Lung Cancer Screening, towards a Multidimensional Approach: Why and How?
Abstract
:1. Introduction
2. Lung Cancer Screening Can Be Improved
2.1. Refine Selection Criteria to Improve the Effectiveness and Efficiency of Lung Cancer Screening
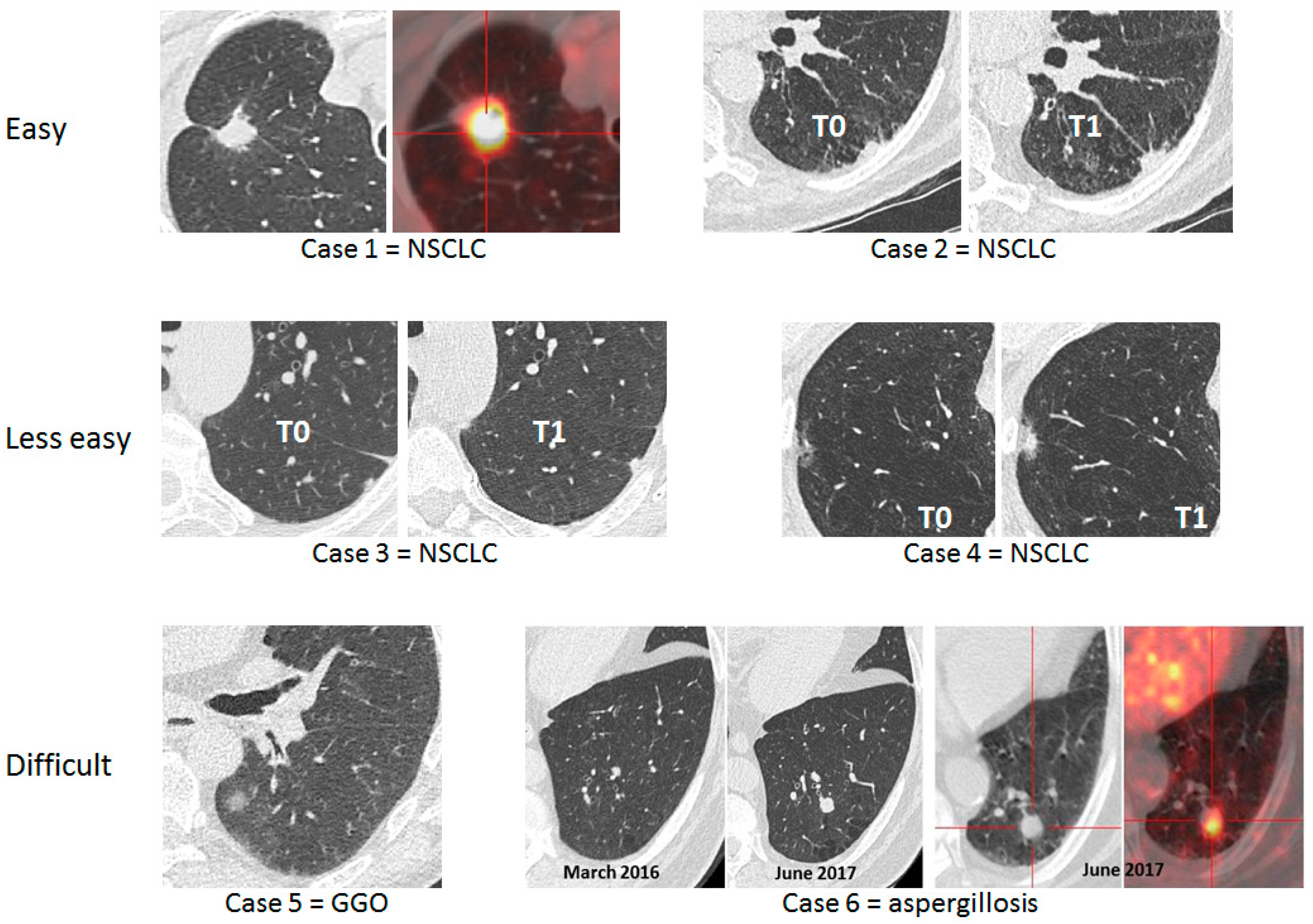
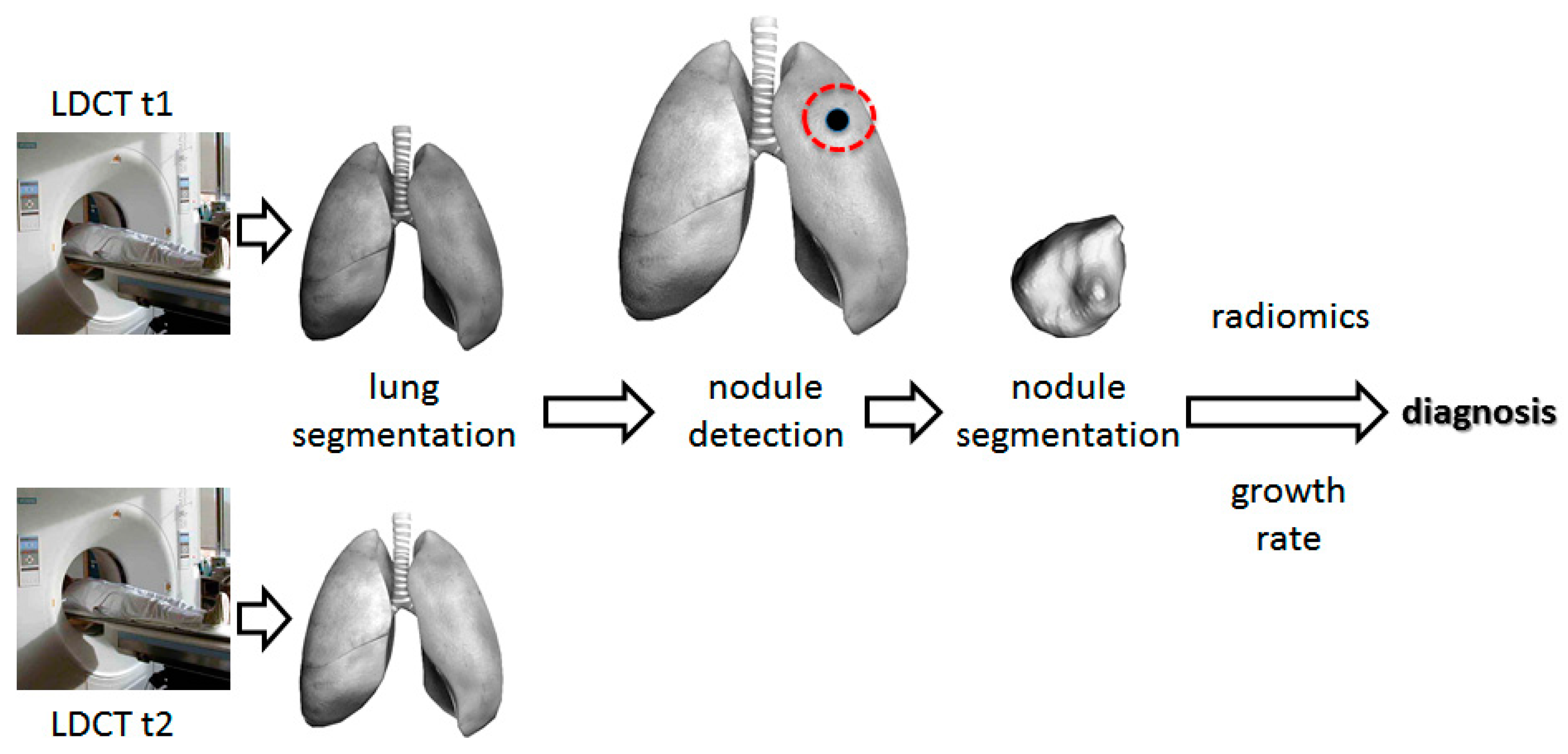
2.2. Use Computer Aided Diagnosis for Low-Dose Computed Tomography Interpretation to Facilitate Lung Cancer Screening and Lessen the False-Positive Rate
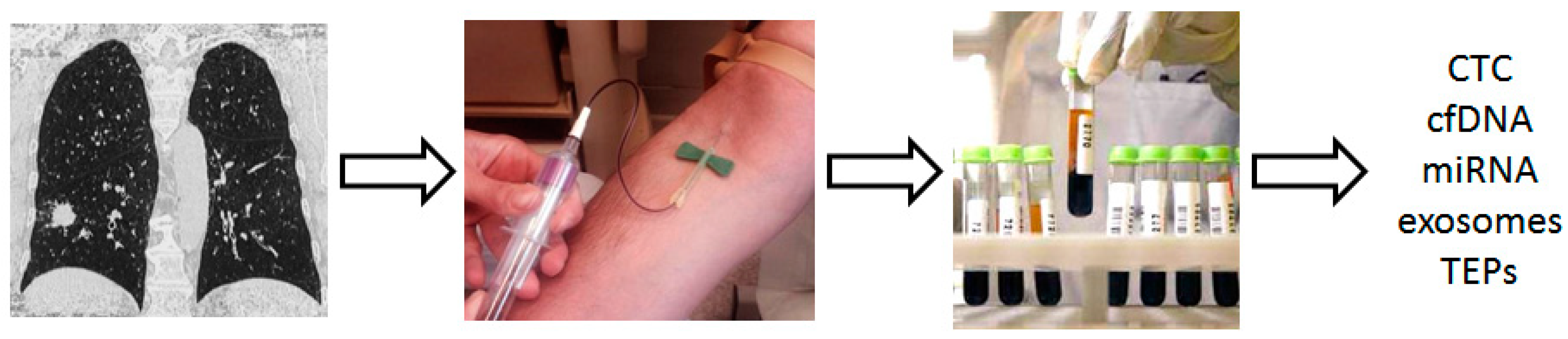
2.3. Use Blood Biomarkers in the Setting of Lung Cancer Screening
2.3.1. Biomarkers to Detect Early-Stage Lung Cancer
2.3.2. Biomarkers to Identify High-Risk Individuals and to Spot the Optimal Target Population for Lung Cancer Screening
