CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1
Abstract
:1. Introduction
2. Results
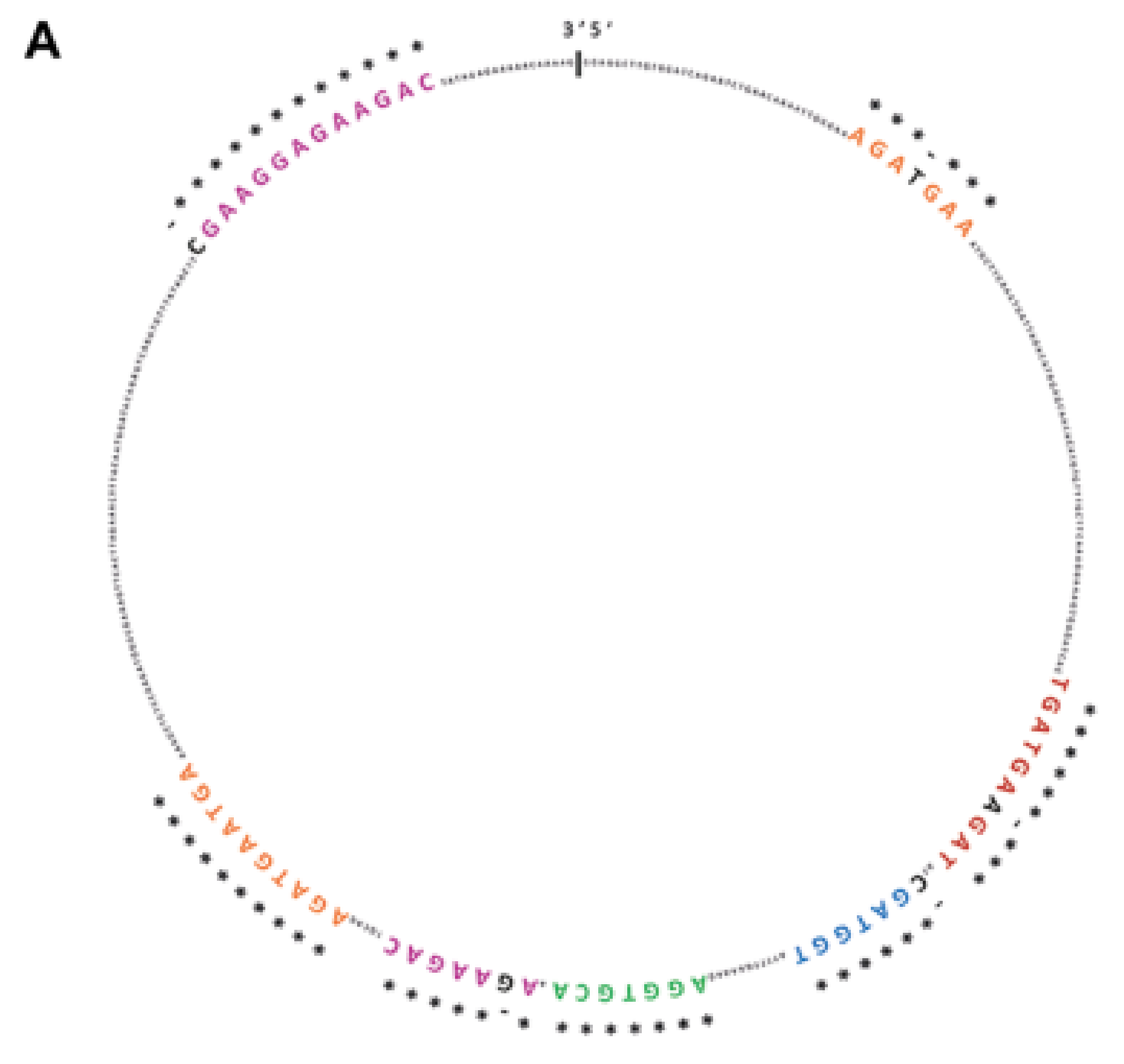
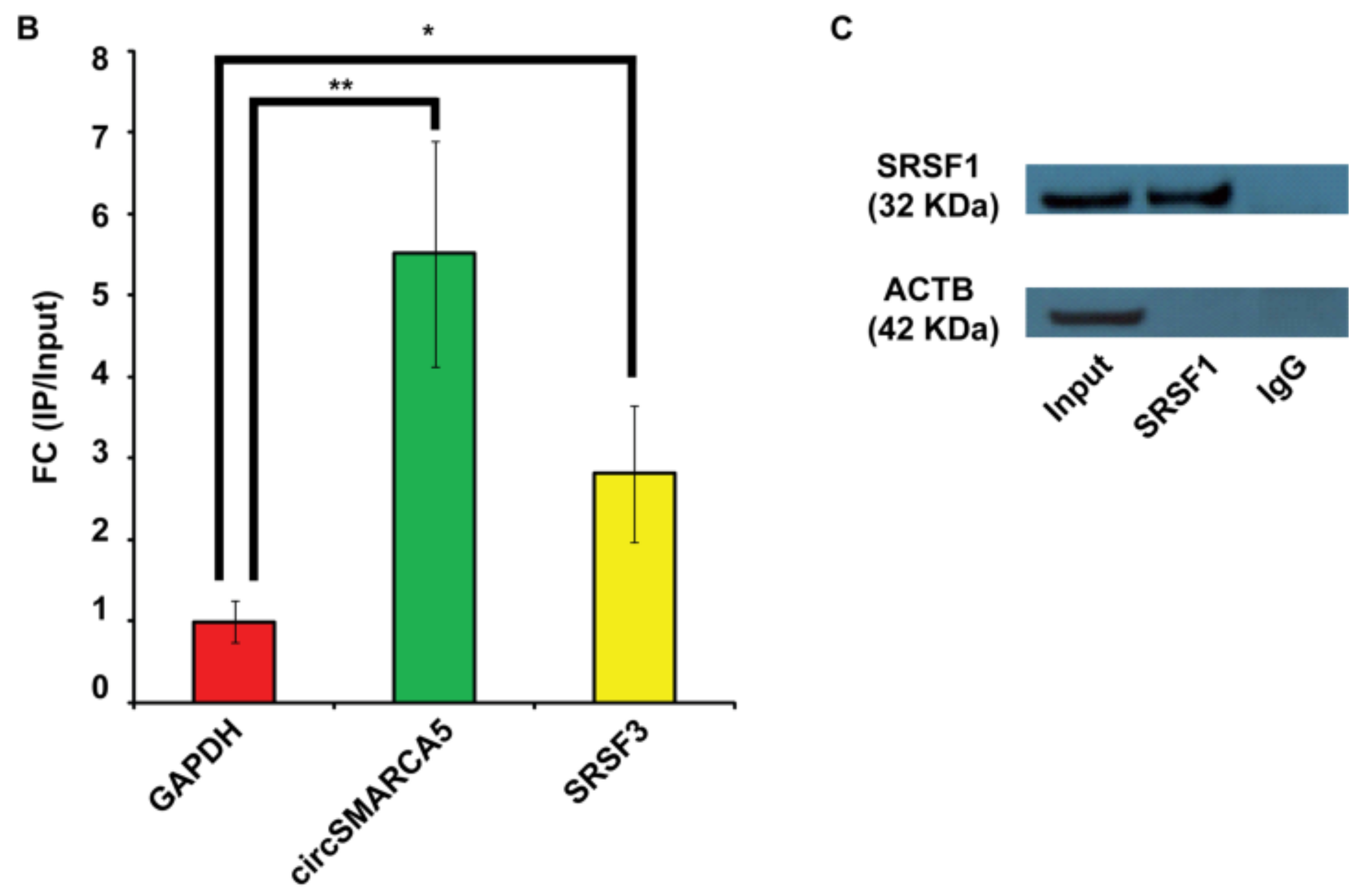
2.1. SRSF1 and CircSMARCA5 Physically Interact within GBM Cells
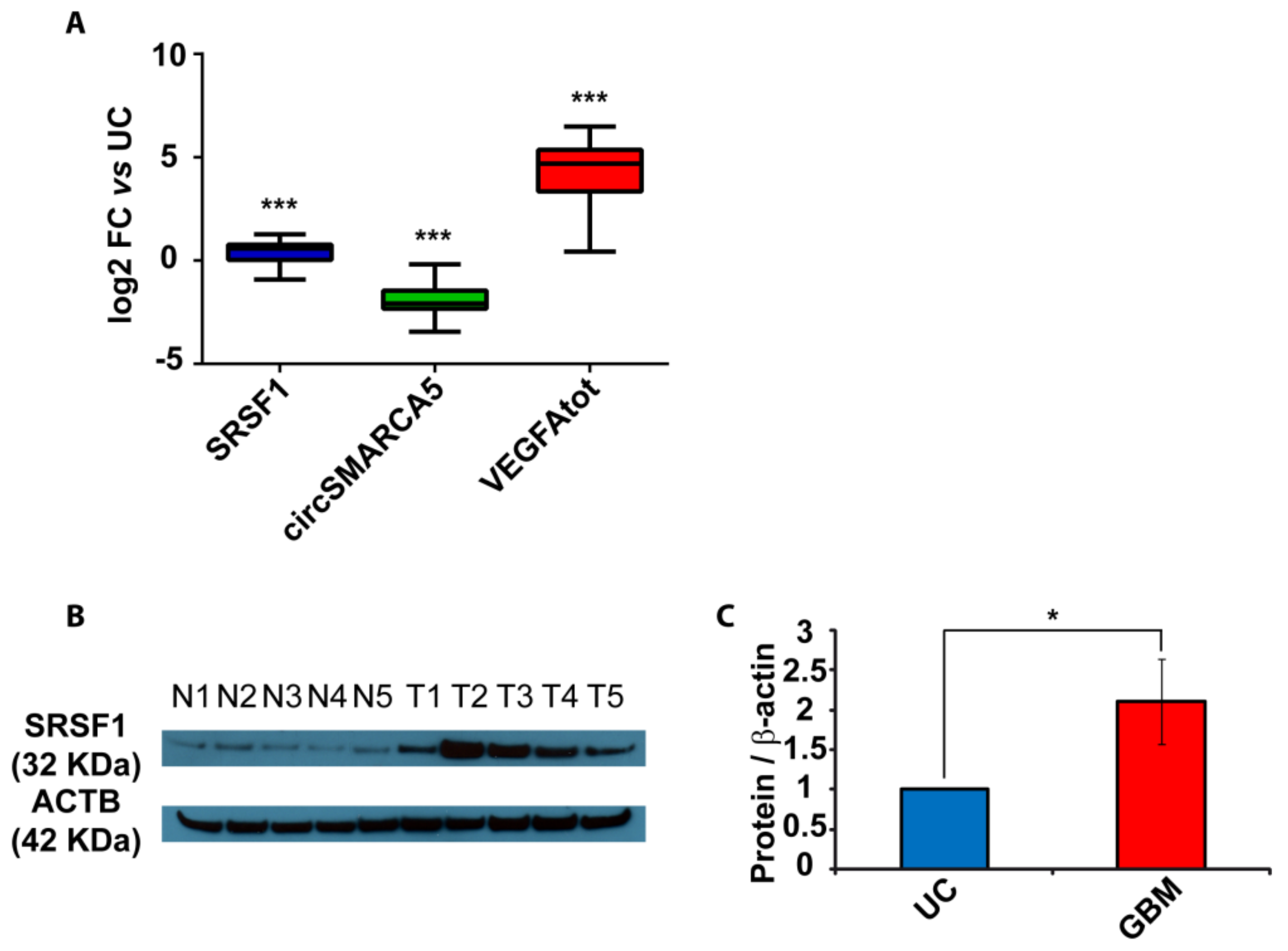
2.2. SRSF1 and VEGFA Are Upregulated While circSMARCA5 Is Downregulated in GBM Biopsies
2.3. The Ratio of Pro- to Anti-Angiogenic VEGFA mRNA Isoforms Increases in GBM Cells as Compared to Unaffected Brain Parenchyma and Decreases in U87-MG Overexpressing circSMARCA5
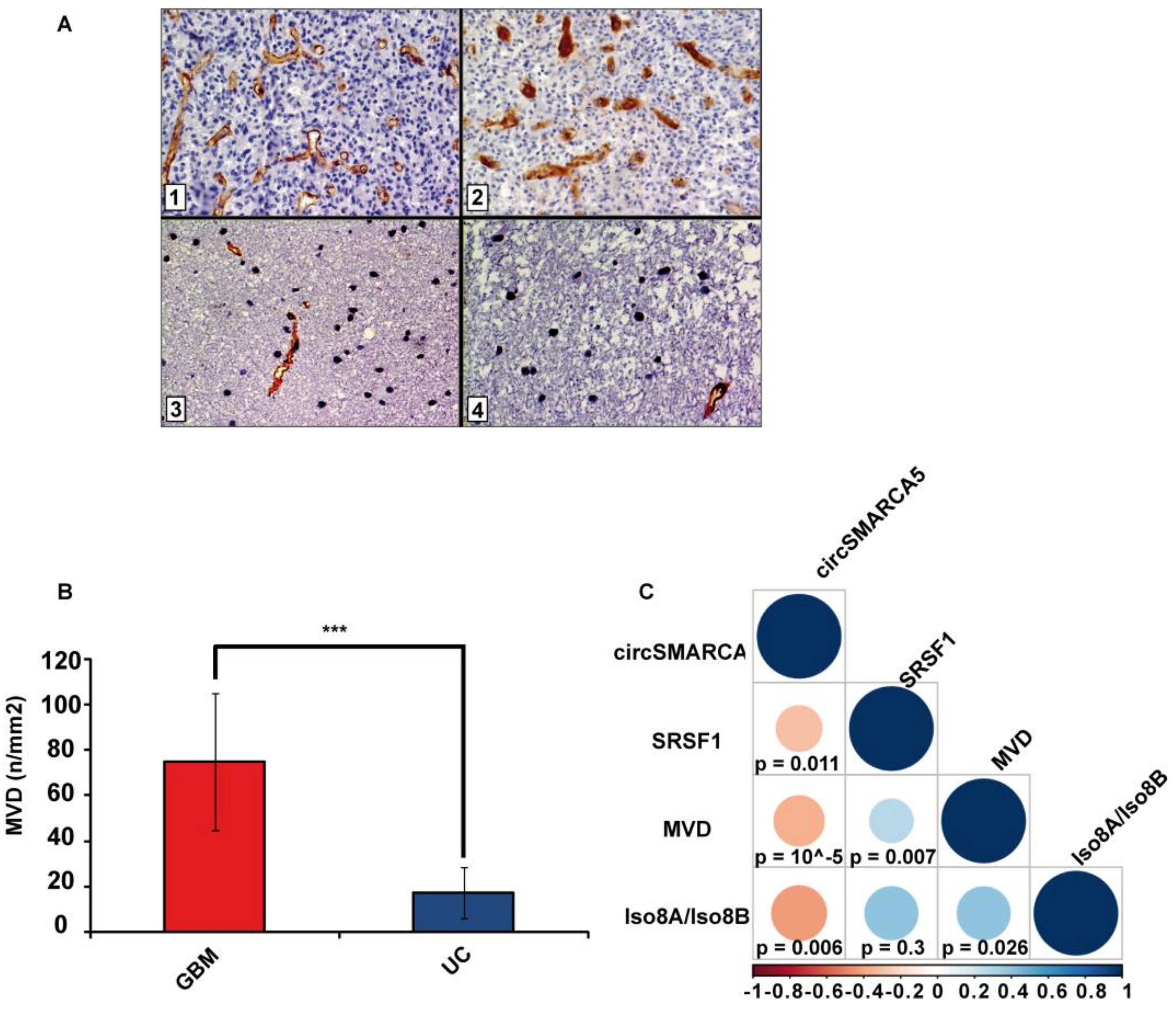
2.4. Blood Vascular Microvessel Density (MVD) Negatively Correlates with the Expression of circSMARCA5, While Positively Correlating with SRSF1 mRNA Expression and Pro/Anti-Angiogenic VEGFA Isoforms Ratio
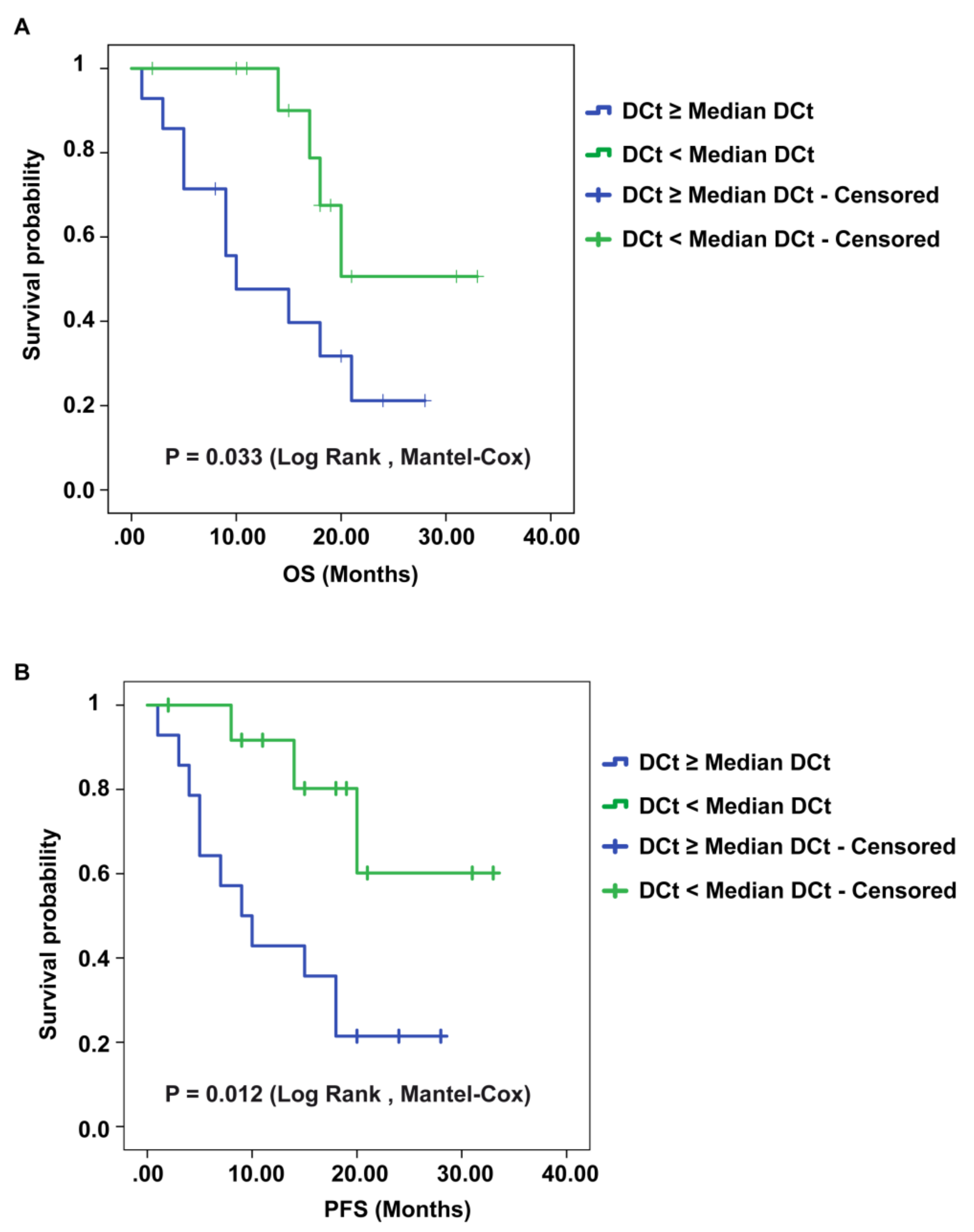
2.5. CircSMARCA5 Expression Negatively Correlates with GBM Patients’ Overall Survival (OS) and Progression-Free Survival (PFS)
3. Discussion
4. Materials and Methods
4.1. GBM Biopsies
4.2. Cell Cultures and Transfection
4.3. RNA Immunoprecipitation (RIP)
4.4. RNA Extraction and Real-Time PCR
4.5. Protein Extraction and Immunoblotting
4.6. Immunohistochemistry
4.7. Assessment of Blood Vascular Microvessel Density (MVD)
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgements
Conflicts of Interest
References
