Impact of ATM and DNA-PK Inhibition on Gene Expression and Individual Response of Human Lymphocytes to Mixed Beams of Alpha Particles and X-Rays
Abstract
1. Introduction
2. Results
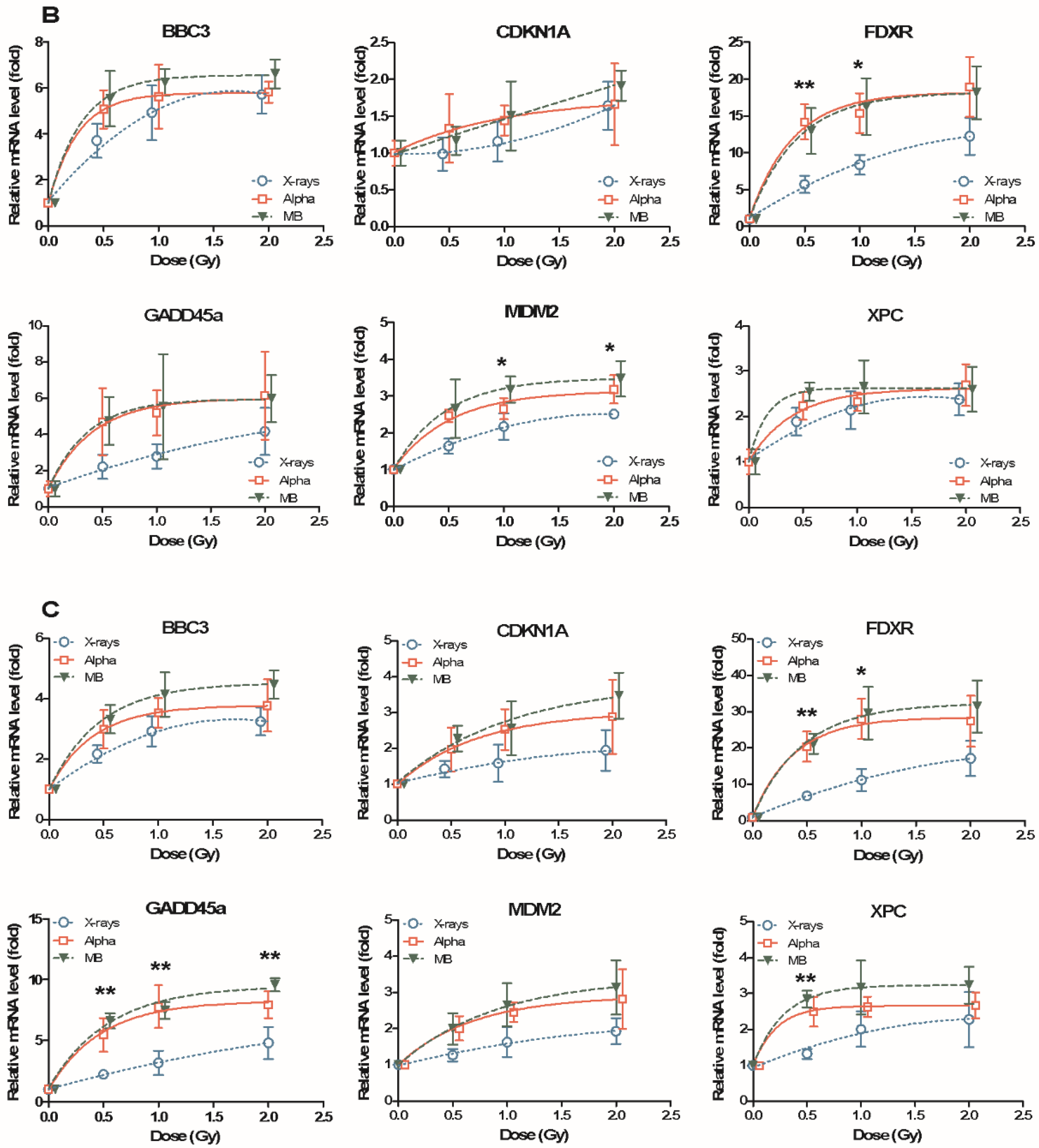
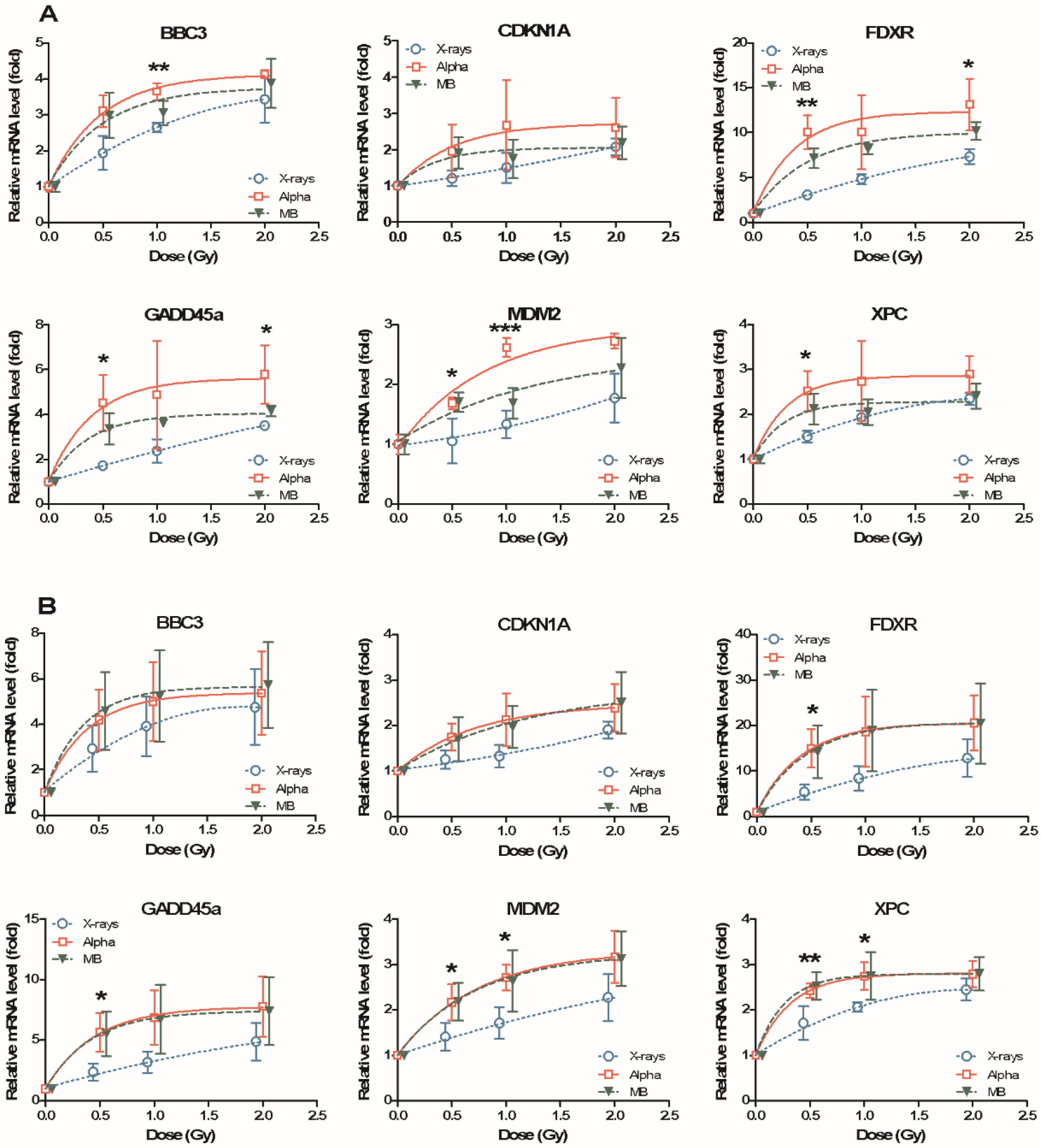
2.1. Dose Response of Radiation-Induced mRNAs after Exposure to X-rays, Alpha Particles and Mixed Beams
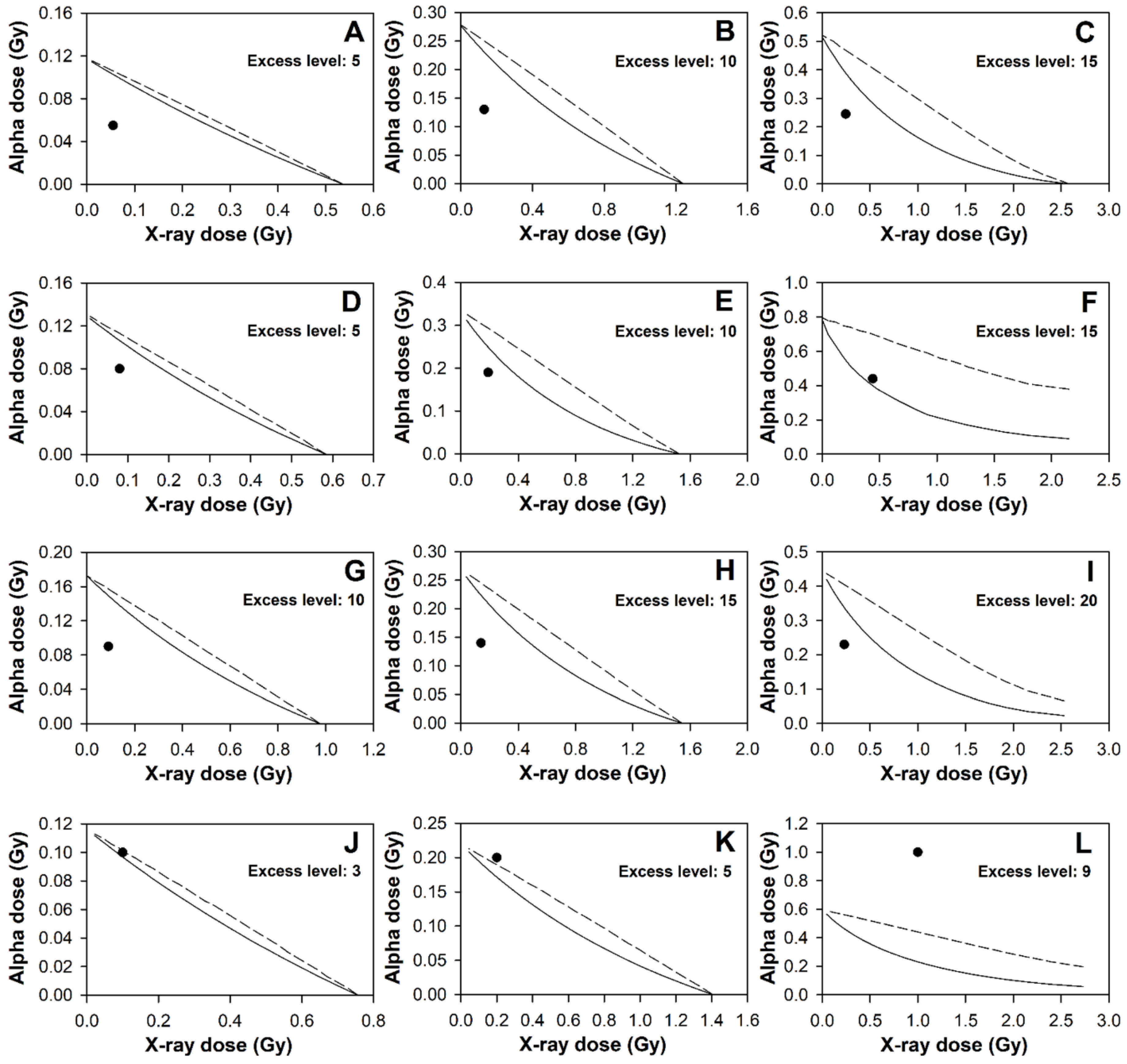
2.2. Mixed Beam Effect on Individuals Analysed Using Envelopes of Additivity
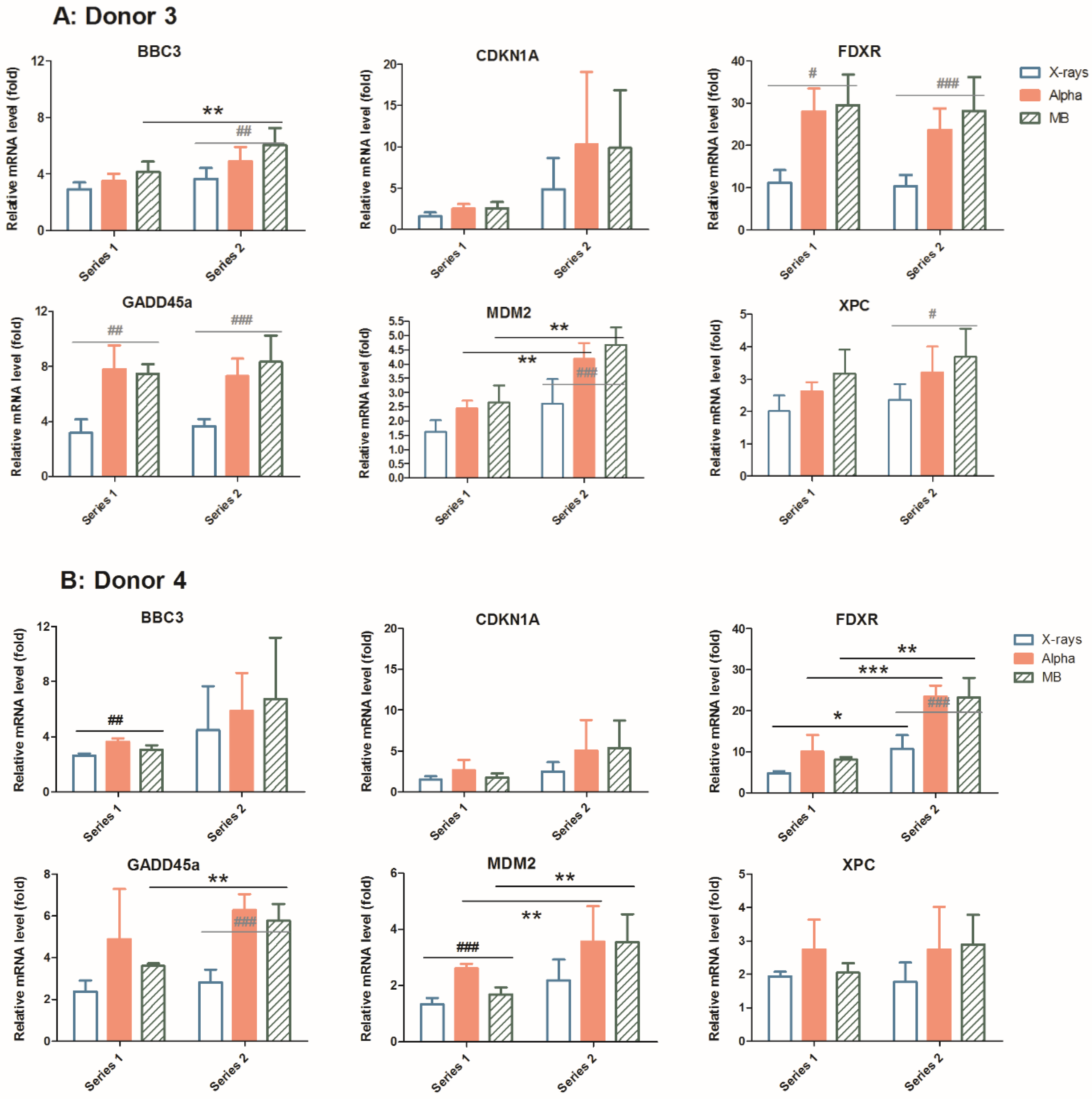
2.3. Mixed Beam Effect Changes in the Same Donor at Different Sampling Times
2.4. Micronucleus (MN) Assay in Donor 3 and Donor 4
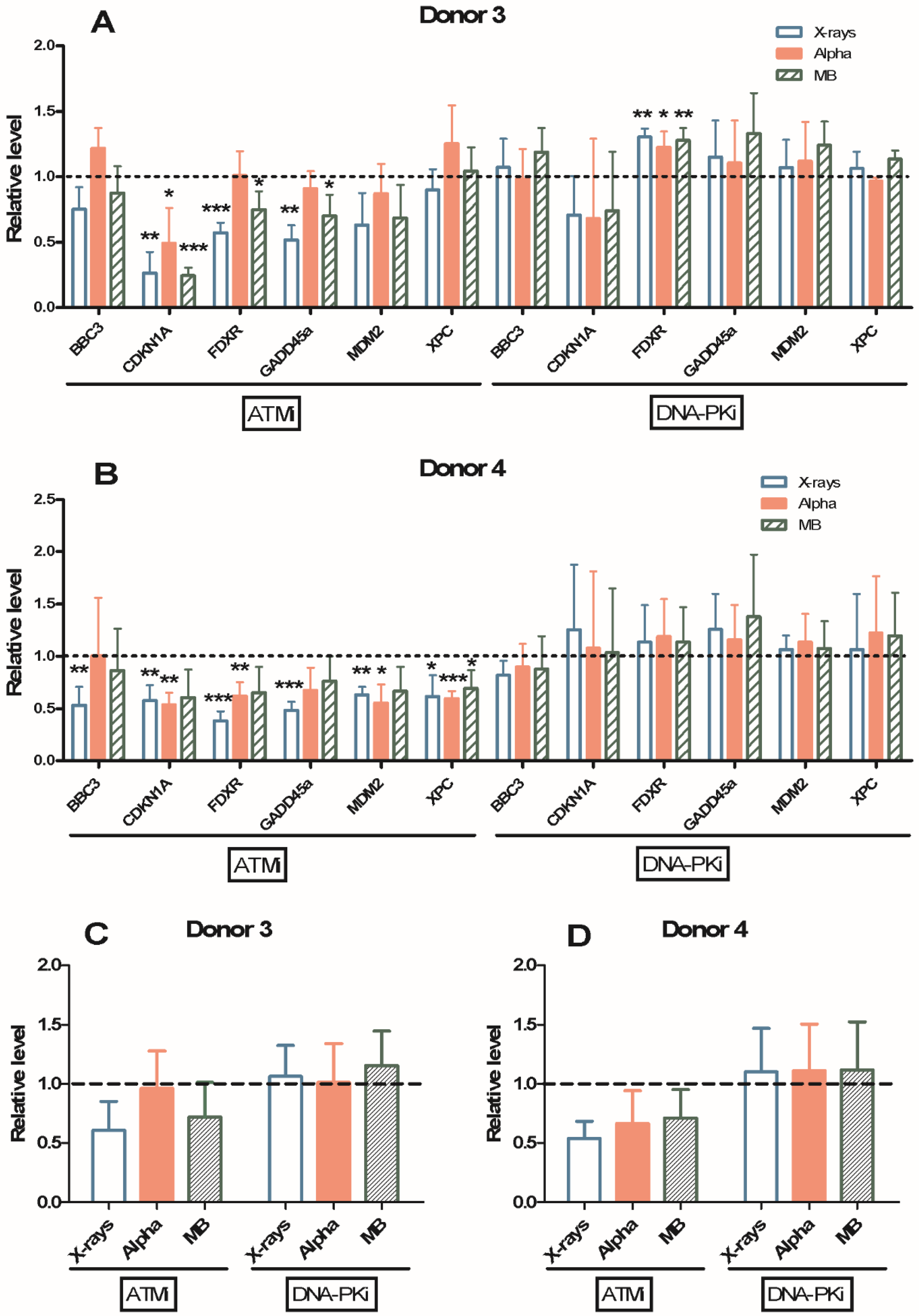
2.5. Gene Expression Modulation Using ATM or DNA-PK Inhibitor
3. Discussion
4. Materials and Methods
4.1. Blood Collection and Irradiation for Gene Expression Analysis
4.2. Gene Expression Analysis by qPCR
4.3. Irradiation and Micronucleus Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pouget, J.P.; Mather, S.J. General aspects of the cellular response to low-and high-LET radiation. Eur. J. Nucl. Med. 2001, 28, 541–561. [Google Scholar] [CrossRef] [PubMed]
- Hada, M.; Georgakilas, A.G. Formation of clustered DNA damage after high-LET irradiation: A review. J. Radiat. Res. 2008, 49, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.F. Radiation mutagenesis: The initial DNA lesions responsible. Radiat. Res. 1995, 142, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Mavragani, I.V.; Nikitaki, Z.; Souli, M.P.; Aziz, A.; Nowsheen, S.; Aziz, K.; Rogakou, E.; Georgakilas, A.G. Complex DNA damage: A route to radiation-induced genomic instability and carcinogenesis. Cancers 2017, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Kakarougkas, A.; Jeggo, P.A. DNA DSB repair pathway choice: An orchestrated handover mechanism. Br. J. Radiol. 2014, 87, 20130685. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Jeggo, P. A historical reflection on our understanding of radiation-induced DNA double strand break repair in somatic mammalian cells; interfacing the past with the present. Int. J. Radiat. Biol. 2019, 95, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef]
- Choi, S.; Gamper, A.M.; White, J.S.; Bakkenist, C.J. Inhibition of ATM kinase activity does not phenocopy ATM protein disruption: Implications for the clinical utility of ATM kinase inhibitors. Cell Cycle 2010, 9, 4052–4057. [Google Scholar] [CrossRef]
- Finzel, A.; Grybowski, A.; Strasen, J.; Cristiano, E.; Loewer, A. Hyperactivation of ATM upon DNA-PKcs inhibition modulates p53 dynamics and cell fate in response to DNA damage. Mol. Biol. Cell 2016, 27, 2360–2367. [Google Scholar] [CrossRef]
- Thorne, M.C. Background radiation: Natural and man-made. J. Radiol. Prot. 2003, 23, 29–42. [Google Scholar] [CrossRef]