2.3.3. Biomarkers to Help Classify Indeterminate Lung Nodules
2.4. Highly Sensitive Bronchoscopic Techniques to Enhance the Detection Rate of Central Airway Lesions
3. Deep Learning for Early Cancer Detection
4. Conclusions
Funding
Conflicts of Interest
References
- National Lung Screening Trial Research Team; Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [PubMed]
- De Koning, H.J.; Van Der Aalst, C.M.; Ten Haaf, K.; Oudkerk, M. PL02.05. Effects of Volume CT Lung Cancer Screening: Mortality Results of the NELSON Randomised-Controlled Population Based Trial. J. Thorac. Oncol. 2018, 13, S185. [Google Scholar] [CrossRef]
- Moyer, V.A.; On behalf of U.S. Preventive Services Task Force. Screening for lung cancer: U.S. preventive services task force recommendation statement. Ann. Intern. Med. 2014, 160, 330–338. [Google Scholar] [CrossRef] [PubMed]
- CMS.gov. Available online: https://www.cms.gov/medicare-coverage-database/details/nca-decision-memo.aspx?NCAId=274 (accessed on 3 February 2019).
- Oudkerk, M.; Devaraj, A.; Vliegenthart, R.; Henzler, T.; Prosch, H.; Heussel, C.P.; Bastarrika, G.; Sverzellati, N.; Mascalchi, M.; Delorme, S.; et al. European position statement on lung cancer screening. Lancet Oncol. 2017, 18, e754–e766. [Google Scholar] [CrossRef]
- IASLC. Available online: https://www.iaslc.org/news/iaslc-issues-statement-lung-cancer-screening-low-dose-computed-tomography?fbclid=IwAR3LdKR99ek3D2cGKlII4fA8s7IcaW6jhefdxHSYVPJAgkYn1ER8RW0MWOk (accessed on 3 February 2019).