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Ebbesen, K.K.; Hansen, T.B.; Kjems, J. Insights into circular RNA biology. RNA Biol. 2017, 14, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, T.; Xiao, J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018, 34, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.; Daugaard, I.; Andersen, M.S.; Hansen, T.B.; Gronbaek, K.; Kjems, J.; Kristensen, L.S. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Invest. 2018, 98, 1657–1669. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Hansen, T.B.; Veno, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef]
- Dragomir, M.; Calin, G.A. Circular RNAs in Cancer—Lessons Learned From microRNAs. Front. Oncol. 2018, 8, 179. [Google Scholar] [CrossRef]
- Floris, G.; Zhang, L.; Follesa, P.; Sun, T. Regulatory Role of Circular RNAs and Neurological Disorders. Mol. Neurobiol. 2017, 54, 5156–5165. [Google Scholar] [CrossRef]
- Altesha, M.A.; Ni, T.; Khan, A.; Liu, K.; Zheng, X. Circular RNA in cardiovascular disease. J. Cell Physiol. 2018. [Google Scholar] [CrossRef]
- Barbagallo, D.; Condorelli, A.; Ragusa, M.; Salito, L.; Sammito, M.; Banelli, B.; Caltabiano, R.; Barbagallo, G.; Zappala, A.; Battaglia, R.; et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget 2016, 7, 4746–4759. [Google Scholar] [CrossRef]
- Pajares, M.J.; Ezponda, T.; Catena, R.; Calvo, A.; Pio, R.; Montuenga, L.M. Alternative splicing: An emerging topic in molecular and clinical oncology. Lancet Oncol. 2007, 8, 349–357. [Google Scholar] [CrossRef]
- Wang, B.D.; Lee, N.H. Aberrant RNA Splicing in Cancer and Drug Resistance. Cancers (Basel) 2018, 10, 458. [Google Scholar] [CrossRef] [PubMed]
