; Light, A.; Biswas, A.; Hayes, E.; Lee, H.J.; Barad, D.; Gleicher, N.; Hammes, S.R. Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. Proc. Natl. Acad. Sci. USA 2014, 111, 3008–3013. [Google Scholar] [CrossRef] [PubMed]
- McFall, T.; McKnight, B.; Rosati, R.; Kim, S.; Huang, Y.; Viola-Villegas, N.; Ratnam, M. Progesterone receptor A promotes invasiveness and metastasis of luminal breast cancer by suppressing regulation of critical microRNAs by estrogen. J. Biol. Chem. 2018, 293, 1163–1177. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010, 39, 493–506. [Google Scholar] [CrossRef]
- Di Girolamo, D.; Ambrosio, R.; De Stefano, M.A.; Mancino, G.; Porcelli, T.; Luongo, C.; Di Cicco, E.; Scalia, G.; Vecchio, L.D.; Colao, A.; et al. Reciprocal interplay between thyroid hormone and microRNA-21 regulates hedgehog pathway-driven skin tumorigenesis. J. Clin. Investig. 2016, 126, 2308–2320. [Google Scholar] [CrossRef]
- Aucher, A.; Rudnicka, D.; Davis, D.M. MicroRNAs Transfer from Human Macrophages to Hepato-Carcinoma Cells and Inhibit Proliferation. J. Immunol. 2013, 191, 6250–6260. [Google Scholar] [CrossRef]
- Bovy, N.; Blomme, B.; Freres, P.; Dederen, S.; Nivelles, O.; Lion, M.; Carnet, O.; Martial, J.A.; Noel, A.; Thiry, M.; et al. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget 2015, 6, 10253–10266. [Google Scholar] [CrossRef]
- Nishida-Aoki, N.; Ochiya, T. Interactions between cancer cells and normal cells via miRNAs in extracellular vesicles. Cell. Mol. Life Sci. 2015, 72, 1849–1861. [Google Scholar] [CrossRef]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.H.; Ketova, T.; Hoshino, D.; Zijlstra, A.; Weaver, A.M. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 2015, 6, 7164. [Google Scholar] [CrossRef] [PubMed]
- Josson, S.; Gururajan, M.; Sung, S.Y.; Hu, P.; Shao, C.; Zhau, H.E.; Liu, C.; Lichterman, J.; Duan, P.; Li, Q.; et al. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 2015, 34, 2690–2699. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M. MicroRNAs and miRceptors: A new mechanism of action for intercellular communication. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Liu, D.Q.; Chen, X.; Li, J.; Li, L.M.; Bian, Z.; Sun, F.; Lu, J.W.; Yin, Y.A.; Cai, X.; et al. Secreted Monocytic miR-150 Enhances Targeted Endothelial Cell Migration. Mol. Cell 2010, 39, 133–144. [Google Scholar] [CrossRef]
- Huang-Doran, I.; Zhang, C.Y.; Vidal-Puig, A. Extracellular Vesicles: Novel Mediators of Cell Communication In Metabolic Disease. Trends Endocrinol. Metab. 2017, 28, 3–18. [Google Scholar] [CrossRef]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, L.; Mei, H.; Zhang, J.; Zhu, Y.; Han, X.; Zhu, D. Inflamed macrophage microvesicles induce insulin resistance in human adipocytes. Nutr. Metab. (Lond.) 2015, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Le, M.T.N.; Hamar, P.; Guo, C.Y.; Basar, E.; Perdigao-Henriques, R.; Balaj, L.; Lieberman, J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Investig. 2014, 124, 5109–5128. [Google Scholar] [CrossRef] [PubMed]
- Pigati, L.; Yaddanapudi, S.C.; Iyengar, R.; Kim, D.J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 2010, 5, e13515. [Google Scholar] [CrossRef]
- Anfossi, S.; Fu, X.; Nagvekar, R.; Calin, G.A. MicroRNAs, Regulatory Messengers Inside and Outside Cancer Cells. Adv. Exp. Med. Biol. 2018, 1056, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.C.; Chen, Y.F.; Chen, H.B.; Fei, S.R.; Chen, D.D.; Cai, X.N.; Liu, L.; Lin, B.C.; Su, H.F.; Zhao, L.H.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef]
- Matsumura, T.; Sugimachi, K.; Iinuma, H.; Takahashi, Y.; Kurashige, J.; Sawada, G.; Ueda, M.; Uchi, R.; Ueo, H.; Takano, Y.; et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 2015, 113, 275–281. [Google Scholar] [CrossRef]