- Kinsinger, L.S.; Anderson, C.; Kim, J.; Larson, M.; Chan, S.H.; King, H.A.; Rice, K.L.; Slatore, C.G.; Tanner, N.T.; Pittman, K.; et al. Implementation of lung cancer screening in the Veterans Health Administration. JAMA Intern. Med. 2017, 177, 399–406. [Google Scholar] [CrossRef]
- Pham, D.; Bhandari, S.; Oechsli, M.; Pinkston, C.M.; Kloecker, G.H. Lung cancer screening rates: Data from the lung cancer screening registry. J. Clin. Oncol. 2018, 36, 6504. [Google Scholar] [CrossRef]
- Tonge, J.E.; Atack, M.; Crosbie, P.A.; Barber, P.V.; Booton, R.; Colligan, D. “To know or not to know…?” Push and pull in ever smokers lung screening uptake decision-making intentions. Health Expect. 2018, 13, S968. [Google Scholar] [CrossRef]
- Haute Autorité de Santé. Available online: https://www.has-sante.fr/portail/jcms/c_2001613/fr/pertinence-du-depistage-du-cancer-broncho-pulmonaire-en-france-point-de-situation-sur-les-donnees-disponibles-analyse-critique-des-etudes-controlees-randomisees (accessed on 3 February 2019).
- Katki, H.A.; Kovalchik, S.A.; Berg, C.D.; Cheung, L.C.; Chaturvedi, A.K. Development and validation of risk models to select ever-smokers for ct lung cancer screening. JAMA J. Am. Med. Assoc. 2016, 315, 2300–2311. [Google Scholar] [CrossRef]
- Caverly, T.J.; Fagerlin, A.; Wiener, R.S.; Slatore, C.G.; Tanner, N.T.; Yun, S.; Hayward, R. Comparison of Observed Harms and Expected Mortality Benefit for Persons in the Veterans Health Affairs Lung Cancer Screening Demonstration Project. JAMA Intern. Med. 2018, 178, 426–428. [Google Scholar] [CrossRef]
- Bach, P.B.; Elkin, E.B.; Pastorino, U.; Kattan, M.W.; Mushlin, A.I.; Begg, C.B.; Parkin, D.M. Benchmarking lung cancer mortality rates in current and former smokers. Chest 2004, 126, 1742–1749. [Google Scholar] [CrossRef]
- Wood, D.E.; Kazerooni, E.A.; Baum, S.L.; Eapen, G.A.; Ettinger, D.S.; Hou, L.; Jackman, D.M.; Klippenstein, D.; Kumar, R.; Lackner, R.P.; et al. Lung cancer screening, version 3.2018. JNCCN J. Natl. Compr. Cancer Netw. 2018, 16, 412–441. [Google Scholar] [CrossRef] [PubMed]
- El-Baz, A.; Beache, G.M.; Gimel’Farb, G.; Suzuki, K.; Okada, K.; Elnakib, A.; Soliman, A.; Abdollahi, B. Computer-aided diagnosis systems for lung cancer: Challenges and methodologies. Int. J. Biomed. Imaging 2013, 2013, 942353. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.; Skalski, M.; Gaillard, F. The utilisation of convolutional neural networks in detecting pulmonary nodules: A review. Br. J. Radiol. 2018, 91, 20180028. [Google Scholar] [CrossRef] [PubMed]
- Armato, S.G., 3rd; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Kaggle. Kaggle Data Science Bowl 2017. 2017. Available online: https://www.kaggle.com/www.kaggle.com (accessed on 3 February 2019).