- Montes, M.; Sanford, B.L.; Comiskey, D.F.; Chandler, D.S. RNA Splicing and Disease: Animal Models to Therapies. Trends Genet. 2019, 35, 68–87. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Liao, P.; Vecchione-Koval, T.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro-Oncology 2017, 19, v1–v88. [Google Scholar] [CrossRef]
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.N. Circular RNA Splicing. Adv. Exp. Med. Biol. 2018, 1087, 41–52. [Google Scholar] [CrossRef]
- Qin, M.; Wei, G.; Sun, X. Circ-UBR5: An exonic circular RNA and novel small nuclear RNA involved in RNA splicing. Biochem. Biophys. Res. Commun. 2018, 503, 1027–1034. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Caponnetto, A.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; D’Angeli, F.; Morrone, A.; Caltabiano, R.; Barbagallo, G.M.; Ragusa, M.; et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018, 19, 480. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Krainer, A.R. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 2014, 12, 1195–1204. [Google Scholar] [CrossRef]
- Karni, R.; de Stanchina, E.; Lowe, S.W.; Sinha, R.; Mu, D.; Krainer, A.R. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007, 14, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Ratnadiwakara, M.; Archer, S.K.; Dent, C.I.; Ruiz De Los Mozos, I.; Beilharz, T.H.; Knaupp, A.S.; Nefzger, C.M.; Polo, J.M.; Anko, M.L. SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program. Elife 2018, 7. [Google Scholar] [CrossRef]