- Matsuzaki, Y.; Date, H.; Sutherland, K.L.; Kiyanagi, Y. Nuclear collision processes around the Bragg peak in proton therapy. Radiol. Phys. Technol. 2010, 3, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Kry, S.F.; Salehpour, M.; Followill, D.S.; Stovall, M.; Kuban, D.A.; White, R.A.; Rosen, I.I. Out-of-field photon and neutron dose equivalents from step-and-shoot intensity-modulated radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2005, 62, 1204–1216. [Google Scholar] [CrossRef] [PubMed]
- Liess, M.; Foit, K.; Knillmann, S.; Schafer, R.B.; Liess, H.D. Predicting the synergy of multiple stress effects. Sci. Rep. 2016, 6, 32965. [Google Scholar] [CrossRef] [PubMed]
- Straume, T.; Rugel, G.; Marchetti, A.A.; Ruhm, W.; Korschinek, G.; McAninch, J.E.; Carroll, K.; Egbert, S.; Faestermann, T.; Knie, K.; et al. Measuring fast neutrons in Hiroshima at distances relevant to atomic-bomb survivors. Nature 2003, 424, 539–542. [Google Scholar] [CrossRef] [PubMed]
- UNSCEAR. Health Effects Due to Radiation from the Chernobyl Accident; United Nations Scientific Committee on the Effects of Atomic Radiation: New York, NY, USA, 2008. [Google Scholar]
- WHO. Health Effects of the Chernobyl Accident and Special Health Care Programmes; Report of the UN Chernobyl Forum Expert Group “Health”: Geneva, Switzerland, 2006. [Google Scholar]
- Li, S.; Lu, X.; Feng, J.B.; Tian, M.; Liu, Q.J. Identification and validation of candidate radiation-responsive genes for human biodosimetry. Biomed. Environ. Sci. 2017, 30, 834–840. [Google Scholar] [CrossRef]