- Liao, F.; Liang, M.; Li, Z.; Hu, X.; Song, S. Evaluate the Malignancy of Pulmonary Nodules Using the 3D Deep Leaky Noisy-or Network. IEEE Trans. Cybern. 2017, 14, 1–12. [Google Scholar]
- Santarpia, M.; Liguori, A.; D’Aveni, A.; Karachaliou, N.; Gonzalez-Cao, M.; Daffinà, M.G.; Lazzari, C.; Altavilla, G.; Rosell, R. Liquid biopsy for lung cancer early detection. J. Thorac. Dis. 2018, 10, S882–S897. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P. Liquid biopsy for early detection of lung cancer. Curr. Opin. Oncol. 2017, 29, 73–78. [Google Scholar] [CrossRef]
- Ilie, M.; Hofman, V.; Long-Mira, E.; Selva, E.; Vignaud, J.M.; Padovani, B.; Mouroux, J.; Marquette, C.H.; Hofman, P. “Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with Chronic obstructive pulmonary disease. PLoS ONE 2014, 9, e111597. [Google Scholar] [CrossRef]
- Hofman, V.J.; Ilie, M.; Hofman, P.M. Detection and characterization of circulating tumor cells in lung cancer: Why and how? Cancer Cytopathol. 2016, 124, 380–387. [Google Scholar] [CrossRef]
- Leroy, S.; Benzaquen, J.; Mazzetta, A.; Marchand-Adam, S.; Padovani, B.; Israel-Biet, D.; Pison, C.; Chanez, P.; Cadranel, J.; Mazières, J.; et al. Circulating tumour cells as a potential screening tool for lung cancer (the AIR study): Protocol of a prospective multicentre cohort study in France. BMJ Open 2017, 7, e018884. [Google Scholar] [CrossRef]
- Wozniak, M.B.; Scelo, G.; Muller, D.C.; Mukeria, A.; Zaridze, D.; Brennan, P. Circulating MicroRNAs as Non-Invasive Biomarkers for Early Detection of Non-Small-Cell Lung Cancer. PLoS ONE 2015, 10, e0125026. [Google Scholar] [CrossRef] [PubMed]
- Sanfiorenzo, C.; Ilie, M.I.; Belaid, A.; Barlési, F.; Mouroux, J.; Marquette, C.H.; Brest, P.; Hofman, P. Two Panels of Plasma MicroRNAs as Non-Invasive Biomarkers for Prediction of Recurrence in Resectable NSCLC. PLoS ONE 2013, 8, e54596. [Google Scholar] [CrossRef] [PubMed]
- Boeri, M.; Verri, C.; Conte, D.; Roz, L.; Modena, P.; Facchinetti, F.; Calabro, E.; Croce, C.M.; Pastorino, U.; Sozzi, G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 3713–3718. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, G.; Boeri, M.; Rossi, M.; Verri, C.; Suatoni, P.; Bravi, F.; Roz, L.; Conte, D.; Grassi, M.; Sverzellati, N.; et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: A correlative MILD trial study. J. Clin. Oncol. 2014, 32, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Montani, F.; Marzi, M.J.; Dezi, F.; Dama, E.; Carletti, R.M.; Bonizzi, G.; Bertolotti, R.; Bellomi, M.; Rampinelli, C.; Maisonneuve, P.; et al. MiR-test: A blood test for lung cancer early detection. J. Natl. Cancer Inst. 2015, 107, djv063. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Billatos, E.; Vick, J.L.; Lenburg, M.E.; Spira, A.E. The airway transcriptome as a biomarker for early lung cancer detection. Clin. Cancer Res. 2018, 24, 2984–2992. [Google Scholar] [CrossRef] [PubMed]