- Madhavan, S.; Zenklusen, J.C.; Kotliarov, Y.; Sahni, H.; Fine, H.A.; Buetow, K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res. 2009, 7, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwinska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Pozn) 2015, 19, A68–A77. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, E.L.; Pratt, G.A.; Shishkin, A.A.; Gelboin-Burkhart, C.; Fang, M.Y.; Sundararaman, B.; Blue, S.M.; Nguyen, T.B.; Surka, C.; Elkins, K.; et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 2016, 13, 508–514. [Google Scholar] [CrossRef]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2018. [Google Scholar] [CrossRef]
- Charley, P.A.; Wilusz, J. Sponging of cellular proteins by viral RNAs. Curr. Opin. Virol. 2014, 9, 14–18. [Google Scholar] [CrossRef]
- Barbagallo, C.; Brex, D.; Caponnetto, A.; Cirnigliaro, M.; Scalia, M.; Magnano, A.; Caltabiano, R.; Barbagallo, D.; Biondi, A.; Cappellani, A.; et al. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol. Ther. Nucleic Acids 2018, 12, 229–241. [Google Scholar] [CrossRef]
- Kim, J.; Abdelmohsen, K.; Yang, X.; De, S.; Grammatikakis, I.; Noh, J.H.; Gorospe, M. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016, 44, 2378–2392. [Google Scholar] [CrossRef]
- Anczukow, O.; Akerman, M.; Clery, A.; Wu, J.; Shen, C.; Shirole, N.H.; Raimer, A.; Sun, S.; Jensen, M.A.; Hua, Y.; et al. SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol. Cell. 2015, 60, 105–117. [Google Scholar] [CrossRef]