- Cheng, L.; Brzozowska, B.; Sollazzo, A.; Lundholm, L.; Lisowska, H.; Haghdoost, S.; Wojcik, A. Simultaneous induction of dispersed and clustered DNA lesions compromises DNA damage response in human peripheral blood lymphocytes. PLoS ONE 2018, 13, e0204068. [Google Scholar] [CrossRef]
- Lisowska, H.; Cheng, L.; Sollazzo, A.; Lundholm, L.; Wegierek-Ciuk, A.; Sommer, S.; Lankoff, A.; Wojcik, A. Hypothermia modulates the DNA damage response to ionizing radiation in human peripheral blood lymphocytes. Int. J. Radiat. Biol. 2018, 94, 551–557. [Google Scholar] [CrossRef]
- Sollazzo, A.; Brzozowska, B.; Cheng, L.; Lundholm, L.; Haghdoost, S.; Scherthan, H.; Wojcik, A. Alpha particles and X rays interact in inducing DNA damage in U2OS cells. Radiat. Res. 2017, 188, 400–411. [Google Scholar] [CrossRef]
- Sollazzo, A.; Brzozowska, B.; Cheng, L.; Lundholm, L.; Scherthan, H.; Wojcik, A. Live dynamics of 53BP1 foci following simultaneous induction of clustered and dispersed DNA damage in U2OS cells. Int. J. Mol. Sci. 2018, 19, 519. [Google Scholar] [CrossRef]
- Staaf, E.; Brehwens, K.; Haghdoost, S.; Nievaart, S.; Pachnerova-Brabcova, K.; Czub, J.; Braziewicz, J.; Wojcik, A. Micronuclei in human peripheral blood lymphocytes exposed to mixed beams of X-rays and alpha particles. Radiat. Environ. Biophys. 2012, 51, 283–293. [Google Scholar] [CrossRef]
- Staaf, E.; Deperas-Kaminska, M.; Brehwens, K.; Haghdoost, S.; Czub, J.; Wojcik, A. Complex aberrations in lymphocytes exposed to mixed beams of 241Am alpha particles and X-rays. Mutat. Res. 2013, 756, 95–100. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, G.; Cruz-Garcia, L.; Majewski, M.; Grepl, J.; Abend, M.; Port, M.; Tichy, A.; Sirak, I.; Malkova, A.; Donovan, E.; et al. FDXR is a biomarker of radiation exposure in vivo. Sci. Rep. 2018, 8, 684. [Google Scholar] [CrossRef] [PubMed]
- Staaf, E.; Brehwens, K.; Haghdoost, S.; Pachnerova-Brabcova, K.; Czub, J.; Braziewicz, J.; Nievaart, S.; Wojcik, A. Characterisation of a setup for mixed beam exposures of cells to 241Am alpha particles and X-rays. Radiat. Prot. Dosim. 2012, 151, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Staaf, E.; Brehwens, K.; Haghdoost, S.; Czub, J.; Wojcik, A. Gamma-H2AX foci in cells exposed to a mixed beam of X-rays and alpha particles. Genome Integr. 2012, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Pollard, J.M.; Gatti, R.A. Clinical radiation sensitivity with DNA repair disorders: An overview. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, C.N.; Alsner, J.; Overgaard, J. Does variability in normal tissue reactions after radiotherapy have a genetic basis—Where and how to look for it? Radiother. Oncol. 2002, 64, 131–140. [Google Scholar] [CrossRef]
- Burga, A.; Lehner, B. Beyond genotype to phenotype: Why the phenotype of an individual cannot always be predicted from their genome sequence and the environment that they experience. FEBS J. 2012, 279, 3765–3775. [Google Scholar] [CrossRef]
- Vral, A.; Thierens, H.; Baeyens, A.; De Ridder, L. Chromosomal aberrations and in vitro radiosensitivity: Intra-individual versus inter-individual variability. Toxicol. Lett. 2004, 149, 345–352. [Google Scholar] [CrossRef]