- Sugimachi, K.; Matsumura, T.; Hirata, H.; Uchi, R.; Ueda, M.; Ueo, H.; Shinden, Y.; Iguchi, T.; Eguchi, H.; Shirabe, K.; et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer 2015, 112, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Schoepp, M.; Strose, A.J.; Haier, J. Dysregulation of miRNA Expression in Cancer Associated Fibroblasts (CAFs) and Its Consequences on the Tumor Microenvironment. Cancers 2017, 9, 54. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Berindan-Neagoe, I.; Calin, G.A. Molecular pathways: MicroRNAs, cancer cells, and microenvironment. Clin. Cancer Res. 2014, 20, 6247–6253. [Google Scholar] [CrossRef]
- Baroni, S.; Romero-Cordoba, S.; Plantamura, I.; Dugo, M.; D’Ippolito, E.; Cataldo, A.; Cosentino, G.; Angeloni, V.; Rossini, A.; Daidone, M.G.; et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016, 7, e2312. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef]
- Eiring, A.M.; Harb, J.G.; Neviani, P.; Garton, C.; Oaks, J.J.; Spizzo, R.; Liu, S.; Schwind, S.; Santhanam, R.; Hickey, C.J.; et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 2010, 140, 652–665. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef]
- Patel, S.A.; Gooderham, N.J. IL6 Mediates Immune and Colorectal Cancer Cell Cross-talk via miR-21 and miR-29b. Mol. Cancer Res. 2015, 13, 1502–1508. [Google Scholar] [CrossRef]
- Challagundla, K.B.; Wise, P.M.; Neviani, P.; Chava, H.; Murtadha, M.; Xu, T.; Kennedy, R.; Ivan, C.; Zhang, X.; Vannini, I.; et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef] [PubMed]
- EL Andaloussi, S.; Mager, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef]
- Donnarumma, E.; Fiore, D.; Nappa, M.; Roscigno, G.; Adamo, A.; Iaboni, M.; Russo, V.; Affinito, A.; Puoti, I.; Quintavalle, C.; et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 2017, 8, 19592–19608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, X.; Liu, J.; Dong, L.; Chen, Q.; Kong, H.; Zhang, Q.; Qi, X.; Hou, D.; Zhang, L.; et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res 2015, 25, 39–49. [Google Scholar] [CrossRef]
- Li, Z.; Xu, R.; Li, N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. (Lond.) 2018, 15, 68. [Google Scholar] [CrossRef]
- Xie, W.; Weng, A.; Melzig, M.F. MicroRNAs as New Bioactive Components in Medicinal Plants. Planta Med. 2016, 82, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Kraus, W.L. From Discovery to Function: The Expanding Roles of Long Non-Coding RNAs in Physiology and Disease. Endocr. Rev. 2015. [Google Scholar] [CrossRef]
- Ferdin, J.; Nishida, N.; Wu, X.; Nicoloso, M.S.; Shah, M.Y.; Devlin, C.; Ling, H.; Shimizu, M.; Kumar, K.; Cortez, M.A.; et al. HINCUTs in cancer: Hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ. 2013, 20, 1675–1687. [Google Scholar] [CrossRef] [PubMed]
- Redis, R.S.; Vela, L.E.; Lu, W.; Ferreira de Oliveira, J.; Ivan, C.; Rodriguez-Aguayo, C.; Adamoski, D.; Pasculli, B.; Taguchi, A.; Chen, Y.; et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol. Cell 2016, 61, 640. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.Y.; Ferracin, M.; Pileczki, V.; Chen, B.; Redis, R.; Fabris, L.; Zhang, X.; Ivan, C.; Shimizu, M.; Rodriguez-Aguayo, C.; et al. Cancer-associated rs6983267 SNP and its accompanying long noncoding RNA CCAT2 induce myeloid malignancies via unique SNP-specific RNA mutations. Genome Res. 2018, 28, 432–447. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Wu, W.; Wagner, E.K.; Hao, Y.; Rao, X.; Dai, H.; Han, J.; Chen, J.; Storniolo, A.M.; Liu, Y.; He, C. Tissue-specific Co-expression of Long Non-coding and Coding RNAs Associated with Breast Cancer. Sci. Rep. 2016, 6, 32731. [Google Scholar] [CrossRef]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Dragomir, M.; Chen, B.; Calin, G.A. Exosomal lncRNAs as new players in cell-to-cell communication. Transl. Cancer Res. 2018, 7, S243–S252. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef]
- Qu, L.; Ding, J.; Chen, C.; Wu, Z.J.; Liu, B.; Gao, Y.; Chen, W.; Liu, F.; Sun, W.; Li, X.F.; et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016, 29, 653–668. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Lanz, R.B.; McKenna, N.J.; Onate, S.A.; Albrecht, U.; Wong, J.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999, 97, 17–27. [Google Scholar] [CrossRef]