- Martinez-Terroba, E.; Ezponda, T.; Bertolo, C.; Sainz, C.; Remirez, A.; Agorreta, J.; Garmendia, I.; Behrens, C.; Pio, R.; Wistuba, I.I.; et al. The oncogenic RNA-binding protein SRSF1 regulates LIG1 in non-small cell lung cancer. Lab. Invest. 2018, 98, 1562–1574. [Google Scholar] [CrossRef] [PubMed]
- Amin, E.M.; Oltean, S.; Hua, J.; Gammons, M.V.; Hamdollah-Zadeh, M.; Welsh, G.I.; Cheung, M.K.; Ni, L.; Kase, S.; Rennel, E.S.; et al. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell 2011, 20, 768–780. [Google Scholar] [CrossRef]
- Mavrou, A.; Brakspear, K.; Hamdollah-Zadeh, M.; Damodaran, G.; Babaei-Jadidi, R.; Oxley, J.; Gillatt, D.A.; Ladomery, M.R.; Harper, S.J.; Bates, D.O.; et al. Serine-arginine protein kinase 1 (SRPK1) inhibition as a potential novel targeted therapeutic strategy in prostate cancer. Oncogene 2015, 34, 4311–4319. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Abou-Rayyah, Y.; Bischoff, J.; Ritchie, A.; Sebire, N.J.; Watts, P.; Churchill, A.J.; Bates, D.O. Altered ratios of pro- and anti-angiogenic VEGF-A variants and pericyte expression of DLL4 disrupt vascular maturation in infantile haemangioma. J. Pathol. 2016, 239, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Hamdollah Zadeh, M.A.; Amin, E.M.; Hoareau-Aveilla, C.; Domingo, E.; Symonds, K.E.; Ye, X.; Heesom, K.J.; Salmon, A.; D’Silva, O.; Betteridge, K.B.; et al. Alternative splicing of TIA-1 in human colon cancer regulates VEGF isoform expression, angiogenesis, tumour growth and bevacizumab resistance. Mol. Oncol. 2015, 9, 167–178. [Google Scholar] [CrossRef]