- Guida, F.; Sun, N.; Bantis, L.E.; Muller, D.C.; Li, P.; Taguchi, A.; Dhillon, D.; Kundnani, D.L.; Patel, N.J.; Yan, Q.; et al. and the Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Consortium for Early Detection of Lung Cancer. Assessment of Lung Cancer Risk on the Basis of a Biomarker Panel of Circulating Proteins. JAMA Oncol. 2018, 4, e182078. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Ren, S.; Zhou, C. Role of circulating-tumor DNA analysis in non-small cell lung cancer. Lung Cancer 2015, 90, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Lehmann-Werman, R.; Neiman, D.; Zemmour, H.; Moss, J.; Magenheim, J.; Vaknin-Dembinsky, A.; Rubertsson, S.; Nellgård, B.; Blennow, K.; Zetterberg, H.; et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E1826–E1834. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, M.; Amati, M.; Neuzil, J.; Santarelli, L. Circulating epigenetic biomarkers in lung malignanciesFrom early diagnosis to therapy. Lung Cancer 2017, 107, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Hosgood, H.D.; Chen, B.; Absher, D.; Assimes, T.; Horvath, S. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging 2015, 7, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Warton, K.; Samimi, G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front. Mol. Biosci. 2015, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- Hulbert, A.; Jusue-Torres, I.; Stark, A.; Chen, C.; Rodgers, K.; Lee, B.; Griffin, C.; Yang, A.; Huang, P.; Wrangle, J.; et al. Early detection of lung cancer using DNA promoter hypermethylation in plasma and sputum. Clin. Cancer Res. 2017, 23, 1998–2005. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, G.A.; Tanner, N.T.; Kearney, P.; Vachani, A.; Massion, P.P.; Porter, A.; Springmeyer, S.C.; Fang, K.C.; Midthun, D.; Mazzone, P.J. and PANOPTIC Trial Team. Assessment of Plasma Proteomics Biomarker’s Ability to Distinguish Benign from Malignant Lung Nodules: Results of the PANOPTIC (Pulmonary Nodule Plasma Proteomic Classifier) Trial. Chest 2018, 154, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Massion, P.P.; Healey, G.F.; Peek, L.J.; Fredericks, L.; Sewell, H.F.; Murray, A.; Robertson, J.F.R. Autoantibody Signature Enhances the Positive Predictive Power of Computed Tomography and Nodule-Based Risk Models for Detection of Lung Cancer. J. Thorac. Oncol. 2017, 12, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, F.M.; Farmer, E.; Mair, F.S.; Treweek, S.; Kendrick, D.; Jackson, C.; Robertson, C.; Briggs, A.; McCowan, C.; Bedford, L.; et al. Detection in blood of autoantibodies to tumour antigens as a case-finding method in lung cancer using the EarlyCDT®-Lung Test (ECLS): Study protocol for a randomized controlled trial. BMC Cancer 2017, 17, 187. [Google Scholar] [CrossRef] [PubMed]
- Ajona, D.; Okrój, M.; Pajares, M.J.; Agorreta, J.; Lozano, M.D.; Zulueta, J.J.; Verri, C.; Roz, L.; Sozzi, G.; Pastorino, U.; et al. Complement C4d-specific antibodies for the diagnosis of lung cancer. Oncotarget 2018, 9, 6346–6355. [Google Scholar] [CrossRef]
- Andolfi, M.; Potenza, R.; Capozzi, R.; Liparulo, V.; Puma, F.; Yasufuku, K. The role of bronchoscopy in the diagnosis of early lung cancer: A review. J. Thorac. Dis. 2016, 8, 3329–3337. [Google Scholar] [CrossRef]
- McWilliams, A.; Mayo, J.; MacDonald, S.; leRiche, J.C.; Palcic, B.; Szabo, E. Lung Cancer Screening. Am. J. Respir. Crit. Care Med. 2003, 168, 1167–1173. [Google Scholar] [CrossRef]