- Kushiro, J.; Nakamura, N.; Kyoizumi, S.; Nishiki, M.; Dohi, K.; Akiyama, M. Absence of correlations between radiosensitivities of human T-lymphocytes in G0 and skin fibroblasts in log phase. Radiat. Res. 1990, 122, 326–332. [Google Scholar] [CrossRef]
- Brzoska, K.; Kruszewski, M. Toward the development of transcriptional biodosimetry for the identification of irradiated individuals and assessment of absorbed radiation dose. Radiat. Environ. Biophys. 2015, 54, 353–363. [Google Scholar] [CrossRef]
- Manning, G.; Kabacik, S.; Finnon, P.; Bouffler, S.; Badie, C. High and low dose responses of transcriptional biomarkers in ex vivo X-irradiated human blood. Int. J. Radiat. Biol. 2013, 89, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, A.A.; Jeggo, P.A. The heterochromatic barrier to DNA double strand break repair: How to get the entry visa. Int. J. Mol. Sci. 2012, 13, 11844–11860. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Yu, D.; Furusawa, Y.; Okayasu, R.; Tong, J.; Cao, J.; Fan, S. Regulation of ATM in DNA double strand break repair accounts for the radiosensitivity in human cells exposed to high linear energy transfer ionizing radiation. Mutat. Res. 2009, 670, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, P.; Foiani, M.; Kumar, A. ATM and ATR signaling at a glance. J. Cell Sci. 2015, 128, 4255–4262. [Google Scholar] [CrossRef]