- Gilbert, M.R.; Dignam, J.J.; Armstrong, T.S.; Wefel, J.S.; Blumenthal, D.T.; Vogelbaum, M.A.; Colman, H.; Chakravarti, A.; Pugh, S.; Won, M.; et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N. Engl. J. Med. 2014, 370, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Circular RNAs as Therapeutic Agents and Targets. Front. Physiol. 2018, 9, 1262. [Google Scholar] [CrossRef]
- Fukuhara, T.; Hosoya, T.; Shimizu, S.; Sumi, K.; Oshiro, T.; Yoshinaka, Y.; Suzuki, M.; Yamamoto, N.; Herzenberg, L.A.; Herzenberg, L.A.; et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. USA 2006, 103, 11329–11333. [Google Scholar] [CrossRef]
- Peritz, T.; Zeng, F.; Kannanayakal, T.J.; Kilk, K.; Eiriksdottir, E.; Langel, U.; Eberwine, J. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006, 1, 577–580. [Google Scholar] [CrossRef]
- Di Pietro, C.; Vento, M.; Guglielmino, M.R.; Borzi, P.; Santonocito, M.; Ragusa, M.; Barbagallo, D.; Duro, L.R.; Majorana, A.; De Palma, A.; et al. Molecular profiling of human oocytes after vitrification strongly suggests that they are biologically comparable with freshly isolated gametes. Fertil. Steril. 2010, 94, 2804–2807. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Urbano, F.; Filippello, A.; Di Pino, A.; Barbagallo, D.; Di Mauro, S.; Pappalardo, A.; Rabuazzo, A.M.; Purrello, M.; Purrello, F.; Piro, S. Altered expression of uncoupling protein 2 in GLP-1-producing cells after chronic high glucose exposure: Implications for the pathogenesis of diabetes mellitus. Am. J. Physiol. Cell Physiol. 2016, 310, C558–C567. [Google Scholar] [CrossRef]
- Caltabiano, R.; Puzzo, L.; Barresi, V.; Ieni, A.; Loreto, C.; Musumeci, G.; Castrogiovanni, P.; Ragusa, M.; Foti, P.; Russo, A.; et al. ADAM 10 expression in primary uveal melanoma as prognostic factor for risk of metastasis. Pathol. Res. Pract. 2016, 212, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Weidner, N.; Semple, J.P.; Welch, W.R.; Folkman, J. Tumor angiogenesis and metastasis—Correlation in invasive breast carcinoma. N. Engl. J. Med. 1991, 324, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, V.E.; Stensjoen, A.L.; Granli, U.S.; Berntsen, E.M.; Salvesen, O.; Solheim, O.; Torp, S.H. Angiogenesis and radiological tumor growth in patients with glioblastoma. BMC Cancer 2018, 18, 862. [Google Scholar] [CrossRef] [PubMed]
- Burton, A.L.; Egger, M.E.; Gilbert, J.E.; Stromberg, A.J.; Hagendoorn, L.; Martin, R.C.; Scoggins, C.R.; McMasters, K.M.; Callender, G.G. Assessment of mitotic rate reporting in melanoma. Am. J. Surg. 2012, 204, 969–974. [Google Scholar] [CrossRef] [PubMed]

| Sample | N | Mean Age (Years ± Std. Dev.) | Sex | Mean OS (Months ± Std. Dev.) | Mean PFS (Months ± Std. Dev.) | |
|---|---|---|---|---|---|---|
| M | F | |||||
| Fresh frozen GBM biopsies | 31 | 63.6 ± 10.9 | 15 | 16 | 15 ± 8.2 | 13.8 ± 8.7 |