- McWilliams, A.M.; Mayo, J.R.; Ahn, M.I.; MacDonald, S.L.S.; Lam, S.C. Lung cancer screening using multi-slice thin-section computed tomography and autofluorescence bronchoscopy. J. Thorac. Oncol. 2006, 1, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, A.; Taghizadeh, N.; McWilliams, A.M.; MacEachern, P.; Stather, D.R.; Soghrati, K.; Puksa, S.; Goffin, J.R.; Yasufuku, K.; Amjadi, K.; et al. Low Prevalence of High-Grade Lesions Detected with Autofluorescence Bronchoscopy in the Setting of Lung Cancer Screening in the Pan-Canadian Lung Cancer Screening Study. Chest 2016, 150, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Epelbaum, O.; Aronow, W.S. Autofluorescence bronchoscopy for lung cancer screening: A time to reflect. Ann. Transl. Med. 2016, 4. [Google Scholar] [CrossRef] [PubMed]



| NLST | NELSON | ||||
|---|---|---|---|---|---|
| Country | USA | BE/NL | |||
| Enrollment | 2002–2004 | 2003–NR | |||
| Number of Centers | 33 | 4 | |||
| Number of screens | 3 | ||||
| Screening planned at years | 1, 2 and 3 | 1, 2 and 4 | |||
| Comparison | LDCT vs. Xray | LDCT vs. usual care | |||
| Population | |||||
| Age | 55–74 | 50–69 (50–75) | |||
| Smoking (pack-years) | ≥30 | >15 * | |||
| Sex | both (male 59%) | men º (male 84%) | |||
| Years since quit | ≤15 | ≤10 | |||
| Patients Screened, n | 26,722 vs. 26,732 | 7907 vs. 7915 | |||
| Planned follow-up, y | >7 | 10 | |||
| Nodule Size warranting Follow-up | 2011 |  | 2009 | 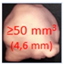 | + VDT |
| 2014 | ≥100 mm3 (≥5 mm) | + VDT | |||
| LC diagnosed at screening, % | 1.02 | 0.9 | |||
| 5 mm Reduction of LC mortality | 20% | 26% a | |||
| Kaggle | Luna16 | NLST | COPDG Gene | LTRC | |
|---|---|---|---|---|---|
| Number | 1397 | 888 | >1000 | >1000 | >1000 |
| Date | 2002–2004 to 2009 | Start 2008 | |||
| Available without registration | Yes | Yes | No | No | No |
| COPD * cases | Yes | Yes | |||
| Ground truth | Cancer/no cancer one year after the CT scan | x, y, z coordinates and diameter of nodules | |||
| Image data | Yes | Yes | Yes | Yes | Yes |
| Cohort level | “high-risk patient” | Age 55–74 >30 years smoking history <15 years since quitting | Age 45–80 >10 pack-years smoking history | “Most donor subjects have interstitial fibrotic lung disease or COPD” Average age 60 | |
| Individual level | Questionnaire: living condition, family history. Cancer diagnosis: location/tumor size | Subject phenotype: living condition, gender, medical history, comorbidities, physical characteristics… | Clinical and pathological diagnoses, pulmonary function tests, living condition, exercises tests… | ||
| Biological data | Lung tissues | SNP genotype | Blood and lung tissues |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benzaquen, J.; Boutros, J.; Marquette, C.; Delingette, H.; Hofman, P. Lung Cancer Screening, towards a Multidimensional Approach: Why and How? Cancers 2019, 11, 212. https://doi.org/10.3390/cancers11020212
Benzaquen J, Boutros J, Marquette C, Delingette H, Hofman P. Lung Cancer Screening, towards a Multidimensional Approach: Why and How? Cancers. 2019; 11(2):212. https://doi.org/10.3390/cancers11020212
Chicago/Turabian StyleBenzaquen, Jonathan, Jacques Boutros, Charles Marquette, Hervé Delingette, and Paul Hofman. 2019. "Lung Cancer Screening, towards a Multidimensional Approach: Why and How?" Cancers 11, no. 2: 212. https://doi.org/10.3390/cancers11020212
APA StyleBenzaquen, J., Boutros, J., Marquette, C., Delingette, H., & Hofman, P. (2019). Lung Cancer Screening, towards a Multidimensional Approach: Why and How? Cancers, 11(2), 212. https://doi.org/10.3390/cancers11020212