- Cmielova, J.; Havelek, R.; Kohlerova, R.; Soukup, T.; Bruckova, L.; Suchanek, J.; Vavrova, J.; Mokry, J.; Rezacova, M. The effect of ATM kinase inhibition on the initial response of human dental pulp and periodontal ligament mesenchymal stem cells to ionizing radiation. Int. J. Radiat. Biol. 2013, 89, 501–511. [Google Scholar] [CrossRef]
- Cmielova, J.; Havelek, R.; Vavrova, J.; Rezacova, M. Changes in the response of MCF-7 cells to ionizing radiation after the combination of ATM and DNA-PK inhibition. Med. Oncol. 2015, 32, 138. [Google Scholar] [CrossRef]
- Ding, D.; Zhang, Y.; Wang, J.; Zhang, X.; Gao, Y.; Yin, L.; Li, Q.; Li, J.; Chen, H. Induction and inhibition of the pan-nuclear gamma-H2AX response in resting human peripheral blood lymphocytes after X-ray irradiation. Cell Death Discov. 2016, 2, 16011. [Google Scholar] [CrossRef]
- Lundholm, L.; Haag, P.; Juntti, T.; Lewensohn, R.; Viktorsson, K. Phosphoprotein analysis reveals MEK inhibition as a way to target non-small cell lung cancer tumor initiating cells. Int. J. Radiat. Biol. 2014, 90, 718–726. [Google Scholar] [CrossRef]
- Cheng, L.; Lisowska, H.; Sollazzo, A.; Wegierek-Ciuk, A.; Stepien, K.; Kuszewski, T.; Lankoff, A.; Haghdoost, S.; Wojcik, A. Modulation of radiation-induced cytogenetic damage in human peripheral blood lymphocytes by hypothermia. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 793, 96–100. [Google Scholar] [CrossRef]
- Steel, G.G.; Peckham, M.J. Exploitable mechanisms in combined radiotherapy-chemotherapy: The concept of additivity. Int. J. Radiat. Oncol. Biol. Phys. 1979, 5, 85–91. [Google Scholar] [CrossRef]