| Fresh frozen unaffected brain parenchyma | 20 | 64 ± 10.3 | 8 | 12 | ||
| FirstChoice® Human Brain Reference RNA | 1 (commercially available) | 68.3 ± 15 | 13 | 10 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbagallo, D.; Caponnetto, A.; Brex, D.; Mirabella, F.; Barbagallo, C.; Lauretta, G.; Morrone, A.; Certo, F.; Broggi, G.; Caltabiano, R.; et al. CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1. Cancers 2019, 11, 194. https://doi.org/10.3390/cancers11020194
Barbagallo D, Caponnetto A, Brex D, Mirabella F, Barbagallo C, Lauretta G, Morrone A, Certo F, Broggi G, Caltabiano R, et al. CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1. Cancers. 2019; 11(2):194. https://doi.org/10.3390/cancers11020194
Chicago/Turabian StyleBarbagallo, Davide, Angela Caponnetto, Duilia Brex, Federica Mirabella, Cristina Barbagallo, Giovanni Lauretta, Antonio Morrone, Francesco Certo, Giuseppe Broggi, Rosario Caltabiano, and et al. 2019. "CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1" Cancers 11, no. 2: 194. https://doi.org/10.3390/cancers11020194
APA StyleBarbagallo, D., Caponnetto, A., Brex, D., Mirabella, F., Barbagallo, C., Lauretta, G., Morrone, A., Certo, F., Broggi, G., Caltabiano, R., Barbagallo, G. M., Spina-Purrello, V., Ragusa, M., Di Pietro, C., Hansen, T. B., & Purrello, M. (2019). CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1. Cancers, 11(2), 194. https://doi.org/10.3390/cancers11020194