- Hatchell, E.C.; Colley, S.M.; Beveridge, D.J.; Epis, M.R.; Stuart, L.M.; Giles, K.M.; Redfern, A.D.; Miles, L.E.; Barker, A.; MacDonald, L.M.; et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol. Cell 2006, 22, 657–668. [Google Scholar] [CrossRef]
- Cooper, C.; Guo, J.; Yan, Y.; Chooniedass-Kothari, S.; Hube, F.; Hamedani, M.K.; Murphy, L.C.; Myal, Y.; Leygue, E. Increasing the relative expression of endogenous non-coding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009, 37, 4518–4531. [Google Scholar] [CrossRef]
- Mayama, T.; Marr, A.K.; Kino, T. Differential Expression of Glucocorticoid Receptor Noncoding RNA Repressor Gas5 in Autoimmune and Inflammatory Diseases. Horm. Metab. Res. 2016, 48, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009, 28, 195–208. [Google Scholar] [CrossRef]
- Sousa, D.; Lima, R.T.; Vasconcelos, M.H. Intercellular Transfer of Cancer Drug Resistance Traits by Extracellular Vesicles. Trends Mol. Med. 2015, 21, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, R.; Ji, H.; Greening, D.W.; Rai, A.; Izumikawa, K.; Ishikawa, H.; Takahashi, N.; Simpson, R.J. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci. Rep. 2016, 6, 38397. [Google Scholar] [CrossRef] [PubMed]
- Koldemir, O.; Ozgur, E.; Gezer, U. Accumulation of GAS5 in exosomes is a marker of apoptosis induction. Biomed. Rep. 2017, 6, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lin, C.; Liu, W.; Zhang, J.; Ohgi, K.A.; Grinstein, J.D.; Dorrestein, P.C.; Rosenfeld, M.G. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011, 147, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, L.H.; Spieker, T.; Koschmieder, S.; Schaffers, S.; Humberg, J.; Jungen, D.; Bulk, E.; Hascher, A.; Wittmer, D.; Marra, A.; et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. 2011, 6, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xia, Y.; Wang, Z.; Zheng, J.; Chen, Y.; Li, X.; Wang, Y.; Ming, H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017, 490, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.L.; Hu, G.W.; Zhang, B.; Kuang, W.; Chen, Y.; Wu, L.; Xu, G.H. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol. Rep. 2017, 38, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Redis, R.S.; Sieuwerts, A.M.; Look, M.P.; Tudoran, O.; Ivan, C.; Spizzo, R.; Zhang, X.; de Weerd, V.; Shimizu, M.; Ling, H.; et al. CCAT2, a novel long non-coding RNA in breast cancer: Expression study and clinical correlations. Oncotarget 2013, 4, 1748–1762. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, T.; Matsuyama, T.; Toiyama, Y.; Takahashi, N.; Ishikawa, T.; Uetake, H.; Yamada, Y.; Kusunoki, M.; Calin, G.; Goel, A. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ‘gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann. Oncol. 2017, 28, 1882–1888. [Google Scholar] [CrossRef] [PubMed]
- Fosselteder, J.; Calin, G.A.; Pichler, M. Long non-coding RNA CCAT2 as a therapeutic target in colorectal cancer. Expert Opin. Ther. Targets 2018, 22, 973–976. [Google Scholar] [CrossRef]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafa, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef] [PubMed]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef]
- Bullock, M.D.; Silva, A.M.; Kanlikilicer-Unaldi, P.; Filant, J.; Rashed, M.H.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. Exosomal Non-Coding RNAs: Diagnostic, Prognostic and Therapeutic Applications in Cancer. Noncoding RNA 2015, 1, 53–68. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardini, B.; Calin, G.A. MicroRNAs and Long Non-Coding RNAs and Their Hormone-Like Activities in Cancer. Cancers 2019, 11, 378. https://doi.org/10.3390/cancers11030378
Pardini B, Calin GA. MicroRNAs and Long Non-Coding RNAs and Their Hormone-Like Activities in Cancer. Cancers. 2019; 11(3):378. https://doi.org/10.3390/cancers11030378
Chicago/Turabian StylePardini, Barbara, and George A. Calin. 2019. "MicroRNAs and Long Non-Coding RNAs and Their Hormone-Like Activities in Cancer" Cancers 11, no. 3: 378. https://doi.org/10.3390/cancers11030378
APA StylePardini, B., & Calin, G. A. (2019). MicroRNAs and Long Non-Coding RNAs and Their Hormone-Like Activities in Cancer. Cancers, 11(3), 378. https://doi.org/10.3390/cancers11030378