- Streffer, C.; Muller, W.U. Dose-effect relationships and general mechanisms of combined exposures. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1987, 51, 961–969. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, L.; Brzozowska-Wardecka, B.; Lisowska, H.; Wojcik, A.; Lundholm, L. Impact of ATM and DNA-PK Inhibition on Gene Expression and Individual Response of Human Lymphocytes to Mixed Beams of Alpha Particles and X-Rays. Cancers 2019, 11, 2013. https://doi.org/10.3390/cancers11122013
Cheng L, Brzozowska-Wardecka B, Lisowska H, Wojcik A, Lundholm L. Impact of ATM and DNA-PK Inhibition on Gene Expression and Individual Response of Human Lymphocytes to Mixed Beams of Alpha Particles and X-Rays. Cancers. 2019; 11(12):2013. https://doi.org/10.3390/cancers11122013
Chicago/Turabian StyleCheng, Lei, Beata Brzozowska-Wardecka, Halina Lisowska, Andrzej Wojcik, and Lovisa Lundholm. 2019. "Impact of ATM and DNA-PK Inhibition on Gene Expression and Individual Response of Human Lymphocytes to Mixed Beams of Alpha Particles and X-Rays" Cancers 11, no. 12: 2013. https://doi.org/10.3390/cancers11122013
APA StyleCheng, L., Brzozowska-Wardecka, B., Lisowska, H., Wojcik, A., & Lundholm, L. (2019). Impact of ATM and DNA-PK Inhibition on Gene Expression and Individual Response of Human Lymphocytes to Mixed Beams of Alpha Particles and X-Rays. Cancers, 11(12), 2013. https://doi.org/10.3390/cancers11122013




